Abstract
Objective
Recent data suggest that adipose triglyceride lipase (ATGL) plays a key role in providing energy substrate from triglyceride pools and that alterations of its expression/activity relate to metabolic disturbances in skeletal muscle. Yet little is known about its regulation. We here investigated the role of the protein G0/G1 Switch Gene 2 (G0S2), recently described as an inhibitor of ATGL in white adipose tissue, in the regulation of lipolysis and oxidative metabolism in skeletal muscle.
Methods
We first examined G0S2 protein expression in relation to metabolic status and muscle characteristics in humans. We next overexpressed and knocked down G0S2 in human primary myotubes to assess its impact on ATGL activity, lipid turnover and oxidative metabolism, and further knocked down G0S2 in vivo in mouse skeletal muscle.
Results
G0S2 protein is increased in skeletal muscle of endurance-trained individuals and correlates with markers of oxidative capacity and lipid content. Recombinant G0S2 protein inhibits ATGL activity by about 40% in lysates of mouse and human skeletal muscle. G0S2 overexpression augments (+49%, p < 0.05) while G0S2 knockdown strongly reduces (−68%, p < 0.001) triglyceride content in human primary myotubes and mouse skeletal muscle. We further show that G0S2 controls lipolysis and fatty acid oxidation in a strictly ATGL-dependent manner. These metabolic adaptations mediated by G0S2 are paralleled by concomitant changes in glucose metabolism through the modulation of Pyruvate Dehydrogenase Kinase 4 (PDK4) expression (5.4 fold, p < 0.001). Importantly, downregulation of G0S2 in vivo in mouse skeletal muscle recapitulates changes in lipid metabolism observed in vitro.
Conclusion
Collectively, these data indicate that G0S2 plays a key role in the regulation of skeletal muscle ATGL activity, lipid content and oxidative metabolism.
Keywords: Lipid metabolism, Skeletal muscle, Lipolysis, Adipose triglyceride lipase, Oxidative metabolism
Highlights
-
•
G0S2 correlates with lipid content and oxidative capacity in skeletal muscle.
-
•
ATGL activity is tightly controlled by basal expression of G0S2 in skeletal muscle.
-
•
G0S2 controls lipid metabolism in a strictly ATGL-dependent manner in muscle.
-
•
G0S2 regulates fuel selection through the transcriptional control of PDK4.
1. Introduction
Obesity is one of the most prevalent diseases worldwide and constitutes a major risk factor for the development of type 2 diabetes [1], [2]. Over the past few decades, efforts have been made to understand how alterations in lipid metabolism can lead to the development of insulin resistance [3]. A common feature of obesity and type 2 diabetes is ectopic lipid storage. Indeed, in conditions of excess body fat and/or dietary lipid intake, fatty acids are stored as triacylglycerols within lipid droplets in adipose and non-adipose tissues such as liver, heart and skeletal muscle [4].
Interestingly, it has been repeatedly observed that intramyocellular triacylglycerol (IMTG) accumulation predicts the development of insulin resistance [5], [6], [7]. However, this relationship has proven much more complex than initially thought. Indeed, the skeletal muscle of endurance-trained athletes is highly insulin-sensitive despite having an elevated IMTG content [8]. This may be explained by a more efficient coupling between fatty acid storage and utilization in muscles from trained subjects. On the contrary, defective lipid handling in skeletal muscle has been reported in obese and type 2 diabetic subjects, associated with the accumulation of lipotoxic lipid species, such as diacylglycerols and ceramides, that impair insulin signaling and action [9], [10]. We and others recently observed that disturbances of lipolysis and lipid droplet dynamics in skeletal muscle relate to lipotoxicity and insulin resistance [11], [12], [13]. The first and rate-limiting step of skeletal muscle lipolysis is catalyzed by adipose triglyceride lipase (ATGL), and elevated expression and activity of ATGL causes insulin resistance in vitro in primary skeletal muscle cells [11]. However, little is known about its regulation in skeletal muscle. We showed in a previous study that Comparative Gene Identification-58 (CGI-58) co-activates ATGL in skeletal muscle [14]. G0/G1 Switch Gene 2 encodes for an 11 KDa protein (i.e. G0S2) discovered in 1991 to be induced during the transition from the G0 to G1 phase of the cell cycle in lymphocytes [15]. In 2005, a metabolic role of G0S2 was suggested when it was identified as a Peroxisome-Proliferator-Activated Receptor (PPAR) target gene [16]. Recently, G0S2 was shown to inhibit ATGL activity in metabolic organs such as adipose tissue [17], [18] and liver [19], [20].
In the present study, we investigated the role of G0S2 in the control of ATGL activity and lipid metabolism in mouse and human skeletal muscle. The functional role of G0S2 in the regulation of lipolysis and energy substrate oxidation was studied through gain and loss of function studies in vitro in human primary muscle cells and in vivo in mouse skeletal muscle.
2. Materials and methods
2.1. Muscle sampling
Data and samples from 50 men aged between 34 and 53 years were available from a prior study [21]. Of these, 11 were normal weight sedentary controls (mean age 44.4 ± 1.1 yrs; mean BMI 23.9 ± 0.5 kg m−2), 11 were normal weight endurance-trained individuals (mean age 47.9 ± 1.8 yrs; mean BMI 23.4 ± 0.4 kg m−2), and others were sedentary obese (mean age 43.2 ± 1.1 yrs; mean BMI 34.0 ± 0.8 kg m−2) (n = 28). The overall study design and subject testing have been partly described in [21]. The study was performed according to the latest version of the Declaration of Helsinki and the Current International Conference on Harmonization (ICH) guidelines. The research protocol was approved by the Université Laval ethics committee, and all subjects provided written informed consent. Samples of vastus lateralis (∼100 mg) were obtained, blotted free of blood, cleaned to remove fat and connective tissue, and snap-frozen in liquid nitrogen for lipid and Western blot analyses. All samples were stored at −80 °C under argon or nitrogen gas until use.
2.2. Skeletal muscle primary cell culture
Chemicals and culture media were from Sigma–Aldrich and Life Technologies.
Satellite cells from rectus abdominis of healthy male subjects (age 34.3 ± 2.5 years, BMI 26.0 ± 1.4 kg/m2, fasting glucose 5.0 ± 0.2 mM) were kindly provided by Prof. Arild C. Rustan (Oslo University, Norway). Satellite cells were isolated by trypsin digestion, preplated on an uncoated petri dish for 1 h to remove fibroblasts, and subsequently transferred to T-25 collagen-coated flasks in Dulbecco's Modified Eagle's Medium (DMEM) low glucose (1 g/L) supplemented with 10% FBS and various factors (human epidermal growth factor, BSA, dexamethasone, gentamycin, fungizone, fetuin) as previously described [22]. Cells from several donors were pooled and grown at 37 °C in a humidified atmosphere of 5% CO2. Differentiation of myoblasts (i.e. activated satellite cells) into myotubes was initiated at ∼80–90% confluence by switching to α-Minimum Essential Medium with 2% penicillin-streptomycin, 2% FBS, and fetuin (0.5 mg/ml). The medium was changed every other day, and cells were grown up to 5 days.
2.3. Overexpression and knockdown studies
For overexpression experiments, adenoviruses expressing in tandem GFP and human G0S2 (hG0S2) were used (Vector Biolabs, Philadelphia, PA). Control was performed using adenoviruses containing GFP gene only. Myotubes were infected with both adenoviruses at day 4 of differentiation and remained exposed to the virus for 24 h in serum-free DMEM containing 100 μM of oleate complexed to BSA (ratio 2/1). For knockdown studies, myoblasts were exposed for 24 h at the beginning of the differentiation to lentiviral particles encoding for hG0S2 shRNA, hATGL shRNA or a scramble shRNA (non-target control) (Sigma–Aldrich, France). Oleate was preferred to palmitate for lipid loading of the cells to favor triacylglycerol (TAG) synthesis and to avoid the intrinsic lipotoxic effect of palmitate [23].
2.4. Animal studies
All experimental procedures were approved by a local ethical committee and performed according to INSERM animal core facility guidelines and to the 2010/63/UE European Directive for the care and use of laboratory animals. Eight-week-old C57BL/6J male mice were housed in a pathogen-free barrier facility (12 h light/dark cycle) and fed normal chow diet (10% calories from fat) (D12450B, Research Diets, New Jersey). After 6 weeks of diet, mice were injected with 1 × 1010 GC (i.e. genome copy) of AAV1 vector (Vector Biolabs, Philadelphia, PA) in tibialis anterior and gastrocnemius muscles. Each mouse had one leg injected with AAV1-shG0S2 and the contralateral leg injected with AAV1-shNT (nontarget) as a control. Six weeks following the injections, mice were killed by cervical dislocation and muscles (i.e. tibialis anterior, gastrocnemius, extensor digitorum longus and soleus) were dissected and either used ex-vivo for palmitate oxidation assay or stored at −80 °C for protein and RNA analyses.
2.5. Real-time RT-qPCR
Total RNA from cultured myotubes or tibialis anterior muscle was isolated using Qiagen RNeasy mini kit according to manufacturer's instructions (Qiagen GmbH, Hilden, Germany). The quantity of RNA was determined on a Nanodrop ND-1000 (Thermo Scientific, Rockford, IL, USA). Reverse transcriptase PCR was performed on a Techne PCR System TC-412 using the Multiscribe Reverse Transcriptase method (Applied Biosystems, Foster City, CA). Real-time quantitative PCR (qPCR) was performed to determine cDNA content. All primers were bought from Applied Biosystems. Primers used were: 18S (Taqman assay ID: Hs99999901_s1), G0S2 (Hs00274783_s1 and Mm00484537_g1), PDK4 (Hs01037712_m1), and PGC1α (Hs00173304_m1). The amplification reaction was performed in duplicate on 10 ng of cDNA in 96-well reaction plates on a StepOnePlus™ system (Applied Biosystems). All expression data were normalized by the 2(ΔCt) method using 18S as internal control.
2.6. Western blot analysis
Muscle tissues and cell extracts were homogenized in a buffer containing 50 mM HEPES, pH 7.4, 2 mM EDTA, 150 mM NaCl, 30 mM NaPPO4, 10 mM NaF, 1% Triton X-100, 1.5 mg/ml benzamidine HCl and 10 μl/ml of each: protease inhibitor, phosphatase I inhibitor, and phosphatase II inhibitor (Sigma–Aldrich). Tissue homogenates were centrifuged for 25 min at 15,000 g, and supernatants were stored at −80 °C. A total of 30 μg of solubilized proteins from muscle tissue and myotubes were run on a 4–12% SDS-PAGE (Bio-Rad), transferred onto nitrocellulose membrane (Hybond ECL, Amersham Biosciences), and blotted with the following primary antibodies: mG0S2 (#sc-133423, Santa Cruz Biotechnology Inc.), hG0S2 (#12091-1-AP, Protein Tech), ATGL (#2138, Cell Signaling Technology Inc.), and PDK4 (#H00005166-A02, Abnova). Subsequently, immunoreactive proteins were blotted with secondary HRP-coupled antibodies (Cell Signaling Technology Inc.) and revealed by enhanced chemiluminescence reagent (SuperSignal West Femto, Thermo Scientific), visualized using the ChemiDoc MP Imaging System, and data analyzed using the ImageLab 4.2 version software (Bio-Rad Laboratories, Hercules, USA). GAPDH (#2118, Cell Signaling Technology Inc.) was used as an internal control.
2.7. ATGL activity assay
ATGL activity was measured on muscle tissue and cell lysates (i.e. myotubes and COS7 cells stably overexpressing hATGL [24]) as previously described [25]. Briefly, tissue and cell lysates were extracted in a lysis buffer containing 0.25 M sucrose, 1 mM EDTA, 1 mM DTT, 20 μg/ml leupeptin, and 2 μg/ml antipain. [9, 10–3H(N)] triolein (PerkinElmer), and cold triolein were emulsified with phospholipids by sonication. The emulsion was incubated for 30 min at 37 °C in the presence of 10–40 μg of total protein from tissue and cell lysates. After incubation, the reaction was terminated by adding 3.25 ml of methanol-chloroform-heptane (10:9:7) and 1.1 ml of 0.1 M potassium carbonate/0.1 M boric acid (pH 10.5). After centrifugation (800 g, 15 min), 0.5 ml of the upper phase was collected for scintillation counting. ATGL activity was measured in the presence of 1 μg of human recombinant G0S2 protein (rhG0S2) (OriGene), CGI-58 (rhCGI-58) (Abnova), or both. The data are expressed in nmol of oleic acid released per h per mg of protein.
2.8. Determination of glucose metabolism
Cells were pre-incubated with a glucose- and serum-free medium for 90 min, then exposed to DMEM supplemented with D[U-14C] glucose (1 μCi/ml; PerkinElmer, Boston, MA). Following incubation, glucose oxidation was determined by counting of 14CO2 released into the culture medium. The cells were then solubilized in KOH 30%, and glycogen synthesis was determined as previously described [26].
2.9. Determination of fatty acid metabolism
Cells were pulsed overnight for 18 h with [1–14C] oleate (1 μCi/ml; PerkinElmer, Boston, MA) and cold oleate (100 μM) to prelabel the endogenous TAG pool. Oleate was coupled to FA-free BSA in a molar ratio of 5:1. Following the pulse, myotubes were chased for 3 h in DMEM containing 0.1 mM glucose, 0.5% FA-free BSA, and 10 μM triacsin C to block FA recycling into the TAG pool as described elsewhere [27]. TAG-derived FA oxidation was measured by the sum of 14CO2 and 14C-ASM (acid soluble metabolites) in the absence of triacsin C as previously described [22]. Myotubes were harvested in 0.2 ml SDS 0.1% at the end of the pulse and of the chase period to determine oleate incorporation into TAG, diacylglycerol (DAG), monoacylglycerol, FA, and protein content. The lipid extract was separated by TLC using heptane-isopropylether-acetic acid (60:40:4, v/v/v) as developing solvent. All assays were performed in duplicates, and data were normalized to cell protein content. Palmitate oxidation rate was measured as previously described [28].
2.10. Determination of mitochondrial content
We determined mitochondrial mass in myotubes using Mitotracker Green FM (Invitrogen, Carlsbad, CA), which stains mitochondrial matrix protein irrespective of the membrane potential and thus provides an accurate assessment of mitochondrial mass. Similarly, we measured mitochondrial membrane potential using a Mitotracker Red CMX-Ros (Invitrogen, Carlsbad, CA), which stains mitochondria according to their membrane potential. Briefly, cells were washed with PBS and incubated at 37 °C for 30 min with 100 nM of each Mitotracker. Cells were then harvested using trypsin/EDTA and resuspended in PBS. Fluorescence intensity was measured on a fluorometer and values expressed as relative fluorescence units (RFU).
2.11. Tissue-specific [2–3H] deoxyglucose uptake in vivo
Muscle-specific glucose uptake was assessed in response to an intraperitoneal bolus injection of 2-[1,2–3H(N)]deoxy-d-Glucose (PerkinElmer, Boston, Massachusetts) (0.4 μCi/g body weight) and insulin (3 mU/g body weight). The dose of insulin was determined in preliminary studies to reach a nearly maximal stimulation of insulin signaling and glucose uptake in all muscle types and metabolic tissues. Mice were fasted 2 h before injection, killed 30 min after injection, and tissues were extracted by precipitation of 2-deoxyglucose-6-phosphate as previously described [29].
2.12. Determination of triacylglycerol and ceramide content
Triacylglycerol and ceramide species content was determined by high-performance liquid chromatography-mass spectrometry after lipid extraction as described elsewhere [29], [30].
2.13. Statistical analyses
All statistical analyses were performed using GraphPad Prism 5.0 for Windows (GraphPad Software Inc., San Diego, CA). Normal distribution and homogeneity of variance of the data were tested using Shapiro–Wilk and F tests, respectively. One-way ANOVA followed by Tukey's post hoc tests and Student's t-tests were performed to determine differences between treatments. Two-way ANOVA and Bonferroni's post hoc tests were used when appropriate. All values in figures and tables are presented as mean ± SEM. Statistical significance was set at p < 0.05.
3. Results
3.1. G0S2 is associated with lipid content and oxidative capacity in human skeletal muscle
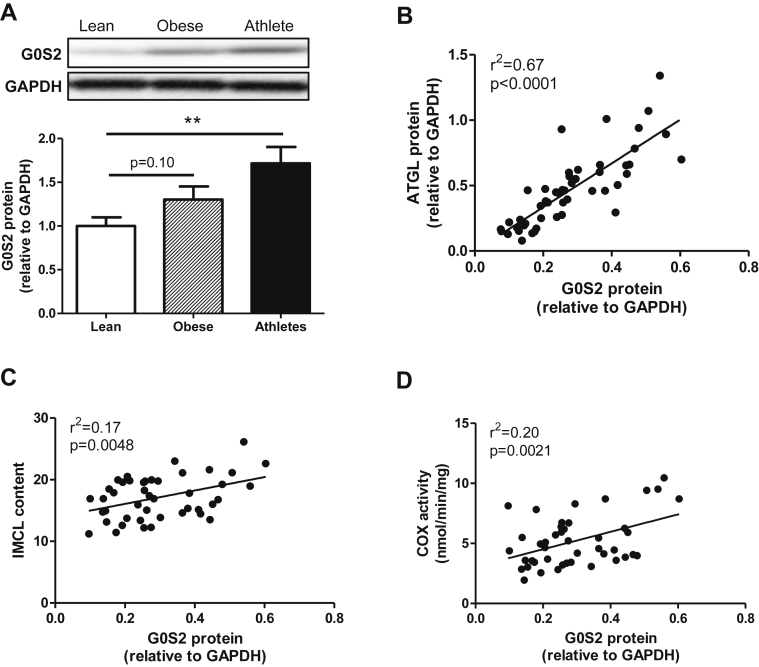
Muscle G0S2 protein expression was assessed in human vastus lateralis samples in healthy lean, obese, and athlete volunteers. We observed that G0S2 protein content was increased in endurance-trained individuals compared to lean, while a trend was observed in obese sedentary volunteers (Figure 1A). Interestingly, muscle G0S2 protein content tightly correlates with ATGL protein (r2 = 0.67, p < 0.0001) (Figure 1B). The ratio of G0S2 to ATGL protein content was not statistically different between lean, obese and athlete (data not shown). We also noted that G0S2 protein was positively associated with IMCL content (r2 = 0.17, p = 0.0048) (Figure 1C), and cytochrome oxidase activity, a marker of muscle oxidative capacity (r2 = 0.20, p = 0.0021) (Figure 1D). Together, these data indicate that G0S2 is significantly expressed in skeletal muscle and may play a role in the regulation of lipid storage and metabolism.
Figure 1.
G0S2 is associated with lipid content and oxidative capacity in human skeletal muscle. (A) Representative blot and quantification of G0S2 protein content measured in vastus lateralis muscle of healthy lean, obese, and endurance-trained volunteers (n = 11 per group). Correlations between muscle G0S2 protein and ATGL protein (B), IMCL content (C), and cytochrome oxidase (COX) activity (D) (n = 50). **p < 0.01 versus lean.
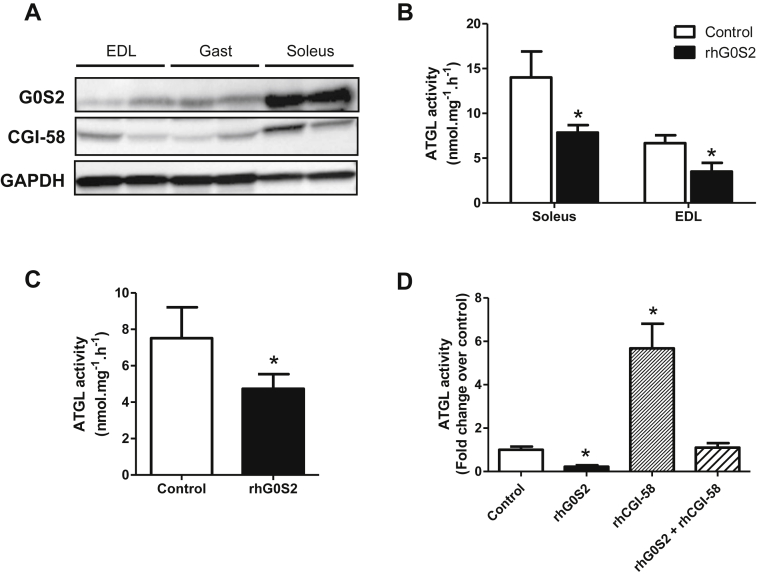
3.2. G0S2 inhibits ATGL activity in mouse and human skeletal muscle
Muscle G0S2 protein expression was measured in different types of mouse skeletal muscles. We observed that G0S2 protein content was higher in the oxidative soleus muscle compared to mixed gastrocnemius and glycolytic extensor digitorum longus muscles (Figure 2A). A similar expression pattern was observed for CGI-58 as previously described [14]. ATGL activity measured in lysates of both soleus and EDL muscles was significantly reduced by the addition of G0S2 recombinant protein (−44% and −47% respectively, p < 0.05) (Figure 2B). We observed a similar effect in lysates of human vastus lateralis muscle (−37%, p < 0.05) (Figure 2C). Interestingly, G0S2 strongly inhibited ATGL activity in COS-7 cell lysates stably overexpressing ATGL but also completely abolished its activation by the ATGL co-activator CGI-58 (Figure 2D). Collectively, these data suggest that G0S2 is a potent inhibitor of ATGL activity in mouse and human skeletal muscle.
Figure 2.
G0S2 inhibits ATGL activity in mouse and human skeletal muscle. (A) Representative blots of G0S2 and CGI-58 protein content in mouse extensor digitorum longus (EDL), gastrocnemius (Gast), and soleus skeletal muscles. Triacylglycerol hydrolase activity (TAGH) was measured in absence (control) or presence of recombinant human G0S2 (rhG0S2) in (B) mice soleus and EDL (n = 7) and (C) human vastus lateralis muscle (n = 5). (D) TAGH activity was measured in absence (control) or presence of recombinant human G0S2 (rhG0S2), CGI-58 (rhCGI-58), or both (rhG0S2+rhCGI-58) in COS7 cell extracts overexpressing human ATGL (n = 4). *p < 0.05 versus control.
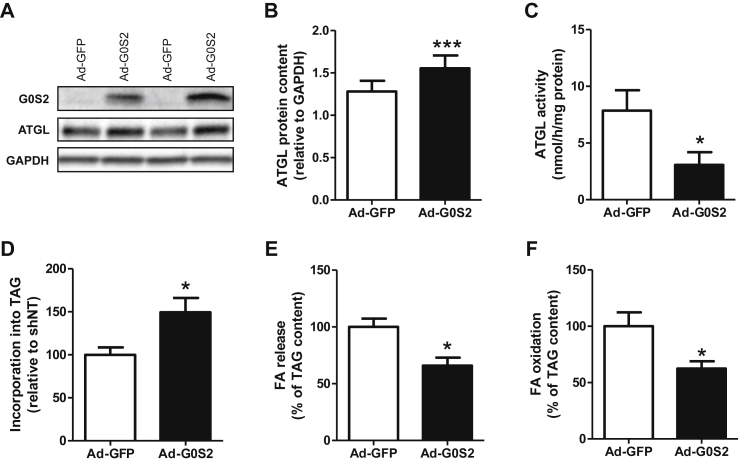
3.3. G0S2 overexpression in human primary myotubes inhibits lipolysis and FA oxidation
G0S2 was overexpressed using an adenovirus containing G0S2 cDNA. An adenovirus containing GFP cDNA was used as a control. Adenovirus-mediated G0S2 overexpression led to a 4.3-fold increase of G0S2 gene expression (p < 0.001) (data not shown) and a 4.6-fold increase of protein content (p < 0.001) (Figure 3A) in 5 days differentiated human myotubes. Surprisingly, this was accompanied by a 21% increase of ATGL protein content in these cells (p < 0.001) (Figure 3B). As expected, ATGL activity was reduced by 61% in cells overexpressing G0S2 (p = 0.026) (Figure 3C).
Figure 3.
G0S2 overexpression in human primary myotubes inhibits lipolysis and FA oxidation. Representative blots (A) and quantification (B) of G0S2 and ATGL protein content measured in control myotubes (Ad-GFP) and myotubes overexpressing G0S2 (Ad-G0S2) (n = 8). (C) ATGL enzyme activity in control myotubes (Ad-GFP) and myotubes overexpressing G0S2 (Ad-G0S2) (n = 3). Pulse-Chase studies using [1–14C] oleate were performed to determine the rate of (D) incorporation of radiolabeled oleate into TAG (Ad-GFP = 41 ± 11 nmol/3 h/mg protein), (E) FA release (Ad-GFP = 26.3 ± 2.5 nmol/3 h/mg protein), and (F) oleate oxidation (Ad-GFP = 2.74 ± 0.19 nmol/3 h/mg protein) in control myotubes (Ad-GFP) and myotubes overexpressing G0S2 (Ad-G0S2) (n = 6). *p < 0.05, ***p < 0.001 versus Ad-GFP.
We next examined the effect of G0S2 overexpression on lipolysis and FA metabolism. We observed that G0S2 overexpression increased TAG accumulation (+49%, p = 0.031) (Figure 3D), which was concomitantly accompanied by a decrease of FA release into the culture medium (−34%, p = 0.01) (Figure 3E) and FA oxidation (−38%, p = 0.038) (Figure 3F). Taken together, these data show that G0S2 inhibits ATGL activity in human primary myotubes, leading to a reduction of lipolysis rate and FA oxidation.
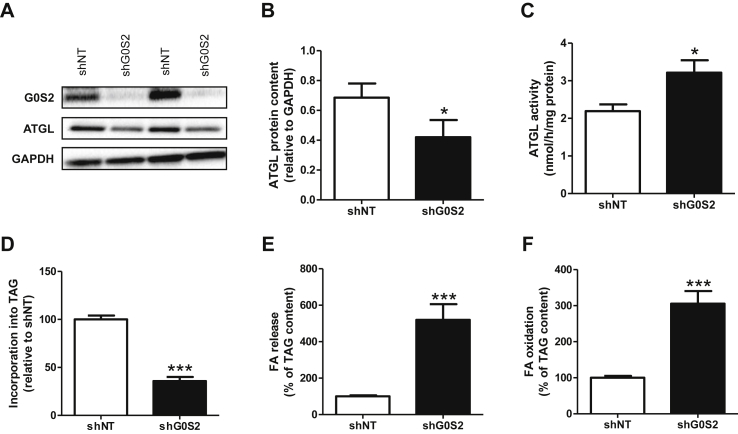
3.4. G0S2 knockdown in human primary myotubes promotes lipolysis and FA oxidation
G0S2 silencing in human primary skeletal muscle cells was realized using lentivirus containing an shRNA directed against G0S2 (shG0S2), and a scramble shRNA (shNT) was used as a control. No significant change in cell viability was observed between shNT and shG0S2 conditions (data not shown). Lentivirus-mediated G0S2 knockdown strongly reduced G0S2 gene expression (−61%, p = 0.009, data not shown), leading to a virtually complete loss of G0S2 protein (Figure 4A). Interestingly, G0S2 downregulation was accompanied by a significant decrease of ATGL protein (−39%, p = 0.026) (Figure 4B), despite a nearly 50% increase of ATGL activity (p = 0.027) (Figure 4C).
Figure 4.
G0S2 knockdown in human primary myotubes promotes lipolysis and FA oxidation. Representative blots (A) and quantification (B) of G0S2 and ATGL protein content in control myotubes (shNT) and myotubes downregulated for G0S2 (shG0S2) (n = 5). (C) ATGL enzyme activity in control myotubes (shNT) and myotubes downregulated for G0S2 (shG0S2) (n = 3). Pulse-Chase studies using [1–14C] oleate were performed to determine the rate of (D) incorporation of radiolabeled oleate into TAG (shNT = 41.8 ± 5.1 nmol/3 h/mg protein), (E) FA release (shNT = 74 ± 6 nmol/3 h/mg protein), and (F) oleate oxidation (shNT = 3.74 ± 0.32 nmol/3 h/mg protein) in control myotubes (shNT) and myotubes with G0S2 silencing (shG0S2). n = 9. *p < 0.05, ***p < 0.001 versus shNT.
G0S2 knockdown was associated with a marked reduction of the TAG pool (−68%, p < 0.001) (Figure 4D) that was accompanied by a robust increase of FA release (4.8 fold, p < 0.001) (Figure 4E) and FA oxidation (3 fold, p < 0.001) (Figure 4F). Collectively, these results show that G0S2 knockdown promotes lipolysis and increases FA oxidation in human primary myotubes.
3.5. G0S2 controls lipolysis in an ATGL-dependent manner in human primary myotubes
Because G0S2 may display effects on lipid metabolism independent of ATGL [31], we performed a double knockdown of G0S2 and ATGL and assessed lipid metabolism (Figure 5A). As previously observed, the TAG pool was strongly reduced in myotubes knocked down for G0S2, but this effect was totally abrogated and the TAG pool increased (+65% vs shNT, p = 0.004) when ATGL was simultaneously ablated. A similar increase in TAG pool was observed when ATGL was down regulated alone (Figure 5B). In line with changes in TAG content, the increase of FA release (Figure 5C) and FA oxidation (Figure 5D) was totally blunted when both G0S2 and ATGL were knocked down, in a similar fashion as when ATGL was ablated alone. Altogether, these data suggest that G0S2 controls lipolysis and lipid metabolism in a strictly ATGL-dependent manner.
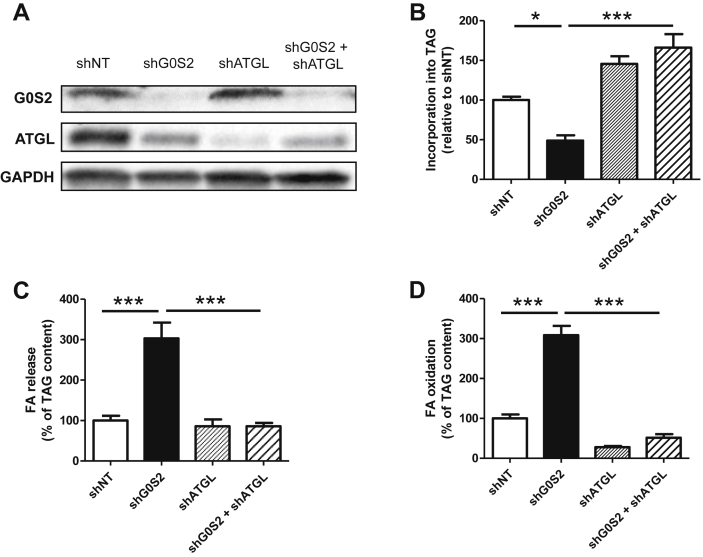
Figure 5.
G0S2 controls lipolysis in an ATGL-dependent manner. (A) Representative blot of G0S2 and ATGL protein content in control myotubes (shNT), myotubes with a downregulation of G0S2 (shG0S2), ATGL (shATGL), or both (shG0S2+shATGL). Pulse-Chase studies using [1–14C] oleate were performed to determine the rate of (B) incorporation of radiolabeled oleate into TAG (shNT = 40.2 ± 1.1 nmol/3 h/mg protein), (C) FA release (shNT = 66.3 ± 5.8 nmol/3 h/mg protein), and (D) oleate oxidation (shNT = 3.09 ± 0.13 nmol/3 h/mg protein) in these cells (n = 3). *p < 0.05, ***p < 0.001.
3.6. G0S2 knockdown reduces glucose metabolism and enhances mitochondrial function
FA and glucose are the main energy substrates in skeletal muscle, and a finely tuned regulation occurs for the use of these two substrates. Considering that G0S2 knockdown has an important impact on IMTG content and lipolysis, we investigated its effect on glucose metabolism. We observed that G0S2 knockdown decreased glucose oxidation (−34%, p = 0.003) (Figure 6A) and glycogen synthesis (−63%, p < 0.001) (Figure 6B). Because glucose oxidative metabolism is under the control of pyruvate dehydrogenase kinase 4 (PDK4), we measured expression of PDK4 gene, a well-known canonical target gene of Peroxisome Proliferator Associated Receptor β (PPARβ), a master regulator of muscle oxidative metabolism. Interestingly, we observed that PDK4 was highly induced by G0S2 knockdown (5.4 fold, p < 0.001), an effect that was strongly abolished when the cells were concomitantly treated with 500 nM of GSK0660, a specific antagonist of PPARβ (−70%, p < 0.001) (Figure 6C).
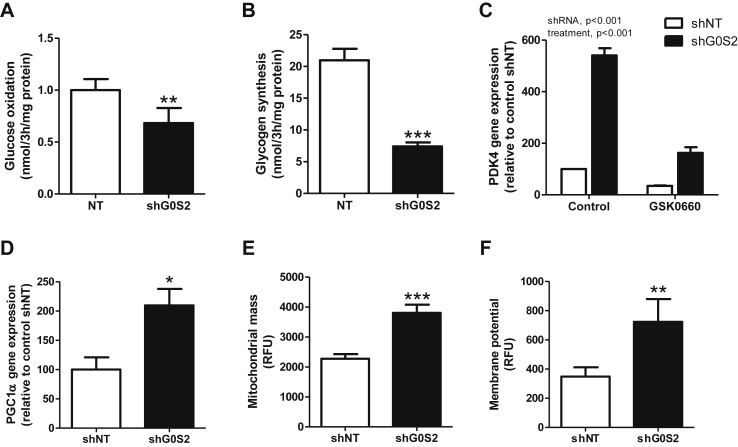
Figure 6.
G0S2 knockdown reduces glucose metabolism and enhances mitochondrial function. (A) Glucose oxidation and (B) glycogen synthesis were measured in control myotubes (shNT) and myotubes knocked down for G0S2 (shG0S2) using [U–14C] glucose. (C) PDK4 gene expression was measured in control myotubes (shNT) and myotubes knocked down for G0S2 (shG0S2) in absence or presence of a selective PPARδ antagonist GSK0660 500 nM PCG1α gene expression (D), mitochondrial mass (E), and mitochondrial membrane potential (F) were measured in control myotubes (shNT) and myotubes knocked down for G0S2 (shG0S2) (n = 9). *p < 0.05, **p < 0.01, ***p < 0.001 versus shNT.
Because PPARγ coactivator 1α (PGC1α), another master regulator of oxidative metabolism is also a PPARβ-target gene, we measured its expression level and showed a 2-fold increase of PGC1α gene expression in myotubes knocked down for G0S2 (p = 0.019) (Figure 6D). This was consistently associated with an increase in mitochondrial mass (+67%, p < 0.001) (Figure 6E) and membrane potential (+108%, p = 0.043) (Figure 6F) in myotubes knocked down for G0S2. The membrane potential to mitochondrial mass ratio was unchanged, suggesting that the increase of mitochondrial membrane potential reflects the increase of total mitochondrial content, instead of a higher oxidative phosphorylation rate per mitochondrion (shNT 0.15 ± 0.02 vs shG0S2 0.19 ± 0.04, NS). We also noted a nearly significant increase of ATP synthase subunit alpha protein of complex V (p = 0.05) (data not shown). Altogether, these data show that G0S2 controls fuel selection (FA vs glucose) in skeletal muscle cells partly through the modulation of lipid ligand availability for PPARβ.
3.7. G0S2 knockdown in vivo increases lipolysis and induces ceramide accumulation in skeletal muscle
To assess the physiological role of G0S2 in vivo, we knocked down its expression by injecting an AAV1 containing an shRNA directed against G0S2 in tibialis anterior and gastrocnemius muscles into 14-week old C57BL/6J mice. Intramuscular AAV1-shRNA-G0S2 injection significantly reduced G0S2 mRNA and protein expression compared to the contralateral leg injected with an AAV1 containing a non-targeted shRNA (Supplemental Figure 1). Interestingly, and in line with in vitro data, knockdown of G0S2 tended to reduce ATGL protein content (−38%, p = 0.056) (Supplemental Figure 1) and decreased IMTG content (−28%, p = 0.047) (Figure 7A). This was accompanied by increased palmitate oxidation rate to CO2 (2.7-fold, p < 0.05), palmitate oxidation to ASM (acid soluble metabolites) (p = 0.052) and total oxidation tended to increase (1.4 fold, p < 0.05) (Figure 7B). Again, as observed in vitro, PDK4 protein was increased in G0S2 knocked down muscles (+23%, p = 0.03) (Figure 7C). Because an elevated rate of lipolysis in muscle may cause lipotoxicity and reduce insulin-mediated glucose uptake in a resting state, we next examined lipotoxic lipid species and glucose uptake. Interestingly, we observed a significant accumulation of different ceramide species (+60%) in muscles knocked down for G0S2 (Figure 7D), and this was associated with a non-significant trend for a lower insulin-stimulated glucose uptake (−19%, p = 0.08) (Figure 7E). We further observed a downregulation of muscle G0S2 protein concomitant with reduced insulin-stimulated glucose uptake in HFD-fed mice with marked body weight gain (Supplemental Figure 2). Together, these in vivo data recapitulate what has been observed in vitro with G0S2 knockdown in myotubes, and strengthen the important metabolic role of G0S2 in skeletal muscle.
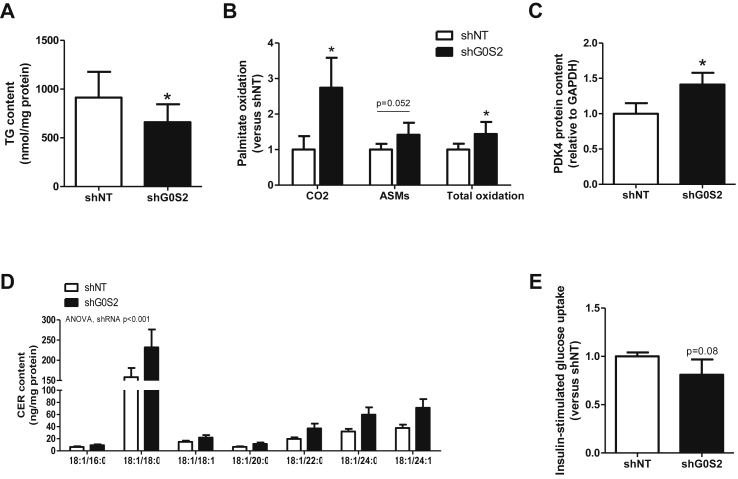
Figure 7.
G0S2 knockdown increases FA oxidation and induces CER accumulation in skeletal muscle in vivo. (A) IMTG content was measured in control (shNT) and G0S2 silenced (shG0S2) mouse tibialis anterior homogenate (n = 7). (B) Palmitate oxidation rate was measured using [1–14C] palmitate in control (shNT) and G0S2 silenced (shG0S2) muscle homogenates. Palmitate oxidation (i.e. CO2), acid soluble metabolites accumulation (i.e. ASMs) and total oxidation (i.e. the sum of CO2 release and ASMs accumulation, shNT = 965 ± 126 nmol/h/g tissue) were measured (n = 6). (C) PDK4 protein content was quantified by western blot (n = 6). (D) Ceramide (CER) subspecies content (n = 7), and (E) insulin-stimulated glucose uptake in control (shNT) and G0S2 knockdown (shG0S2) muscles (shNT = 13 ± 3 μmol/min/100 g tissue). (n = 6). *p < 0.05 versus shNT.
4. Discussion
Obesity and type 2 diabetes are associated with an accumulation of ectopic lipids in insulin-sensitive “non-adipose” tissues such as skeletal muscle [4]. A growing body of data suggests that an altered handling of these lipids may be responsible for the impairment of insulin action [3]. As skeletal muscle acts as a metabolic sink for glucose in postprandial conditions [32], [33], impaired insulin action in this organ has an important impact on whole-body glucose homeostasis. It is therefore important to understand how the IMTG pool is regulated. In this study, we identified G0S2 as an important regulator of lipid metabolism in mouse and human skeletal muscle. We showed that G0S2 is preferentially expressed in oxidative muscles, is associated with oxidative capacity and lipid content in human skeletal muscle, and inhibits ATGL activity. We further demonstrated that G0S2 plays a pivotal role in the regulation of intramyocellular lipolysis and fatty acid oxidation. Finally, we highlighted a potential role for G0S2 in providing lipolysis-derived fatty acids to activate PPARβ and downstream expression of its target genes to regulate energy metabolism in skeletal muscle.
We observed a higher expression of G0S2 in skeletal muscle of endurance-trained individuals compared to lean subjects. This is in line with a recent report showing that G0S2 protein is increased in rat skeletal muscle following 8 weeks of endurance training [34]. It is well known that athletes display high muscle oxidative capacity and IMCL content [8]. Interestingly, we found that G0S2 protein is associated with skeletal muscle oxidative capacity and IMCL levels. Of importance, we observed that G0S2 strongly correlates with ATGL protein content in human skeletal muscle, suggesting a role for G0S2 in the regulation of muscle lipid metabolism.
We observed a higher expression of G0S2 in an oxidative muscle (soleus) compared to more glycolytic muscles (gastrocnemius and extensor digitorum longus), which goes along with a higher expression of CGI-58 and of all the proteins involved in the lipolytic machinery [35]. This is consistent with a high capacity to store, mobilize and oxidize lipids in oxidative muscles [36] and suggests that G0S2 may be essentially expressed in type I oxidative fibers, as observed for ATGL [37]. Studies from different groups report that G0S2 inhibits ATGL activity in adipose tissue [17], [18] and in liver [19], [20], and we show for the first time that G0S2 inhibits ATGL activity in human and mouse skeletal muscle. Mutagenesis studies have described that the hydrophobic domain of G0S2 can directly bind to the patatin-like domain of ATGL and that this interaction is required for the inhibitory action of G0S2 [17], [38]. Interestingly, we observed in COS7 cells overexpressing ATGL that rhG0S2 is able to suppress ATGL activation induced by rhCGI-58. This suggests that G0S2 can bind ATGL independently of the presence of CGI-58, by a non-competing mechanism. However, the molecular mechanisms of ATGL inhibition by G0S2 are still poorly understood and clearly need to be further investigated.
To gain further insight into the functional role of G0S2 in skeletal muscle, G0S2 was downregulated in human primary myotubes. G0S2 silencing induced an increase of ATGL activity and lipolysis, reflected by a strong reduction of TAG content and concomitant increase of FA release. Surprisingly, this was also accompanied by a significant decrease of ATGL protein content. Conversely, G0S2 overexpression led to decreased lipolytic rate, associated with an increase of ATGL protein content. Taken together, these results suggest that a compensatory mechanism occurs when ATGL activity is modulated. Similar findings have been observed when CGI-58 was overexpressed and knocked-down in this model [14]. As no change in ATGL gene expression was observed, either in knockdown or overexpression studies (data not shown), we suggest that this regulation may take place at the post-transcriptional level, although the precise underlying mechanism still remains unknown. In agreement with the increased lipolytic flux in G0S2 silenced cells, we observed an increase in IMTG-derived fatty acid oxidation, the opposite being observed in G0S2 overexpressing cells. These results highlight a pivotal role for G0S2 in providing substrates for mitochondrial β-oxidation. Interestingly, we noted an increased mitochondrial mass and membrane potential in G0S2 silenced cells. However, the membrane potential to mitochondrial mass ratio remained unchanged, suggesting that the increase in membrane potential was due to the presence of a higher number and/or size of mitochondria. Consistently, the gene expression of PGC1α, a master regulator of oxidative metabolism and mitochondrial biogenesis, increased in these cells. A recent study reported that G0S2 positively regulates OXPHOS activity by directly interacting with ATP synthase [31]. However, double G0S2 and ATGL knockdown experiments demonstrated that the effects of G0S2 knockdown on TAG content, FA release, and oxidation were totally dependent of ATGL.
FA and glucose are the main energy sources in skeletal muscle, and it is now well established that a finely tuned regulation occurs for the use of these two substrates, the so called fuel selection [39]. We observed that, concomitantly with an increased lipolytic flux in G0S2 silenced cells, oxidative (i.e. glucose oxidation) and non-oxidative (i.e. glycogen synthesis) glucose metabolism was blunted. This was accompanied by a strong induction of the “switch-gene” PDK4. PDK4 is a mitochondrial protein inhibiting glucose oxidation in response to a high FA availability [40]. As PDK4 is a PPARβ-target gene [41], and considering that FA act as endogenous ligands of PPARβ [42], we hypothesized that IMTG-derived FA could activate PPARβ and induce PDK4 expression in G0S2 silenced cells [14]. Thus activation of lipolysis by G0S2 knockdown robustly induced PDK4 in a PPARβ-dependent manner.
Most importantly, we further observed that G0S2 knockdown in mouse tibialis anterior muscle reduced IMTG content, enhanced FA oxidation in vivo, and induced PDK4 expression. However, perhaps because of lower down regulation of G0S2 in vivo compared to in vitro, the magnitude of change in FA oxidation was moderate and no significant effect was observed on glucose oxidation (data not shown). Interestingly, we also observed an accumulation of various ceramide species, suggesting that an elevated rate of lipolysis in skeletal muscle can lead to lipotoxicity in the resting state despite a slightly elevated FA oxidation rate. A similar metabolic change is classically observed during high fat feeding in which upregulation of mitochondrial FA oxidation cannot prevent ceramide accumulation and insulin resistance [29], [43], [44]. In this study, G0S2 knockdown-mediated ceramide accumulation was associated with a non-significant decrease of insulin-stimulated glucose uptake as would be expected from the negative action of ceramide on insulin signaling [45]. Interestingly, data from mouse models highlight a tissue-specific role for G0S2 in the control of insulin sensitivity. Of importance, G0S2 knockout mice are more insulin-sensitive and glucose tolerant than wild type littermates when fed a high fat diet [46]. These mice are protected against liver steatosis, and liver-specific G0S2 deletion induces a similar phenotype as G0S2 global knockout mice [20]. On the other hand, G0S2 overexpression specifically in adipose tissue leads to an improvement of insulin and glucose tolerance and reduces circulating fatty acids level [47]. In line with these studies, we show that G0S2 knockdown in skeletal muscle causes a lipotoxic injury and reduces insulin action similarly to what is observed in skeletal muscle during high fat feeding. Thus the specific role of muscle G0S2 in the regulation of whole body insulin sensitivity and metabolism should be explored in muscle-specific G0S2 knockout mice.
5. Conclusion
Collectively, our data show for the first time that G0S2 inhibits ATGL activity in mouse and human skeletal muscle and plays a central role in regulating lipid metabolism and substrate oxidation. These results also suggest that changes in G0S2 expression may cause accumulation of lipotoxic species in skeletal muscle and subsequent impairment of insulin action. Future studies are needed to further elucidate the potential contribution of skeletal muscle G0S2 to insulin resistance, obesity, and type 2 diabetes.
Acknowledgments
The authors thank Justine Bertrand-Michel and Aude Dupuy (Lipidomic Core Facility, INSERM, UMR1048 [part of Toulouse Metatoul Platform]) for lipidomic analysis, advice, and technical assistance. We also thank Cédric Baudelin and Xavier Sudre from the Animal Care facility. Special thanks to all participants for their time and invaluable cooperation. The authors would also like to thank Josée St-Onge, Marie-Eve Riou, Etienne Pigeon, Erick Couillard, Guy Fournier, Jean Doré, Marc Brunet, Linda Drolet, Nancy Parent, Marie Tremblay, Rollande Couture, Valérie-Eve Julien, Rachelle Duchesne and Ginette Lapierre for their expert technical assistance in the LIME study. This work was supported by grants from the National Research Agency ANR-12-JSV1-0010-01 (CM), Société Francophone du Diabète (CM), Canadian Institutes of Health Research grant CIHR MOP-68846 (DRJ) and a Pfizer/CIHR research Chair on the pathogenesis of insulin resistance and cardiovascular diseases (AM).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.04.004.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Arnlov J., Ingelsson E., Sundstrom J., Lind L. Impact of body mass index and the metabolic syndrome on the risk of cardiovascular disease and death in middle-aged men. Circulation. 2010;121:230–236. doi: 10.1161/CIRCULATIONAHA.109.887521. [DOI] [PubMed] [Google Scholar]
- 2.Arnlov J., Sundstrom J., Ingelsson E., Lind L. Impact of BMI and the metabolic syndrome on the risk of diabetes in middle-aged men. Diabetes Care. 2011;34:61–65. doi: 10.2337/dc10-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samuel V.T., Shulman G.I. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaffer J.E. Lipotoxicity: when tissues overeat. Current Opinion in Lipidology. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Krssak M., Falk Petersen K., Dresner A., DiPietro L., Vogel S.M., Rothman D.L. Intramyocellular lipid concentrations are correlated with insulin sensitivity in humans: a 1H NMR spectroscopy study. Diabetologia. 1999;42:113–116. doi: 10.1007/s001250051123. [DOI] [PubMed] [Google Scholar]
- 6.Perseghin G., Scifo P., De Cobelli F., Pagliato E., Battezzati A., Arcelloni C. Intramyocellular triglyceride content is a determinant of in vivo insulin resistance in humans: a 1H-13C nuclear magnetic resonance spectroscopy assessment in offspring of type 2 diabetic parents. Diabetes. 1999;48:1600–1606. doi: 10.2337/diabetes.48.8.1600. [DOI] [PubMed] [Google Scholar]
- 7.Virkamaki A., Korsheninnikova E., Seppala-Lindroos A., Vehkavaara S., Goto T., Halavaara J. Intramyocellular lipid is associated with resistance to in vivo insulin actions on glucose uptake, antilipolysis, and early insulin signaling pathways in human skeletal muscle. Diabetes. 2001;50:2337–2343. doi: 10.2337/diabetes.50.10.2337. [DOI] [PubMed] [Google Scholar]
- 8.Goodpaster B.H., He J., Watkins S., Kelley D.E. Skeletal muscle lipid content and insulin resistance: evidence for a paradox in endurance-trained athletes. Journal of Clinical Endocrinology & Metabolism. 2001;86:5755–5761. doi: 10.1210/jcem.86.12.8075. [DOI] [PubMed] [Google Scholar]
- 9.Coen P.M., Dube J.J., Amati F., Stefanovic-Racic M., Ferrell R.E., Toledo F.G. Insulin resistance is associated with higher intramyocellular triglycerides in type I but not type II myocytes concomitant with higher ceramide content. Diabetes. 2010;59:80–88. doi: 10.2337/db09-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szendroedi J., Yoshimura T., Phielix E., Koliaki C., Marcucci M., Zhang D. Role of diacylglycerol activation of PKCtheta in lipid-induced muscle insulin resistance in humans. Proceedings of the National Academy of Sciences U S A. 2014;111:9597–9602. doi: 10.1073/pnas.1409229111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badin P.M., Louche K., Mairal A., Liebisch G., Schmitz G., Rustan A.C. Altered skeletal muscle lipase expression and activity contribute to insulin resistance in humans. Diabetes. 2011;60:1734–1742. doi: 10.2337/db10-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosma M., Hesselink M.K., Sparks L.M., Timmers S., Ferraz M.J., Mattijssen F. Perilipin 2 improves insulin sensitivity in skeletal muscle despite elevated intramuscular lipid levels. Diabetes. 2012;61:2679–2690. doi: 10.2337/db11-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mason R.R., Mokhtar R., Matzaris M., Selathurai A., Kowalski G.M., Mokbel N. PLIN5 deletion remodels intracellular lipid composition and causes insulin resistance in muscle. Molecular Metabolism. 2014;3:652–663. doi: 10.1016/j.molmet.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badin P.M., Loubiere C., Coonen M., Louche K., Tavernier G., Bourlier V. Regulation of skeletal muscle lipolysis and oxidative metabolism by the co-lipase CGI-58. Journal of Lipid & Research. 2012;53:839–848. doi: 10.1194/jlr.M019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell L., Forsdyke D.R. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA and Cell Biology. 1991;10:581–591. doi: 10.1089/dna.1991.10.581. [DOI] [PubMed] [Google Scholar]
- 16.Zandbergen F., Mandard S., Escher P., Tan N.S., Patsouris D., Jatkoe T. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochemical Journal. 2005;392:313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Lu X., Lombes M., Rha G.B., Chi Y.I., Guerin T.M. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metabolism. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweiger M., Paar M., Eder C., Brandis J., Moser E., Gorkiewicz G. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. Journal of Lipid & Research. 2012;53:2307–2317. doi: 10.1194/jlr.M027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Zhang Y., Qian H., Lu J., Zhang Z., Min X. The g0/g1 switch gene 2 is an important regulator of hepatic triglyceride metabolism. PLoS One. 2013;8:e72315. doi: 10.1371/journal.pone.0072315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., Xie X., Heckmann B.L., Saarinen A.M., Czyzyk T.A., Liu J. Targeted disruption of G0/G1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high-fat diet-induced liver steatosis. Diabetes. 2014;63:934–946. doi: 10.2337/db13-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riou M.E., Pigeon E., St-Onge J., Tremblay A., Marette A., Weisnagel S.J. Predictors of cardiovascular fitness in sedentary men. Applied Physiology, Nutrition, and Metabolism. 2009;34:99–106. doi: 10.1139/H08-144. [DOI] [PubMed] [Google Scholar]
- 22.Ukropcova B., McNeil M., Sereda O., de Jonge L., Xie H., Bray G.A. Dynamic changes in fat oxidation in human primary myocytes mirror metabolic characteristics of the donor. Journal of Clinical Investigation. 2005;115:1934–1941. doi: 10.1172/JCI24332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakke S.S., Moro C., Nikolic N., Hessvik N.P., Badin P.M., Lauvhaug L. Palmitic acid follows a different metabolic pathway than oleic acid in human skeletal muscle cells; lower lipolysis rate despite an increased level of adipose triglyceride lipase. Biochimica et Biophysica Acta. 2012;1821:1323–1333. doi: 10.1016/j.bbalip.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Mairal A., Langin D., Arner P., Hoffstedt J. Human adipose triglyceride lipase (PNPLA2) is not regulated by obesity and exhibits low in vitro triglyceride hydrolase activity. Diabetologia. 2006;49:1629–1636. doi: 10.1007/s00125-006-0272-x. [DOI] [PubMed] [Google Scholar]
- 25.Moro C., Galgani J.E., Luu L., Pasarica M., Mairal A., Bajpeyi S. Influence of gender, obesity, and muscle lipase activity on intramyocellular lipids in sedentary individuals. Journal of Clinical Endocrinology & Metabolism. 2009;94:3440–3447. doi: 10.1210/jc.2009-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurens C., Louche K., Sengenes C., Coue M., Langin D., Moro C. Adipogenic progenitors from obese human skeletal muscle give rise to functional white adipocytes that contribute to insulin resistance. International Journal of Obesity (London) 2015 doi: 10.1038/ijo.2015.193. [DOI] [PubMed] [Google Scholar]
- 27.Igal R.A., Coleman R.A. Acylglycerol recycling from triacylglycerol to phospholipid, not lipase activity, is defective in neutral lipid storage disease fibroblasts. Journal of Biological Chemistry. 1996;271:16644–16651. doi: 10.1074/jbc.271.28.16644. [DOI] [PubMed] [Google Scholar]
- 28.Galgani J.E., Vasquez K., Watkins G., Dupuy A., Bertrand-Michel J., Levade T. Enhanced skeletal muscle lipid oxidative efficiency in insulin-resistant vs insulin-sensitive nondiabetic, nonobese humans. Journal of Endocrinology & Metabolism. 2013;98:E646–E653. doi: 10.1210/jc.2012-3111. [DOI] [PubMed] [Google Scholar]
- 29.Badin P.M., Vila I.K., Louche K., Mairal A., Marques M.A., Bourlier V. High-fat diet-mediated lipotoxicity and insulin resistance is related to impaired lipase expression in mouse skeletal muscle. Endocrinology. 2013;154:1444–1453. doi: 10.1210/en.2012-2029. [DOI] [PubMed] [Google Scholar]
- 30.Coue M., Badin P.M., Vila I.K., Laurens C., Louche K., Marques M.A. Defective natriuretic peptide receptor signaling in skeletal muscle links obesity to type 2 diabetes. Diabetes. 2015;64:4033–4045. doi: 10.2337/db15-0305. [DOI] [PubMed] [Google Scholar]
- 31.Kioka H., Kato H., Fujikawa M., Tsukamoto O., Suzuki T., Imamura H. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proceedings of the National Academy of Sciences U S A. 2014;111:273–278. doi: 10.1073/pnas.1318547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeFronzo R.A., Tripathy D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care. 2009;32(Suppl. 2):S157–S163. doi: 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrannini E., Simonson D.C., Katz L.D., Reichard G., Jr., Bevilacqua S., Barrett E.J. The disposal of an oral glucose load in patients with non-insulin-dependent diabetes. Metabolism. 1988;37:79–85. doi: 10.1016/0026-0495(88)90033-9. [DOI] [PubMed] [Google Scholar]
- 34.Turnbull P.C., Longo A.B., Ramos S.V., Roy B.D., Ward W.E., Peters S.J. Increases in skeletal muscle ATGL and its inhibitor G0S2 following 8 weeks of endurance training in metabolically different rat skeletal muscles. American Journal of Physiology, Regulatory, Integrative and Comparative. 2016;310:R125–R133. doi: 10.1152/ajpregu.00062.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurens C., Moro C. Intramyocellular fat storage in metabolic diseases. Hormone Molecular Biology and Clinical Investigation. 2015 doi: 10.1515/hmbci-2015-0045. [DOI] [PubMed] [Google Scholar]
- 36.Dyck D.J., Peters S.J., Glatz J., Gorski J., Keizer H., Kiens B. Functional differences in lipid metabolism in resting skeletal muscle of various fiber types. American Journal of Physiology. 1997;272:E340–E351. doi: 10.1152/ajpendo.1997.272.3.E340. [DOI] [PubMed] [Google Scholar]
- 37.Jocken J.W., Smit E., Goossens G.H., Essers Y.P., van Baak M.A., Mensink M. Adipose triglyceride lipase (ATGL) expression in human skeletal muscle is type I (oxidative) fiber specific. Histochemistry & Cell Biology. 2008;129:535–538. doi: 10.1007/s00418-008-0386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornaciu I., Boeszoermenyi A., Lindermuth H., Nagy H.M., Cerk I.K., Ebner C. The minimal domain of adipose triglyceride lipase (ATGL) ranges until leucine 254 and can be activated and inhibited by CGI-58 and G0S2, respectively. PLoS One. 2011;6:e26349. doi: 10.1371/journal.pone.0026349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Randle P.J., Garland P.B., Hales C.N., Newsholme E.A. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–789. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 40.Spriet L.L., Tunstall R.J., Watt M.J., Mehan K.A., Hargreaves M., Cameron-Smith D. Pyruvate dehydrogenase activation and kinase expression in human skeletal muscle during fasting. Journal of Applied Physiology. 1985;2004(96):2082–2087. doi: 10.1152/japplphysiol.01318.2003. [DOI] [PubMed] [Google Scholar]
- 41.Ehrenborg E., Krook A. Regulation of skeletal muscle physiology and metabolism by peroxisome proliferator-activated receptor delta. Pharmacological Review. 2009;61:373–393. doi: 10.1124/pr.109.001560. [DOI] [PubMed] [Google Scholar]
- 42.Bindesboll C., Berg O., Arntsen B., Nebb H.I., Dalen K.T. Fatty acids regulate perilipin5 in muscle by activating PPARdelta. Journal of Lipid & Research. 2013;54:1949–1963. doi: 10.1194/jlr.M038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancock C.R., Han D.H., Chen M., Terada S., Yasuda T., Wright D.C. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proceedings of the National Academy of Sciences U S A. 2008;105:7815–7820. doi: 10.1073/pnas.0802057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oakes N.D., Kjellstedt A., Thalen P., Ljung B., Turner N. Roles of fatty acid oversupply and impaired oxidation in lipid accumulation in tissues of obese rats. Journal of Lipids. 2013;2013:420754. doi: 10.1155/2013/420754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chavez J.A., Summers S.A. A ceramide-centric view of insulin resistance. Cell Metabolism. 2012;15:585–594. doi: 10.1016/j.cmet.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 46.El-Assaad W., El-Kouhen K., Mohammad A.H., Yang J., Morita M., Gamache I. Deletion of the gene encoding G0/G 1 switch protein 2 (G0s2) alleviates high-fat-diet-induced weight gain and insulin resistance, and promotes browning of white adipose tissue in mice. Diabetologia. 2015;58:149–157. doi: 10.1007/s00125-014-3429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heckmann B.L., Zhang X., Xie X., Saarinen A., Lu X., Yang X. Defective adipose lipolysis and altered global energy metabolism in mice with adipose overexpression of the lipolytic inhibitor G0/G1 switch gene 2 (G0S2) Journal of Biological Chemistry. 2014;289:1905–1916. doi: 10.1074/jbc.M113.522011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.