Abstract
Pathogenic bacteria could adjust gene expression to enable their survival in the distinct host environment. However, the mechanism by which bacteria adapt to the host environment is not well described. In this study, we demonstrated that nucleoside diphosphate kinase (Ndk) of Pseudomonas aeruginosa is critical for adjusting the bacterial virulence determinants during infection. Ndk expression was down-regulated in the pulmonary alveoli of a mouse model of acute pneumonia. Knockout of ndk up-regulated transcription factor ExsA-mediated T3S regulon expression and decreased exoproduct-related gene expression through the inhibition of the quorum sensing hierarchy. Moreover, in vitro and in vivo studies demonstrated that the ndk mutant exhibits enhanced cytotoxicity and host pathogenicity by increasing T3SS proteins. Taken together, our data reveal that ndk is a critical novel host-responsive gene required for coordinating P. aeruginosa virulence upon acute infection.
Pseudomonas aeruginosa is a versatile Gram-negative opportunistic pathogen responsible for causing acute and chronic infections. It has the largest bacterial genome (more than 6.2 million base pairs) in which 8.4% of the genome encodes transcriptional regulators and environmental sensors, which renders the bacteria highly adaptive to overcome hostile conditions1. The capacity of P. aeruginosa to perceive the host environmental stresses and to make adaptive responses enables the bacteria to survive and establish infection2. Nevertheless, the investigations of how bacteria sense host environmental stimuli and make adjustments are limited.
Acute lung infection caused by P. aeruginosa is a great threat for human health and is associated with poor clinical outcome. The secreted virulence products, such as elastase (LasA, LasB), alkaline proteinase, exotoxin A, and phospholipase C, play important roles in P. aeruginosa-mediated acute lung epithelial injury3. The type III secretion system (T3SS) is a vital virulence determinant in promoting acute lung infection via P. aeruginosa4.The T3SS is a complex protein secretion and translocation machinery composed of five protein components, including secretion apparatus (termed the injectisome), translocation apparatus, regulatory proteins, effector proteins and chaperones5. The underlying mechanism of T3SS-mediated virulence relies on an arsenal of toxic effector proteins that are directly injected into the cytoplasms of eukaryotic cells by the T3SS syringe-like injectisome5,6. Four effectors, termed ExoS, ExoT, ExoY and ExoU, have been identified in P. aeruginosa6. These effectors are involved in a variety of bacterial virulence associated activities, such as the induction of host cell death, the inhibition of cell proliferation, the preclusion of cell phagocytosis, and the disturbance of the host innate immune response5,6. In addition, T3SS exhibits an exotoxin S/T/Y-independent pathogenic role during acute lung infection7. The T3SS translocation apparatus (constituted by translocators PopB, PopD, and PcrV), which aids in delivering the effector proteins across the host cell plasma membrane, is sufficient to cause eukaryotic cell death by directly increasing the membrane permeability response5.
It has been reported that P. aeruginosa T3SS regulation is involved in a series of complicated mechanisms, such as the direct effect on T3SS-encoding gene expression and type III secretion activity8. Transcription factors, such as ExsA9 and PsrA10, are direct activators that promote the transcription of T3SS-encoding genes. Other proteins, such as RNA-binding protein Crc11 and DeaD (PA2840) (an Escherichia coli RNA-helicase homologue)12 fine tune P. aeruginosa T3SS gene expression by regulating ExsA. Apart from the direct regulation of T3SS gene transcription, T3SS activation is coupled with type III secretion activity in different conditions, such as host cell contact and low Ca2+ signaling, that activate the ExsCEDA regulatory cascade by promoting ExsE secretion8.
Quorum sensing (QS) is a cell-density-dependent mechanism of communication among microorganisms. In P. aeruginosa, three QS regulatory circuits have been reported, including two N-acylhomoserine lactone (AHL) regulatory circuits (las and rhl systems) and a quinolone signaling circuit (pqs system)13. In the las, rhl and pqs systems, the LasI and RhlI synthetases and pqsABCD gene products catalyze the synthesis of signal molecules N-(3-oxododecanoyl)-l-homoserine lactone (3-oxo-C12-HSL), N-butanoyl-l-homoserine lactone (C4-HSL) and 2-heptyl-3-hydroxy-4-quinolone (PQS), respectively13. When 3-oxo-C12-HSL, C4-HSL and PQS interact with their cognate transcriptional regulators LasR, RhlR and PqsR, respectively, the complex modulates the expression levels of its down-stream genes involved in T3SS protein synthesis and the production of elastase, pyocyanin, LasA protease, alkaline protease, exotoxin A and siderophores13,14,15. Multiple factors are involved in the regulation of the P. aeruginosa QS system, such as the regulator proteins QteE16, VqsM17 and QsrO (QS-repressing ORF2226)18, and the quorum quenching acylases PvdQ and PA030519,20.
Nucleoside diphosphate kinase (Ndk), a vital NTP-generating kinase in the maintenance of intracellular NTP pools via its terminal phosphotransfer activity from NTPs to NDPs, shares highly homology among microorganisms, humans, animals, and plants21. Ndk is secreted outside of the bacteria; thus, its direct host pathogenicity has been extensively studied. In Mycobacterium tuberculosis, secreted Ndk promotes bacterial survival within host macrophages by arresting phagosome maturation through GTPase activating protein (GAP) activity or by disturbing host gene expression via its DNAse activity22. Ndk can be transported into host cells by the T3SS apparatus in P. aeruginosa23, and it functions as a T3SS effector in inducing the host inflammatory response via stimulating the secretion of interleukin-1α (IL-1α), IL-1β, and IL-824,25. In spite of previous reports that showed that Ndk plays critical roles in host pathogenicity and the regulation of bacterial virulence-associated traits, such as DNA/RNA synthesis, cell division and alginate synthesis21,26, the available data showing that Ndk contributes to the regulation of bacterial virulence, particularly the virulence of the notorious P. aeruginosa, are still limited.
In this study, it was found that ndk is a novel host-responsive regulator that is down-regulated in the pulmonary alveoli of a mouse model of acute pneumonia. An ndk-deleted mutant promoted P. aeruginosa cytotoxicity and host pathogenicity by activating T3SS. Further analysis revealed that Ndk inhibits T3SS through the QS-ExsA circuit. Therefore, a better understanding of Ndk and its regulatory role in bacterial virulence will maximize opportunities for the development of strategies to treat P. aeruginosa infection.
Results
Transcription of T3SS components was activated whereas ndk was inhibited in P. aeruginosa in a mouse model of acute pneumonia
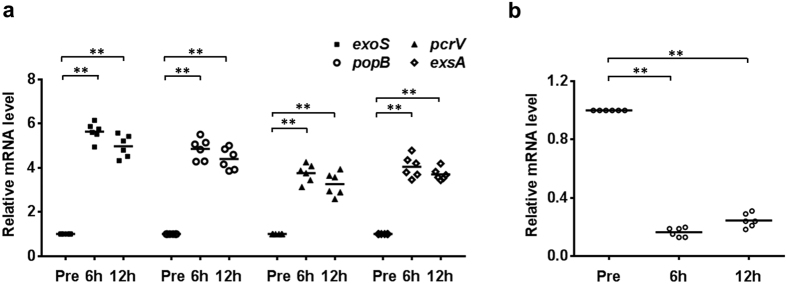
Upon acute infection, the activation of type III secretion machinery is coupled with increased T3SS gene transcription8, which compelled us to investigate the expression levels of T3SS-encoding genes in P. aeruginosa during acute pneumonia. Initially, mice were intranasally challenged with PAO1. At 6 and 12 h post-infection, the bronchoalveolar lavage fluid (BALF) was collected, and the expression levels of T3SS genes were evaluated by real-time PCR. The genes encoding the T3SS effector protein ExoS, the translocation apparatus (PcrV and PopB) and the transcription factor ExsA were drastically up-regulated in P. aeruginosa during infection compared to those of P. aeruginosa cultured in vitro (Fig. 1a).
Figure 1. Transcription of T3SS components was activated whereas ndk was inhibited in P. aeruginosa in a mouse model of acute pneumonia.
Mice were intranasally challenged with 2 × 108 CFU of PAO1. At 6 and 12 h post-infection, the BALFs were collected from the infected mice and the expression levels of T3SS genes (a) and ndk (b) in BALFs were evaluated by real-time PCR. The 50S ribosomal protein-coding gene rplU was used as an internal control. Black bars represent medians for the group of mice. In each time point, six mice were used. *Represents P < 0.05 compared to bacteria in vitro (Pre).
In addition, to test whether ndk might function as a responsive gene during P. aeruginosa acute infection, ndk expression in BALF was investigated. As shown in Fig. 1b, ndk expression was significantly down-regulated during P. aeruginosa infection. These results hint that a connection between ndk and T3SS might exist during P. aeruginosa acute infection.
Given that host cell contact is a crucial factor to activate T3SS gene expression upon P. aeruginosa infection8, we further examined whether the inhibition of ndk expression during pulmonary infection of P. aeruginosa is in a cell contact-dependent manner. To verify this speculation, the PAO1 strain was utilized to infect paraformaldehyde fixed or non-fixed human alveolar basal epithelial adenocarcinomic A549 cells and murine RAW264.7 macrophage cell lines. Unexpectedly, the ndk expression level did not show apparent change in the non-fixed and fixed cells compared to PAO1 cultured in DMEM (Supplementary Fig. 1), indicating that the inhibition of ndk expression during pulmonary infection of P. aeruginosa is not dependent on the host cell contact.
The ndk null-mutant of P. aeruginosa exacerbates host pathogenicity in vivo by T3SS activation
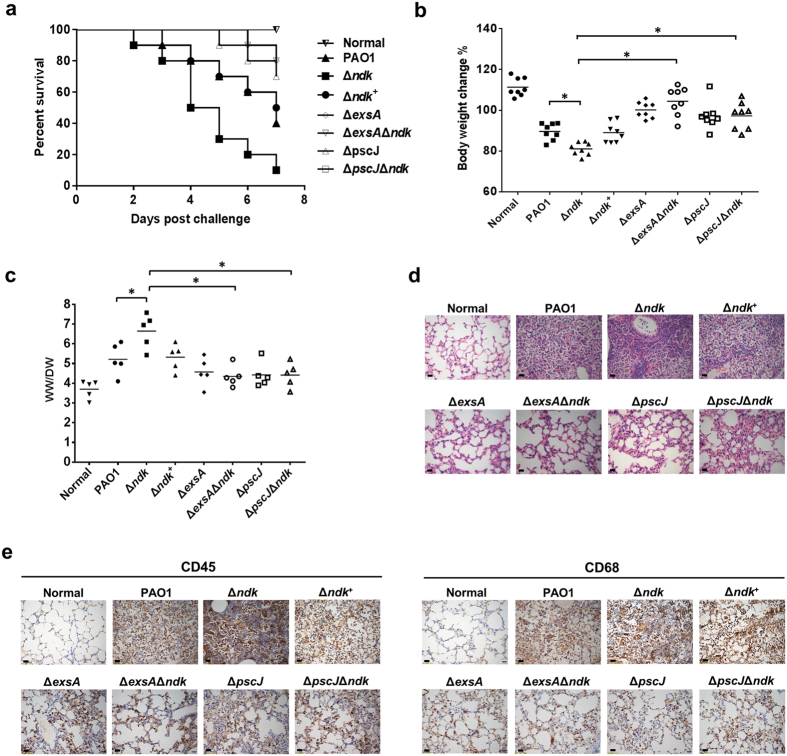
To address whether ndk expression is connected with T3SS in host pathogenicity, the mouse survival rate, body weight, lung edema and histopathology were evaluated in mice intranasally challenged with P. aeruginosa strain PAO1, a chromosomal ndk deletion mutant (Δndk), and an ndk complementary strain (Δndk+). As shown in Fig. 2a, Δndk caused 90% mortality, which was much higher than that caused by PAO1 and Δndk+ (60% and 50%, respectively) at 7 days post-infection. In addition, the mice infected with Δndk suffered a greater loss of body weight (Fig. 2b) and displayed increased lung edema (Fig. 2c) compared to the PAO1- and Δndk+-infected mice. Histopathology determined by hematoxylin and eosin (H&E) staining showed that lungs from Δndk-infected mice displayed more obvious inflammatory cell infiltration than the PAO1- and Δndk+-infected groups (Fig. 2d). Further analysis by immunohistochemical (IHC) staining showed that more leukocytes and macrophages infiltrated the lungs of Δndk-infected mice compared to the PAO1- and Δndk+-infected groups (Fig. 2e).
Figure 2. The ndk null-mutant of P. aeruginosa exacerbates host pathogenicity in vivo by T3SS activation.
Mice were intranasally challenged with the indicated strains (0.5 × 108 CFU per mouse). The subsequent analyses were evaluated at 7 days post-infection. (a) Mouse survival rate. Ten mice were included in each group. Survival curves were estimated using the Kaplan-Meier analysis with a log-rank test. (b) Body weight change. Eight mice were included in each group. (c) Lung edema. Lung edema was evaluated by calculating the lung wet-to-dry weight ratio. Five mice were included in each group. *Represents P < 0.05. Black bars represent medians for the group of mice. (d) Photomicrographs of mouse lung. The lung tissues embedded in paraffin were cut in 5 μm thickness and were subjected to H&E staining. Scale bar: 20 μm. (e) IHC analysis of inflammatory cell infiltration. The leukocyte common antigen CD45 was used for the detection of leukocytes, and CD68 was used for the visualization of macrophage. Scale bar: 20 μm.
To further investigate whether the increased pathogenicity of Δndk is related to activated T3SS upon P. aeruginosa infection, the mice were intranasally challenged with the reduced T3SS expression strain ΔexsA, the T3SS effector deficient strain ΔpscJ (defective in type III secretion27), and the double knock-out strains ΔexsAΔndk and ΔpscJΔndk. Compared to PAO1-infected mice, the ΔexsA- and ΔpscJ-infected mice showed significantly decreased mortality (Fig. 2a), body weight loss (Fig. 2b), lung edema (Fig. 2c), and lung inflammatory cell infiltration (Fig. 2d). Similar results were also observed in ΔexsAΔndk- and ΔpscJΔndk-infected mice when compared to Δndk-infected mice (Fig. 2), suggesting that the inhibition of ndk expression might lead to increased host pathogenicity through T3SS activation.
The ndk null-mutant enhances P. aeruginosa virulence in A549 cells through T3SS activation
Based on the in vivo study, the influence of ndk expression on P. aeruginosa virulence was further investigated in vitro. A549 cells were infected with PAO1, Δndk, and Δndk+, and cell viability was analyzed with a Cell Counting Kit 8 (CCK8) assay and calcein-AM staining. The CCK8 assay demonstrated that Δndk caused a much higher cytotoxicity than PAO1 and Δndk+ (Fig. 3a). Calcein-AM is a fluorescence stain for viable cells; the cell viability was estimated by calculating the green fluorescence intensity28. In accordance with the CCK8 assay, the result from calcein-AM staining also showed a similar trend (Fig. 3b,c). Given that P. aeruginosa secretes a variety of exoproducts that are toxic to cells, we examined whether the virulence products secreted by the bacteria were also involved in cytotoxicity during infection. To verify this speculation, the supernatants from PAO1, Δndk, and Δndk+ cultures were utilized to infect A549 cells. The cytotoxicity of the Δndk culture medium was significantly lower than those from PAO1 and Δndk+ media (Fig. 3a,b), indicating that the increased cytotoxicity of Δndk might be due to the changes of virulence-related determinants other than secretion products.
Figure 3. The ndk null-mutant enhances P. aeruginosa virulence in A549 cells through T3SS activation.
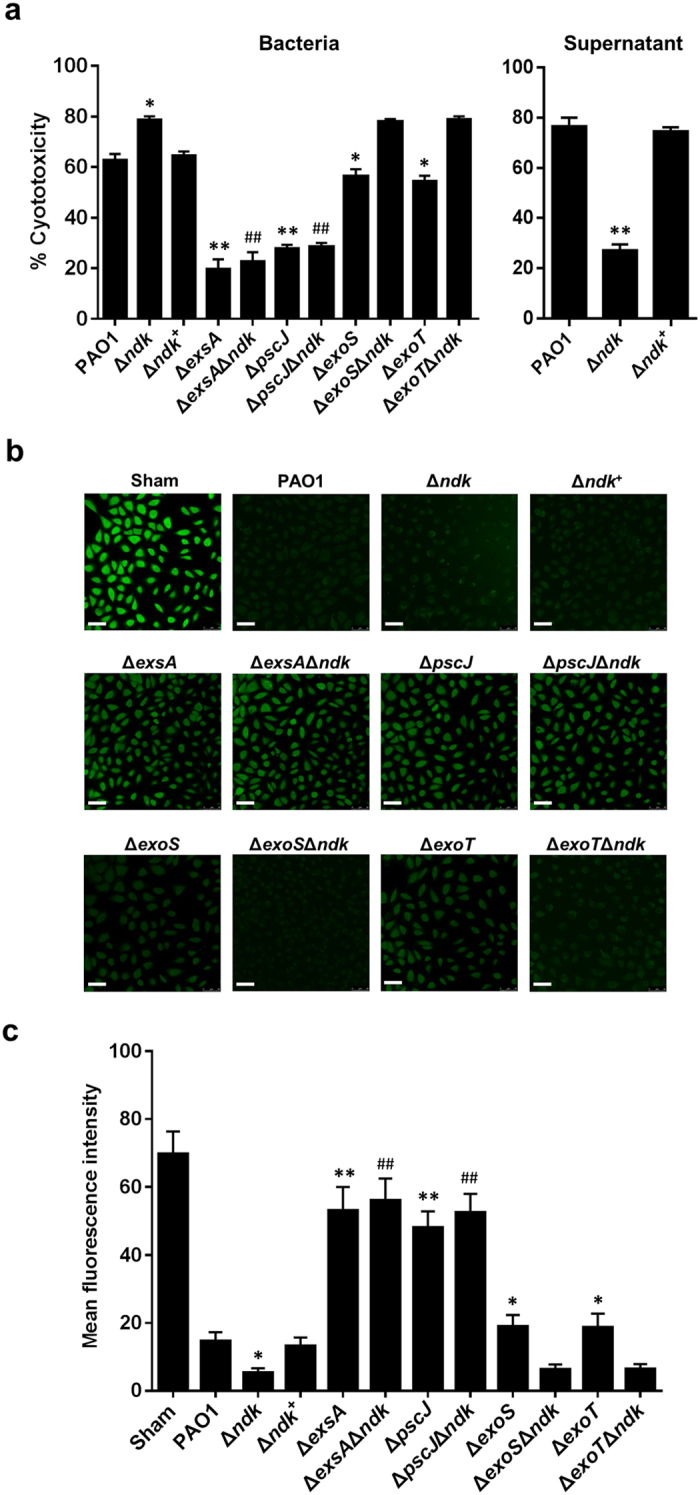
Prior to cell infection, the indicated bacterial strains were cultured in LB broth for 12 h. The bacterial pellets and culture supernatants were collected after centrifugation. A549 cells were infected with the indicated strains (MOI = 50). In addition, A549 cells were treated with bacterial culture media 5-fold diluted in DMEM. At 2 h post-infection, the cell viability was detected. (a) CCK-8 assay. The percentage of cytotoxicity was calculated according to the following formula: cytotoxicity % = 1 − (OD450 infected cells/OD450 sham-infected control) × 100%. (b) Calcein-AM staining. Scale bar: 50 μm. Representative images from triplicate experiments are shown. (c) Fluorescence intensity per cell from (b) was analyzed by Image pro-Plus software (IPP, edition 6.0) (ncell = 90). Data are expressed as the mean ± SD from three independent experiments. *Indicates P < 0.05 compared to PAO1 strain. #Indicates P < 0.05 compared to Δndk strain.
In view of the above observations, we sought to determine whether T3SS is involved in the cytotoxicity of Δndk. Therefore, T3SS gene knockout strains were applied to infect A549 cells. Compared to PAO1-infected cells, the cytotoxicity was much lower in ΔexsA- and ΔpscJ-infected cells (Fig. 3). Moreover, decreased cytotoxicity was also observed in the ΔexsAΔndk- and ΔpscJΔndk-infected cells compared to Δndk-infected ones (Fig. 3), further indicating that T3SS might be involved in Ndk mediated P. aeruginosa virulence. Nevertheless, the cytotoxicity of ΔexoS and ΔexoT strains was decreased in comparison to that of PAO1 strain, while the cytotoxicity of ΔexoSΔndk and ΔexoTΔndk strains displayed no significant change compared to that of Δndk strain (Fig. 3), indicating that the increased cytotoxicity of Δndk was due to the combined cytotoxic effect of T3SS proteins.
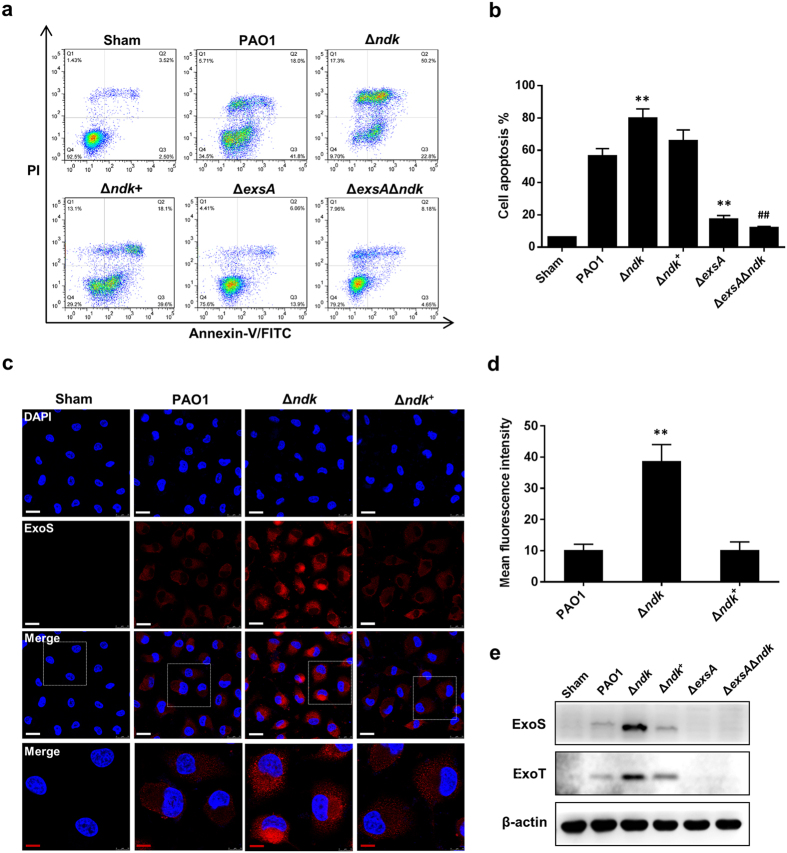
It has been shown that P. aeruginosa T3SS promotes cell apoptosis by injecting toxic effectors (such as ExoS) into the cytoplasm of host cells29; therefore, we detected cell apoptosis by flow cytometry and intracellularly translocated ExoS by immunofluorescence (IF) staining and western blotting in PAO1-, Δndk- or/and ΔexsAΔndk-infected cells. As shown in Fig. 4a,b, the Δndk strain promoted cell apoptosis compared to the PAO1 and Δndk+ strains, whereas apoptosis in the ΔexsAΔndk-infected cells decreased. Furthermore, the IF and western blot assays demonstrated that the intracellular translocation of ExoS was enhanced in Δndk-infected cells compared to PAO1- and Δndk+-infected cells (Fig. 4c–e), whereas the translocation of ExoS was decreased in the ΔexsA- and ΔexsAΔndk-infected cells (Fig. 4e).These results suggested that the null-mutation of ndk promotes cell apoptosis in a T3SS effector-dependent manner.
Figure 4. The ndk null-mutant promotes P. aeruginosa T3SS-mediated cell apoptosis.
A549 cells were challenged with the indicated bacteria (MOI = 50) for 2 h. (a) Cell apoptosis was analyzed by flow cytometry using Annexin V-FITC-PI apoptosis detection kit. (b) Histogram represents the percentage of cell apoptosis from (a). (c) Examination of effector protein ExoS translocation by IF staining. White scale bar: 25 μm. Red scale bar: 10 μm. (d) Fluorescence intensity per cell from (c) was analyzed by IPP software (ncell = 30). (e) Western blot analysis for intracellular translocation of ExoS. Representative images from triplicate experiments are shown. Data are expressed as mean ± SD from three independent experiments. *Indicates P < 0.05 compared to PAO1 strain. #Indicates P < 0.05 compared to Δndk strain.
In addition, the T3SS translocation apparatus-mediated pore formation on the cell membrane is another determinant for cytotoxicity5,7. Hence, we further observed the cell membrane permeability after infection by observing calcein leakage via calcein AM-staining. This experiment confirmed that ndk deprivation promotes cytotoxicity by elevating cell membrane permeability (Supplementary Fig. 2).
Ndk deficiency leads to up-regulated expression of P. aeruginosa T3SS
Given that T3SS consists of complex protein components5, the gene expression profiles of PAO1 and Δndk strains cultured in LB broth for 12 h were analyzed by mRNA transcriptome sequencing to obtain a comprehensive understanding of the role of Ndk for the T3SS. Ndk deprivation led to massive up-regulation of genes participating in the generation of T3SS components. These genes contain four gene clusters, PA1690-PA1697, PA1698-1709, PA1710-1712, and PA1714-1725, and effector protein genes exoS, exoT, and exoY, and chaperone spcS (Table 1).
Table 1. Altered gene expression in Δndk compared to PAO1.
| Gene locus | Name | Gene function | log2 ratio (Δndk/WT) | P vaule |
|---|---|---|---|---|
| T3SS | ||||
| PA1690 | pscU | translocation protein in type III secretion | 3.02 | 1.05E-20 |
| PA1691 | pscT | translocation protein in type III secretion | 4.84 | 1.06E-23 |
| PA1692 | PA1692 | translocation protein in type III secretion | 4.37 | 1.11E-26 |
| PA1693 | pscR | type III secretion system protein | 1.23 | 1.30E-18 |
| PA1694 | pscQ | type III secretion system protein | 6.87 | 6.27E-106 |
| PA1695 | pscP | translocation protein in type III secretion | 17.11 | 2.30E-204 |
| PA1696 | pscO | translocation protein in type III secretion | 16.91 | 3.66E-77 |
| PA1697 | PA1697 | type III secretion system ATPase | 8.15 | 0 |
| PA1698 | popN | type III secretion outer membrane protein PopN | 4.80 | 8.74E-160 |
| PA1699 | pcr1 | hypothetical protein | 17.30 | 5.92E-59 |
| PA1700 | pcr2 | hypothetical protein | 7.06 | 3.27E-41 |
| PA1701 | pcr3 | hypothetical protein | 16.28 | 2.10E-38 |
| PA1702 | pcr4 | hypothetical protein | 4.21 | 5.90E-19 |
| PA1703 | pcrD | type III secretory apparatus protein PcrD | 2.02 | 9.36E-80 |
| PA1704 | pcrR | transcriptional regulator PcrR | 1.91 | 3.68E-06 |
| PA1705 | pcrG | type III secretion regulator | 5.24 | 1.90E-83 |
| PA1706 | pcrV | type III secretion protein PcrV | 5.64 | 0 |
| PA1707 | pcrH | regulatory protein PcrH | 6.31 | 1.94E-209 |
| PA1708 | popB | translocator protein PopB | 5.50 | 0 |
| PA1709 | popD | translocator outer membrane protein PopD | 5.99 | 0 |
| PA1710 | exsC | exoenzyme S synthesis protein C | 4.05 | 0 |
| PA1711 | exsE | hypothetical protein | 4.12 | 5.97E-235 |
| PA1712 | exsB | exoenzyme S synthesis protein B | 4.42 | 2.33E-175 |
| PA1713 | exsA | transcriptional regulator ExsA | 4.06 | 4.80E-157 |
| PA1714 | exsD | hypothetical protein | 4.78 | 0 |
| PA1715 | pscB | type III export apparatus protein | 5.84 | 2.10E-115 |
| PA1716 | pscC | type III secretion outer membrane protein PscC | 3.51 | 4.36E-184 |
| PA1717 | pscD | type III export protein PscD | 6.02 | 7.01E-76 |
| PA1718 | pscE | type III export protein PscE | 5.21 | 3.45E-31 |
| PA1719 | pscF | type III export protein PscF | 3.92 | 7.80E-50 |
| PA1720 | pscG | type III export protein PscG | 4.01 | 3.03E-46 |
| PA1721 | pscH | type III export protein PscH | 3.95 | 2.41E-33 |
| PA1722 | pscI | type III export protein PscI | 4.28 | 3.92E-39 |
| PA1723 | pscJ | type III export protein PscJ | 3.06 | 2.39E-47 |
| PA1724 | pscK | type III export protein PscK | 3.94 | 9.94E-12 |
| PA1725 | pscL | type III secretion system protein | 2.49 | 2.20E-16 |
| PA3841 | exoS | exoenzyme S | 3.69 | 0 |
| PA0044 | exoT | exoenzyme T | 4.54 | 0 |
| PA2191 | exoY | adenylate cyclase | 2.97 | 2.05E-46 |
| PA3842 | spcS | chaperone | 3.69 | 1.27E-68 |
| QS signal molecule synthetase and regulator | ||||
| PA1432 | lasI | autoinducer synthesis protein LasI | −3.12 | 0 |
| PA3476 | rhlI | autoinducer synthesis protein RhlI | −2.09 | 0 |
| PA0996 | pqsA | coenzyme A ligase | −5.81 | 0 |
| PA0997 | pqsB | alkyl quinolones biosynthesis protein | −6.96 | 0 |
| PA0998 | pqsC | alkyl quinolones biosynthesis protein | −5.91 | 4.61E-260 |
| PA0999 | pqsD | 3-oxoacyl-synthase III | −4.22 | 0 |
| PA1000 | pqsE | Quinolone signal response protein | −4.12 | 8.01E-254 |
| PA2587 | pqsH | FAD-dependent monooxygenase | −4.45 | 0 |
| PA1430 | lasR | transcriptional regulator LasR | −1.03 | 1.42E-240 |
| PA3477 | rhlR | transcriptional regulator RhlR | −3.39 | 0 |
| PA1003 | mvfR | transcriptional regulator MvfR (PqsR) | −1.31 | 6.81E-64 |
| PA1898 | qscR | quorum-sensing control repressor | 3.32 | 7.68E-65 |
| Virulence associated exoproducts | ||||
| PA3724 | lasB | elastase LasB | −6.76 | 0 |
| PA1871 | lasA | LasA protease | −7.03 | 0 |
| PA1246 | aprD | alkaline protease secretion protein AprD | −2.92 | 2.19E-116 |
| PA1247 | aprE | alkaline protease secretion protein AprE | −2.95 | 5.20E-102 |
| PA1248 | aprF | alkaline protease secretion outer membrane protein AprF | −2.50 | 3.21E-55 |
| PA1249 | aprA | alkaline metalloproteinase | −1.25 | 0 |
| PA0026 | plcB | phospholipase C | −2.43 | 1.02E-219 |
| PA4210 | phzA1 | phenazine biosynthesis protein | −0.98 | 0.051979 |
| PA4211 | phzB1 | phenazine biosynthesis protein | −2.21 | 1.12E-53 |
| PA4212 | phzC1 | phenazine biosynthesis protein PhzC | −1.81 | 3.68E-06 |
| PA4214 | phzE1 | phenazine biosynthesis protein PhzE; | −2.42 | 4.68E-08 |
| PA4217 | phzS | flavin-containing monooxygenase | −2.05 | 8.77E-114 |
| PA1899 | phzA2 | phenazine biosynthesis protein | −3.04 | 1.79E-22 |
| PA1900 | phzB2 | phenazine biosynthesis protein | −4.89 | 0 |
| PA1901 | phzC2 | phenazine biosynthesis protein PhzC | −4.26 | 1.81E-58 |
| PA1130 | rhlC | rhamnosyltransferase | −3.18 | 0 |
| PA3478 | rhlB | rhamnosyltransferase subunit B | −7.00 | 0 |
| PA3479 | rhlA | rhamnosyltransferase subunit A | −6.69 | 0 |
| PA2570 | lecA | PA-I galactophilic lectin | −5.82 | 0 |
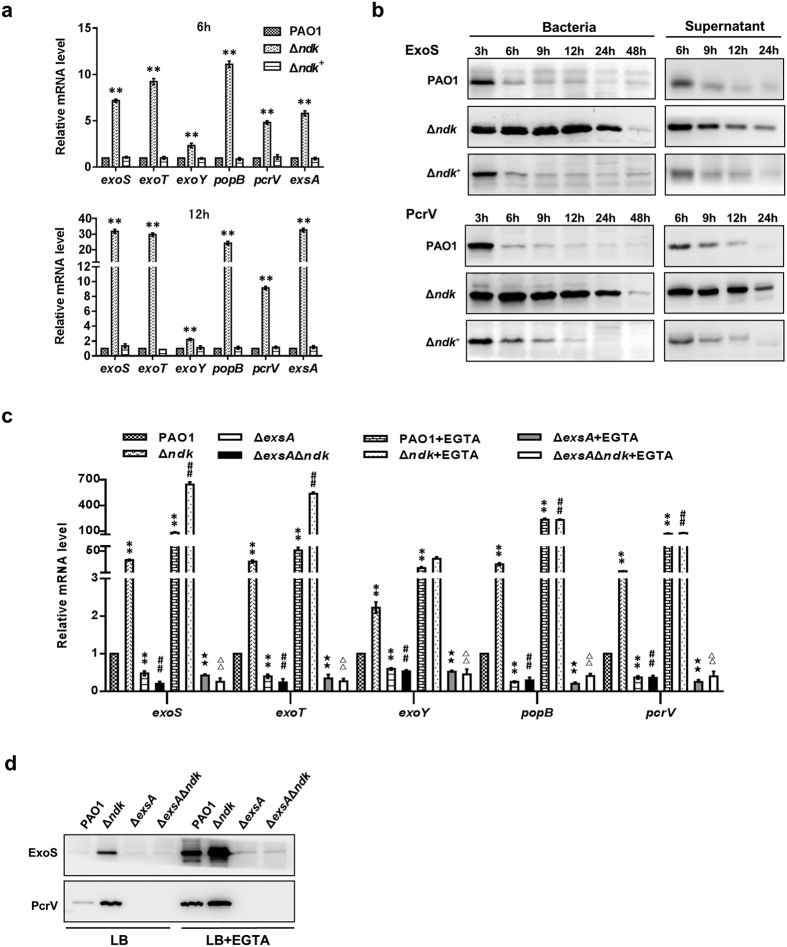
To further verify the results from mRNA transcriptome sequencing, the gene expression levels of T3SS of PAO1, Δndk, and Δndk+ strains cultured for 12 h in LB broth were analyzed by real-time PCR. In accordance with the results from transcriptome sequencing (Table 1), the genes encoding the translocator apparatus (PopB and PcrV), effectors (ExoS and ExoT) and the transcriptional regulator ExsA were all sharply increased in Δndk cells compared to PAO1 and Δndk+ cells at the early (6 h) and middle (12 h) bacterial growth phases (Fig. 5a) (the bacterial growth curve is presented in Supplementary Fig. 3). In addition, western blot analysis revealed that the intracellular and secretory protein levels of ExoS and PcrV of the Δndk strain were significantly higher than those in the PAO1 and Δndk+ strains (Fig. 5b). Taken together, these results indicated that Ndk might serve as a negative regulator controlling T3SS expression in P. aeruginosa.
Figure 5. Ndk deficiency leads to up-regulated expression of P. aeruginosa T3SS.
The indicated bacteria were inoculated into LB broth and a T3SS inducing condition (LB broth supplemented with 5 mM EGTA and 20 mM MgCl2) (initial OD600 of 0.01) and cultured at 37 °C for the indicated time. The bacteria and culture media were harvested for the subsequent analysis. (a) Real-time PCR analysis for T3SS-encoding gene expression. (b) Western blot analysis for the intracellular expression and secretion of T3SS proteins. (c) Real-time PCR analysis for T3SS gene expression with EGTA induction for 12 h. (d) Western blot analysis for intracellular T3SS protein expression with EGTA induction for 12 h. Representative images from triplicate experiments are shown (b,d). Data represent mean ± SD from three independent experiments (a,c). *Indicates P < 0.05 compared to PAO1 strain. #Indicates P < 0.05 compared to Δndk strain. ★ Indicates P < 0.05 compared to PAO1 + EGTA group. Δ Indicates P < 0.05 compared to Δndk + EGTA group.
Ndk negatively regulates T3SS gene expression in an ExsA-dependent manner
ExsA is a master transcription factor for the activation of T3SS gene expression30. In accordance with the results that showed that a lack of ndk resulted in increased expression of exsA (Table 1; Fig. 5a,b), we investigated the role of ExsA in the Ndk-mediated regulation of T3SS. Hence, ΔexsA and ΔexsAΔndk double-knockout strains were generated and cultured for 12 h in LB broth, and gene and protein expression levels were analyzed by real-time PCR and western blot, respectively. As shown in Fig. 5c,d, the ΔexsAΔndk mutant displayed a distinct down-regulation of exoS, exoT, exoY, popB, pcrV and ExoS and PcrV compared to the Δndk mutant. Given that ExsA-dependent activation of T3SS was induced under a Ca2+-limiting growth condition8, the influence of ExsA on T3SS expression in Δndk cells was further investigated by adding a Ca2+ chelating agent (EGTA) to the bacterial culture medium. The addition of 5 mM EGTA markedly stimulated T3SS gene and protein expression levels in the PAO1 and Δndk strains rather than in the ΔexsA and ΔexsAΔndk strains (Fig. 5c,d), demonstrating that ndk negatively regulates T3SS expression in an ExsA-dependent manner.
The deprivation of ndk inhibits the synthesis of exoproducts of P. aeruginosa
In addition to T3SS, the exoproducts secreted by P. aeruginosa are also crucial virulence determinants for acute infection3. To investigate whether Ndk plays a role in regulating these products, the gene expression levels of those factors in PAO1, Δndk and Δndk+ strains cultured in LB broth for 12 h were evaluated. mRNA transcriptome sequencing revealed that the transcription levels of genes encoding exoproducts such as elastase (lasA and lasB), alkaline protease (aprA and aprE-F), phospholipase C (plcB) and phenazine (phzA1-C1, phzE1, phzS and phzA2-C2) were significantly impaired in Δndk cells compared to PAO1 cells (Table 1). In addition, real-time PCR analysis demonstrated a similar trend (Fig. 6a).
Figure 6. The deprivation of ndk inhibits the synthesis of exoproducts of P. aeruginosa.
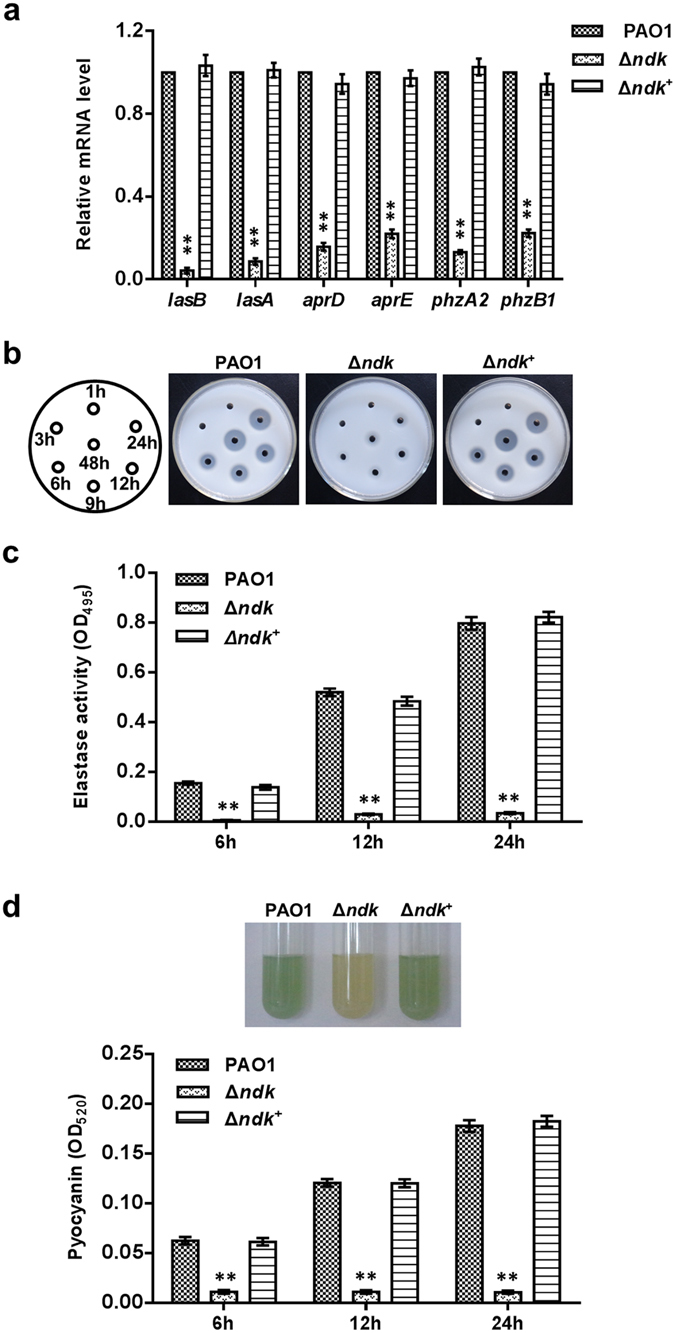
PAO1, Δndk and Δndk+ strains were inoculated into LB broth (initial OD600 of 0.01) and cultured at 37 °C for the indicated time. The bacteria and culture media were separately harvested for the subsequent analysis. (a) Real-time PCR analysis for exoproduct-related gene expression (12 h). (b) Skim milk plate assay for analysis of secreted proteases. (c) ECR assay for detecting elastase. (d) Analysis for pyocyanin production. Data represent mean ± SD from three independent experiments. *Indicates P < 0.05 compared to PAO1 strain.
Subsequently, the secreted proteases from the bacterial culture media were estimated using a skim mild plate assay and an Elastin-Congo Red (ECR) assay. The activities of secreted proteases, including elastase, were significantly inhibited in Δndk cells compared to PAO1 and Δndk+ cells (Fig. 6b,c). Given that phenazine is an intermediate metabolic product for pyocyanin31, it was assumed that the gene changes in phenazine might affect pyocyanin synthesis. Therefore, we determined pyocyanin production in the bacterial culture medium. As shown in Fig. 6d, the Δndk strain produced barely detectable pyocyanin, whereas the Δndk+ strain produced pyocyanin at a level comparable to the PAO1 strain.
Ndk negatively regulates the expression of the ExsA-mediated T3S regulon and positively modulates the expression of exoproducts through the QS system
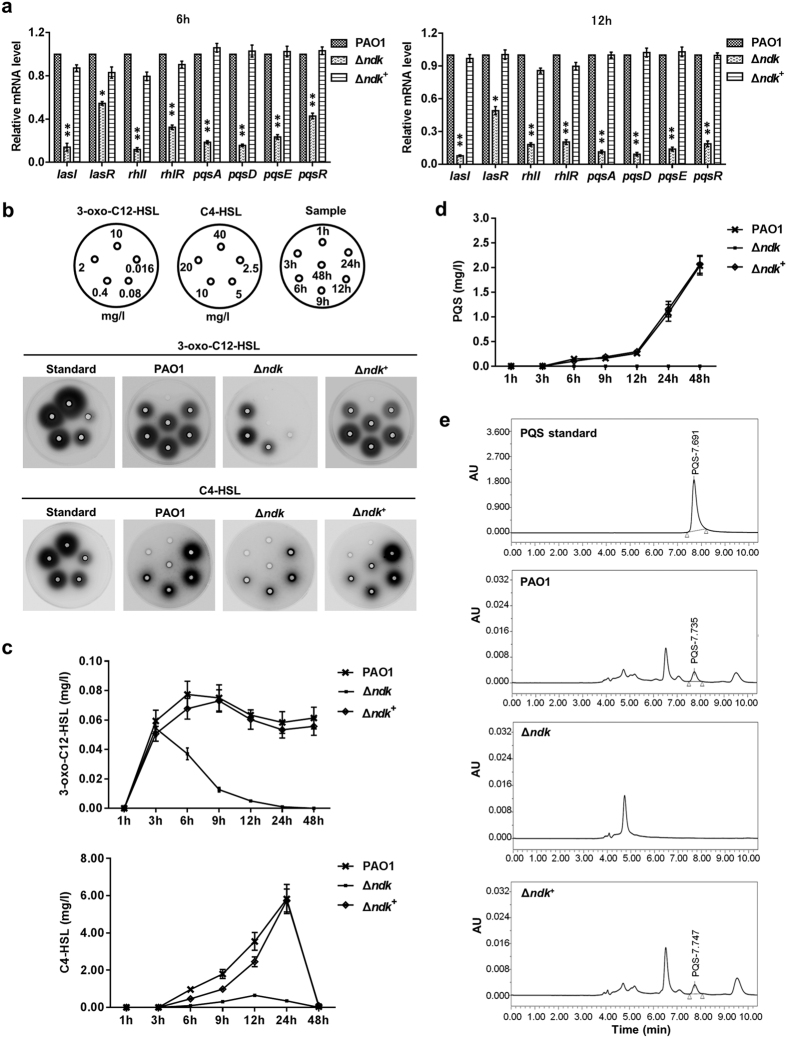
During acute pneumonia caused by P. aeruginosa, ndk gene expression was inhibited, while the expression levels of T3SS-encoding genes were activated in the BALFs of the infected mice (Fig. 1a). Furthermore, the expression levels of lasI, rhlI, and pqsA, which encode the QS synthetases, were also inhibited in that condition (Supplementary Fig. 4). These results suggest that QS might participate in the ndk-mediated regulation of T3SS upon infection. To address this issue, we set out to analyze the expression levels of QS genes in PAO1 and Δndk cells after culture in LB broth. Results from mRNA transcriptome sequencing showed that lasI, rhlI, pqsABCDE, lasR, rhlR and pqsR expression levels were all significantly inhibited in Δndk cells compared to PAO1 cells (Table 1). In addition, gene expression levels verified by real-time PCR also demonstrated decreased trends (Fig. 7a). Afterwards, signaling molecules in the bacterial culture media were further detected with a well-diffusion assay and a high-performance liquid chromatography (HPLC) assay. The null-mutation of ndk resulted in obvious inhibition of 3-oxo-C12-HSL and C4-HSL synthesis, whereas complementation of the ndk gene restored their production (Fig. 7b,c). The HPLC analysis revealed that PQS production in Δndk cells was apparently inhibited in comparison to PAO1 and Δndk+ cells (Fig. 7d,e).
Figure 7. The deprivation of ndk inhibits quorum sensing hierarchy.
PAO1, Δndk and Δndk+ strains were inoculated into LB broth (initial OD600 of 0.01) and cultured at 37 °C for the indicated time. The bacteria harvested at 6 h and 12 h were dissolved in TRIzol for gene expression analysis (a). The 3-oxo-C12-HSL and C4-HSL in the bacterial culture media were detected with a well-diffusion assay (b) and calculated by the measurement of gray value using Image J software (c). The PQS production in the bacterial culture medium was analyzed with HPLC (d) and the representative diagrams (12 h) were presented in (e). Data represent mean ± SD from three independent experiments. *Indicates P < 0.05 compared to PAO1 strain.
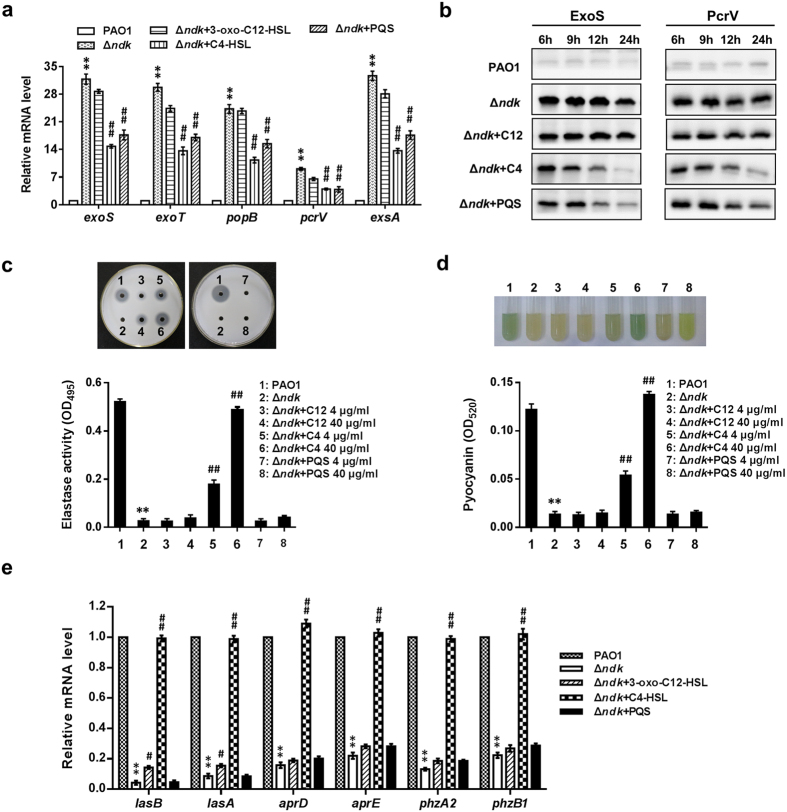
To examine whether decreased QS regulation might contribute to the increased expression of the T3S regulon in Δndk cells, exogenous 3-oxo-C12-HSL, C4-HSL or PQS at different concentrations were added to the culture medium of Δndk cells. The expression levels of exoS, exoT, popB, pcrV, exsA and ExoS and PcrV were reduced in Δndk cells when supplemented with C4-HSLs or PQS at 40 μg/ml (Fig. 8a,b). Supplementing with 40 μg/ml of 3-oxo-C12-HSL only slightly reduced the expression levels in Δndk cells; the protein levels did not significantly change (Fig. 8a,b). These results suggested that the rhl and pqs systems play more important roles than the las system in the regulation process of T3SS by Ndk in P. aeruginosa.
Figure 8. Ndk regulates the expression of the ExsA-mediated T3S regulon and exoproducts through the QS system.
PAO1 and Δndk strains were inoculated into LB broth at an initial OD600 of 0.01. Exogenous 3-oxo-C12-HSL, C4-HSL or PQS (4 μg/ml or 40 μg/ml) was added to the culture medium of Δndk cells. The bacteria and culture supernatants were harvested for subsequent analysis at the indicated culture time. (a) Real-time PCR analysis for T3SS-encoding gene expression (signal molecules at 40 μg/ml, 12 h). (b) Western blot analysis for the intracellular expression of T3SS proteins (signal molecules at 40 μg/ml, 12 h). (c) Skim milk plate and ECR assays for secreted proteases (12 h). (d) Analysis for pyocyanin production (12 h). (e) Real-time PCR analysis for exoproduct-related gene expression (signal molecules at 40 μg/ml, 12 h). C12 indicate 3-oxo-C12-HSL. C4 indicates C4-HSL. Data represent mean ± SD from three independent experiments. *Indicates P < 0.05 compared to PAO1 strain. #Indicates P < 0.05 compared to Δndk strain.
In addition to examining T3SS-encoding genes, we also analyzed the regulatory role of QS in the synthesis of exoproducts by adding exogenous 3-oxo-C12-HSL, C4-HSL or PQS to the culture medium of Δndk cells. Protease activity and pyocyanin production in Δndk cells were partially or completely recovered by the supplementation of C4-HSL at concentrations of 4 and 40 μg/ml, respectively (Fig. 8c,d). The exogenous addition of PQS and 3-oxo-C12HSL could not restore the elastase activity and pyocyanin production of Δndk cells even at a concentration of 40 μg/ml, even though an increase in pyoverdin production was observed in the Δndk +PQS (40 μg/ml) group (Fig. 8c,d). Moreover, gene expression analysis also revealed a similar trend (Fig. 8e). Taken together, our data suggest that ndk may negatively control bacterial virulence through the inhibition of T3SS transcription via the QS hierarchy in P. aeruginosa.
Discussion
During infection, bacteria might alter gene expression to adapt to the host environment32. Adjusting gene expression enables bacterial survival, colonization and dissemination in the host, which might ultimately result in the enhancement of host pathogenesis33,34,35. Nevertheless, the underlying mechanism by which bacteria perceive host environmental stimuli and subsequently adjust remains poorly understood. The activation of bacterial stress response networks, such as the QS systems, two-component systems and regulatory genes may contribute to the bacterial adaptive response and bacterial virulence13,33,34,36. soxR, a gene encoding a superoxide response regulator, is highly induced during P. aeruginosa infection of mice with burn wounds, and increases in its expression promote bacteremia33. In addition, suhB (a metabolism-related gene)34 and PA319135 were reported to be inducible genes of P. aeruginosa in the host pulmonary microenvironment, promoting mouse acute pneumonia through T3SS activation. In this study, we found that Ndk is a novel host-responsive regulator during acute infection and promotes P. aeruginosa virulence by suppressing its own expression, thereby triggering T3SS activation. Host cell contact is a way to modulate bacterial gene expression8. However, result demonstrated the host cell contact did not affect P. aeruginosa ndk gene expression (Supplementary Fig. 1). In the slime mould Dictyostelium discoideum, the sharply decreased transcript level of ndk gene is coinciding with the onset of the organism’s starvation-induced developmental cycle37,38, suggesting that host stress condition might play a role in modulating ndk gene expression. Upon lung infection, P. aeruginosa might encounter diverse pulmonary microenvironments including nutrition, iron, osmotic pressure and oxidative stress39,40. Whether these stimuli might act as the potential host factors that regulate ndk gene expression deserves further investigation.
T3SS consists of multiple protein components. It is reported that at least 42 genes are related to the T3S regulon, including five operons (36 genes) that participate in type III secretory machine generation and six additional genes encoding effectors and effector-specific chaperones41. In this study, we showed that Ndk exerts an extensive regulatory role on the T3SS (Table 1). ExsA, an AraC-like transcriptional regulator, plays a dominant role in the activation of T3S regulon transcription9. It has been shown that the five operons, including exsD-pscL, exsCBA, pscG-popD, popN-pcrR, and pscN-pscU, are all regulated by ExsA41. In addition, ExsA-binding sites have also been identified in the promoter regions of the effectors ExoS, ExoT, ExoY, ExoU, and the chaperone SpcS1. In this study, we demonstrated that ndk knockout results in the up-regulation of exsA and T3SS gene expression levels, whereas a null mutation of exsA from Δndk leads to suppressed expression of T3SS genes, indicating that Ndk regulates the transcription of T3SS genes in an ExsA-dependent manner (Fig. 5c,d). It has been shown that the human Ndk (NM23-H2), which shares highly homology with E. coli Ndk, is capable of binding to a nuclease-hypersensitive c-MYC promoter element and activating the c-MYC transcription42, indicating that bacterial Ndk might play a role in regulating gene transcription. Due to the fact that Ndk regulatesT3S regulon at the transcription level, it hints that Ndk might modulate T3SS in a promoter- or transcription factor-dependent manner.
T3SS activation is fine-tuned by a variety of factors, such as host cell contact, low Ca2+, metabolic signals, DNA damage, hyperosmotic stress and antibiotic exposure8. In addition, the QS hierarchy from P. aeruginosa14 and V. harveyi (Vibrio harveyi)43 also exhibits a negative effect on T3SS. In this study, we found that increased expression of exoS, exoT, pcrV, popB, ExoS and PcrV in Δndk cells was inhibited by C4-HSL or PQS rather than 3-oxo-C12HSL (Fig. 8a,b). Similar findings showed that the exoS gene was induced in the rhlI, rhlR or pqsR mutant, but not in the lasI mutant44,45. Bleves et al. demonstrated that the activity of a T3SS promoter fusion containing the secretion- and translocation-related operons increased in the rhlI mutant of P. aeruginosa but not in the lasR mutant14. In this study, we found that the C4-HSL mediated inhibition of T3SS genes in Δndk was due to exsA down-regulation (Fig. 8a). In V. harveyi, the QS regulator LuxR repressed T3SS gene expression through the inhibition of ExsA by binding to the PexsB promoter region located upstream of the exsBA operon43, but the exact effect of RhlR/I-C4-HSL on ExsA in P. aeruginosa remains unclear.
In contrast to T3SS expression, the gene expression and synthesis of exoproducts in Δndk cells were globally reduced, including elastase (LasA and LasB) and pyocyanin (Fig. 6). This phenomenon was reversed by the addition of C4-HSL but not 3-oxo-C12HSL or PQS (Fig. 8c–e). It had been thought that all three QS systems contribute to the regulation of elastase and pyocyanin in P. aeruginosa13. Due to crosstalk between the three QS systems, that is, the inhibition of the las system might lead to suppressed rhl and pqs regulation46, the decreased elastase activity observed in the lasI mutant47 was considered to be due to the influence of the rhl system. Brint et al. also observed defective synthesis of elastase and pyocyanin in rhlR and rhlI mutants48, suggesting that Ndk mediates the inhibition of exoproducts through the rhl system. In addition, it was found that the deprivation of ndk gene led to deficient swarming, enhanced twitching motility (Supplementary Fig. 6) and decreased biofilm formation compared to PAO1 strain (Supplementary Fig. 7), which indicates that Ndk might act as a multifunctional protein to regulate bacterial virulence.
As an NTP-generating kinase, Ndk maintains NTP balance by catalyzing the reversible transfer of the γ-phosphate of NTP to NDP. In addition, Ndk functions as a histidine kinase to mediate γ-phosphate transfer from NTPs to itself and then to the substrate proteins, such as EnvZ and CheA49,50. The histidine 117 (His117) in the His-Gly-Ser-Asp (HGSD) motif, which is conserved among prokaryotic species, is vital for Ndk-mediated autophosphorylation and phosphotransfer activity50,51. To investigate whether Ndk-mediated virulence regulation is dependent on the Ndk phosphorylation activity, the kinase activity-deficient strain PA (H117Q) was constructed, and its related phenotypes were examined. Unexpectedly, the gene expression levels of T3SS, three QS synthetases and elastase did not show apparent changes in the PA (H117Q) mutant compared to the PAO1 strain (Supplementary Fig. 5a). The T3SS protein levels, AHL production, protease activity and pyocyanin production were also comparable between PAO1 and PA (H117Q) strains (Supplementary Fig. 5b–e), suggesting that Ndk-mediated virulence regulation may not rely on its kinase activity. Consistent with our finding, Neeld et al. showed that the P. aeruginosa Ndk H117Q mutant induces a similar level of cytotoxicity compared to Ndk when expressed inside host cells23.
In this study, it was shown that cytotoxicity were enhanced in Δndk cells, in which T3SS was activated while exoproduct synthesis was inhibited (Figs 2, 3, 4, 5, 6), suggesting that T3SS plays a dominant role in promoting host pathogenicity rather than the toxic exoproducts produced during acute infection. Upon pulmonary infection, T3SS promotes macrophage death and precludes macrophage-mediated phagocytosis to facilitate bacterial survival5, while the exoproducts disrupt epithelial barrier integrity and lyse fibrin to promote bacterial dissemination3. T3SS is activated at a low bacterial density, whereas it is inhibited at high bacterial density43, while the toxic exoproducts are secreted when bacteria reach a high density47. These different bacterial virulence regulatory patterns suggest that bacteria might activate T3SS for their survival in the host. Afterward, the bacterial cells might shut down T3SS and activate virulence product secretion for dissemination when they reach a high density. Because ndk expression is down-regulated upon acute infection, the opposite regulatory effects of Ndk on T3SS and exoproducts observed in this study imply that Ndk plays a critical role in adjusting bacterial host pathogenicity and adaptive responses.
In conclusion, our study showed that ndk of P. aeruginosa is a novel host-responsive gene that is down-regulated during acute lung infection. Ndk down-regulation increased the expression of the T3S regulon in a rhl/pqs circuit-ExsA-dependent manner and suppressed exoproduct synthesis in a rhl system-dependent manner (Fig. 9). The inhibited expression of ndk enhanced cytotoxicity and host pathogenicity, implying that Ndk plays a crucial role in coordinating bacterial virulence and host adaptation during acute P. aeruginosa infection.
Figure 9. Schematic diagram of the proposed mechanism of Ndk-mediated virulence regulation.
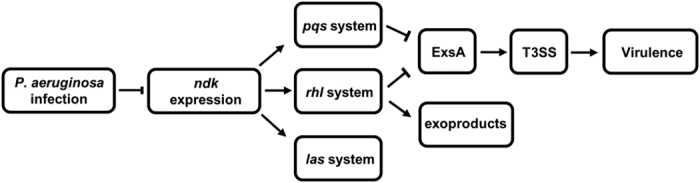
Upon P. aeruginosa infection, ndk expression was down-regulated in the pulmonary alveoli of mouse model of acute pneumonia. Knockout of ndk inhibited the las, rhl and pqs QS system. The inhibited rhl and pqs systems promoted T3SS gene and protein expression levels in an ExsA-dependent manner. The suppressed rhl system decreased exoproduct-related gene expression and product synthesis. In general, the inhibition of ndk promoted P. aeruginosa cytotoxicity and host pathogenicity through T3SS activation.
Methods
Bacterial strains, cells and animals
The details of the bacterial strains used in this study are listed in Supplementary Table 1. The P. aeruginosa strain PAO1 and its derivate mutants were grown in LB broth at 37 °C with shaking at 200 rpm. The AHL biosensor strains E. coli JM109/pSB1075 and JM109/pSB536, used for detection of QS signal molecules 3-oxo-C12-HSL and C4-HSL, respectively, were grown at 30 °C in LB broth. Antibiotics were used at the following concentrations: for E. coli JM109 (pSB1075), tetracycline 10 μg/ml, for E. coli JM109 (pSB536), ampicillin 50 μg/ml, for P. aeruginosa, carbenicillin 300 μg/ml, gentamicin 300 μg/ml, and kanamycin 300 μg/ml.
The A549 cells were purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM, HyClone) supplemented with 10% (vol/vol) fetal bovine serum (FBS, Hyclone) in the presence of 5% CO2 at 37 °C.
BALB/c mice (6 to 8 weeks old, body weights of 20 to 25 g) were purchased from the Animal Center of Third Military Medical University (TMMU). All animal experiments were approved by the animal ethics committees of Xinqiao Hospital, TMMU and complied with the guidelines outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All experiments performed in this study were approved by the Biosafety Committee of Xinqiao Hospital, TMMU.
Plasmid and strain construction
DNA manipulation and plasmid construction were performed using standard molecular cloning technique. The lambda Red-based homologous recombination technique was employed to generate gene modification mutants of P. aeruginosa PAO1, using PCR fragment containing antibiotic cassette flanked by homology regions to the target genome locus52,53. The primers used in plasmid and strain construction were presented in Supplementary Table S2.
Mouse acute pneumonia model
Bacteria cultured in LB broth for 12 h were collected and resuspended in phosphate-buffered saline (PBS) at a concentration of 1 × 1010 CFU/ml. Mice were anesthetized by intraperitoneal administration of pentobarbital sodium. Ten microliters of bacterial cells was then placed in each nostril of the mouse, resulting in inhalation of approximately 0.5 × 108 bacteria per mouse. Control mice were treated with PBS in the same manner. After 7 days of infection, mice were sacrificed, and their lungs were removed for further analysis.
Hematoxylin-eosin and immunohistochemistry staining
The mouse lungs were fixed with paraformaldehyde, embedded in paraffin and stained with H&E. Infiltrating leukocytes and macrophages were identified by IHC staining using antibodies against leukocyte common antigen CD45 (1:200, Santa Cruz) and macrophage antigen CD68 (1:200, Santa Cruz). The tissue sections were visualized using 3,3-diaminobenzidine (Zhongshan) and were observed under a light microscope (Olympus BX63).
Cytotoxicity assay
A total of 1 × 104 A549 cells were seeded in each well of a 96-well culture plate until they reached 80% confluence. The cells were infected with bacteria (MOI = 50) or bacterial culture media for 2 h. Cell viability was detected with a CCK-8 (Beyotime) according to the manufacturer’s instructions. In addition, the cell viability was determined by calcein-AM (Santa Cruz) staining54. A total of 1 × 105 cells were cultured in cell culture dishes (15 mm) and infected with the indicated PAO1 strains (MOI = 50) for 2 h. The cells were loaded with 4 μm calcein-AM for 30 min in D-Hanks buffer at room temperature (RT), followed by 30 min incubation at RT. Image acquisition was performed with a Leica TCS SP5 confocal laser scanning microscope.
Immunofluorescence staining
To determine the cellular location of the effector protein ExoS, A549 cells grown on coverslips were challenged with bacteria (MOI = 50) for 2 h at 37 °C, 5% CO2. Immunostaining was performed with the rat anti-ExoS antibody (generated in our laboratory), followed by hybridization with Alexa Fluor 594-labeled goat anti-rat IgG (1:200, Invitrogen). The cellular nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI) (1:1000, Beyotime). Images were acquired and analyzed using a Leica TCS SP5 laser confocal microscope.
Determination of effector protein translocation
The translocation assay was based on an established protocol55. One day prior to bacterial infection, 1.5 × 106 A549 cells were seeded in 100-mm-diameter tissue culture dishes. Then, the cells were challenged with bacteria (50 MOI) for 2 h at 37 °C, 5% CO2. After washing with PBS, 150 μl of 1% Triton X-100 was added to each plate, and cells were scraped from the dishes. The supernatants were harvested from the cell debris by centrifugation at 14,000 rpm for 15 min. The cellular translocation of the effector ExoS was detected by western blot.
Flow cytometry analysis of cell apoptosis
A549 cells seeded in 6-well plates were infected with the indicated strains at an MOI of 50 for 2 h. Then, 1 × 106 cells were harvested by trypsinization (0.25%) and stained with Annexin V-FITC and propidium iodide (PI) for 15 min at RT using a commercial apoptosis detection kit (BD Pharmingen). Stained cells were analyzed with a flow cytometer (MoFlo XDP, Beckman Coulter).
Real-time PCR for gene expression
For gene expression analysis, the samples were dissolved in TRIzol (Takara Bio Inc.). Total RNA extraction and reverse transcription to cDNA were performed according to the manufacturer’s instructions. The genes were quantified by semi-quantitative real-time PCR. The gene expression levels were normalized to the housekeeping gene, rplU56. The primer sequences are presented in Supplementary Table S3.
Transcriptome sequencing
For transcriptome sequencing, PAO1and Δndk cells were cultured in LB broth medium (initial OD600 of 0.01). After a 12-h culture, the bacterial cells were harvested and then dissolved in TRIzol (Takara Bio, Inc.). The series of experiments, including mRNA extraction, RNA fragmentation, cDNA synthesis and RNA-Seq library construction, were conducted by BGI Biotech Company, and libraries was sequenced using an Illumina HiSeq 2000 platform with the paired-end sequencing module.
Western blot analysis for protein expression
The bacteria were inoculated into 300 ml of LB broth (initial OD600 of 0.01) and cultured at 37 °C for the indicated time. To induce T3SS-encoding gene expression, the bacteria were inoculated into LB broth supplemented with 5 mM EGTA and 20 mM MgCl257. The bacterial proteins were quantitated using a BCA (bicinchonininc acid) protein assay kit. The secreted proteins in bacterial culture supernatants were harvested after precipitation with 30% trichloroacetic acid (TCA). Equal amounts of bacterial proteins or secreted proteins at the indicated times were resolved via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were subjected to western blot analysis using rat anti-ExoS, mouse anti-PcrV or/and mouse anti-Ndk polyclonal antibodies generated in our laboratory. The antibody signals were detected using an ECL plus kit (Thermo Fisher Scientific).
Determination of extracellular virulence products
The P. aeruginosa PAO1 strains were inoculated into 300 ml of LB broth media (an initial OD600 of 0.01) and were cultured at 37 °C with shaking. The activities of extracellular proteases and elastase in the bacterial culture supernatants were determined by skim milk plate and Elastin-Congo Red (ECR) assays, respectively, as described previously47. Pyocyanin production in the supernatant was quantitated using the method described earlier58.
Detection of QS signal molecules
The signal molecules in the supernatants of the strains were extracted with ethyl acetate (acidified with 0.5% formic acid) and dissolved in 50% methanol. For detecting the AHL signal molecules, a well-diffusion assay was performed and prepared as described previously59. The bioluminescent zones, an indicator for AHL concentration, were captured using an ImageQuant LAS 4000 mini (GE Healthcare Biosciences, USA). To detect PQS, the ethyl acetate-extracted signal molecules were subjected to HPLC analysis following a method described elsewhere with some modifications60. Briefly, the PQS was analyzed using a TSKgel ODS-100Z 5 μm HPLC column (4.6 × 250 mm, Tosoh Corporation) and was detected with a Waters 2998 Photodiode Array detector at 325 nm. The PQS was eluted at a flow rate of 1 ml/min in an isocratic elution model using a mobile phase made up of 86% methanol (1% glacial acetic acid acidified) and 14% water (1% glacial acetic acid acidified).
Statistical analysis
Data were analyzed using the SPSS 16.0 software package (SPSS Inc.). The data are expressed as the means ± standard deviations (M ± SD). Statistical analyses were performed using Student’s t-test or analysis of variance (ANOVA) followed by Newman–Keuls test. Survival curves were estimated using the Kaplan-Meier method and were compared with the log-rank statistics. A significant difference was defined as a P value less than or equal to 0.05.
Additional Information
How to cite this article: Yu, H. et al. Ndk, a novel host-responsive regulator, negatively regulates bacterial virulence through quorum sensing in Pseudomonas aeruginosa. Sci. Rep. 6, 28684; doi: 10.1038/srep28684 (2016).
Supplementary Material
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 81471067 and No. 30970115) and the Key Project of the Science Committee of Chongqing, China (No. cstc2014yykfB10009). We thank Paul Williams from Nottingham University for providing the AHL biosensor strains E. coli JM109 (pSB536) and E. coli JM109 (pSB1075) and Ming Li from the Department of Microbiology of Third Military Medical University, Chongqing, China, for providing the P. aeruginosa PAO1 strain (ATCC 15692).
Footnotes
Author Contributions K.Z. and X.R. designed the experiments and analyzed the data. H.Y., J.X., J.Q., X.X., R.X. and X.H. performed the experiments. R.Z. and X.H. participated in experiment design and data analysis. H.Y. and Z.D. wrote the initial manuscript. W.X., H.S., Q.C. and L.Z. participated in revising the manuscript. All authors discussed the results and implications and commented on the manuscript at all stages.
References
- Stover C. K. et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964 (2000). [DOI] [PubMed] [Google Scholar]
- Hogardt M. & Heesemann J. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr Top Microbiol Immunol 358, 91–118 (2013). [DOI] [PubMed] [Google Scholar]
- Sadikot R. T., Blackwell T. S., Christman J. W. & Prince A. S. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med 171, 1209–1223 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser A. R. et al. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med 30, 521–528 (2002). [DOI] [PubMed] [Google Scholar]
- Hauser A. R. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nature reviews. Microbiology 7, 654–665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa T. The molecular mechanism of acute lung injury caused by Pseudomonas aeruginosa: from bacterial pathogenesis to host response. J Intensive Care 2, 10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galle M. et al. The Pseudomonas aeruginosa type III secretion system has an exotoxin S/T/Y independent pathogenic role during acute lung infection. PLos One 7, e41547 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M. R., King J. M. & Yahr T. L. Intrinsic and Extrinsic Regulation of Type III Secretion Gene Expression in Pseudomonas Aeruginosa. Front Microbiol 2, 89 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovey A. K. & Frank D. W. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol 177, 4427–4436 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D. K., Filopon D., Kuhn L., Polack B. & Toussaint B. PsrA is a positive transcriptional regulator of the type III secretion system in Pseudomonas aeruginosa. Infect Immun 74, 1121–1129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y. H., Zhang X. F. & Zhang L. H. The global regulator Crc plays a multifaceted role in modulation of type III secretion system in Pseudomonas aeruginosa. Microbiologyopen 2, 161–172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intile P. J., Balzer G. J., Wolfgang M. C. & Yahr T. L. The RNA Helicase DeaD Stimulates ExsA Translation To Promote Expression of the Pseudomonas aeruginosa Type III Secretion System. J Bacteriol 197, 2664–2674 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., Eberl L. & Tummler B. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol 7, 459–471 (2005). [DOI] [PubMed] [Google Scholar]
- Bleves S., Soscia C., Nogueira-Orlandi P., Lazdunski A. & Filloux A. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J Bacteriol 187, 3898–3902 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. S. & Iglewski B. H. P. aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol 6, 56–60 (2003). [DOI] [PubMed] [Google Scholar]
- Siehnel R. et al. A unique regulator controls the activation threshold of quorum-regulated genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 107, 7916–7921 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Deng X., Li X., Ye Y. & Wu M. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res 42, 10307–10320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T., Ouertatani-Sakouhi H., Cosson P. & van Delden C. QsrO a novel regulator of quorum-sensing and virulence in Pseudomonas aeruginosa. PLos One 9, e87814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sio C. F. et al. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun 74, 1673–1682 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahjudi M. et al. PA0305 of Pseudomonas aeruginosa is a quorum quenching acylhomoserine lactone acylase belonging to the Ntn hydrolase superfamily. Microbiology 157, 2042–2055 (2011). [DOI] [PubMed] [Google Scholar]
- Sundin G. W. et al. Nucleoside diphosphate kinase from Pseudomonas aeruginosa: characterization of the gene and its role in cellular growth and exopolysaccharide alginate synthesis. Mol Microbiol 20, 965–979 (1996). [DOI] [PubMed] [Google Scholar]
- Sun J. et al. Mycobacterial nucleoside diphosphate kinase blocks phagosome maturation in murine RAW 264.7 macrophages. PLos One 5, e8769 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeld D. et al. Pseudomonas aeruginosa injects NDK into host cells through a type III secretion system. Microbiology 160, 1417–1426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J. et al. Nucleoside diphosphate kinase and flagellin from Pseudomonas aeruginosa induce interleukin 1 expression via the Akt/NF-kappaB signaling pathways. Infect Immun 82, 3252–3260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Paek S. H., Jin S., Park B. S. & Ha U. H. A novel Pseudomonas aeruginosa-derived effector cooperates with flagella to mediate the upregulation of interleukin 8 in human epithelial cells. Microb Pathog 66, 24–28 (2014). [DOI] [PubMed] [Google Scholar]
- Sundin G. W., Shankar S. & Chakrabarty A. M. Mutational analysis of nucleoside diphosphate kinase from Pseudomonas aeruginosa: characterization of critical amino acid residues involved in exopolysaccharide alginate synthesis. J Bacteriol 178, 7120–7128 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P. J. et al. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Molecular microbiology 24, 1249–1262 (1997). [DOI] [PubMed] [Google Scholar]
- Pandey M. K. et al. Gambogic acid, a novel ligand for transferrin receptor, potentiates TNF-induced apoptosis through modulation of the nuclear factor-kappaB signaling pathway. Blood 110, 3517–3525 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus A. A., Evans D. J., Barbieri J. T. & Fleiszig S. M. The ADP-ribosylation domain of Pseudomonas aeruginosa ExoS is required for membrane bleb niche formation and bacterial survival within epithelial cells. Infect Immun 78, 4500–4510 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutinel E. D., Vakulskas C. A., Brady K. M. & Yahr T. L. Characterization of ExsA and of ExsA-dependent promoters required for expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol 68, 657–671 (2008). [DOI] [PubMed] [Google Scholar]
- Pierson L. S. 3rd & Pierson E. A. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol 86, 1659–1670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier M. et al. From the environment to the host: re-wiring of the transcriptome of Pseudomonas aeruginosa from 22 degrees C to 37 degrees C. PLos One 9, e89941 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha U. & Jin S. Expression of the soxR gene of Pseudomonas aeruginosa is inducible during infection of burn wounds in mice and is required to cause efficient bacteremia. Infect Immun 67, 5324–5331 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K. et al. SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. MBio 4, e00419–00413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan J. et al. A novel host-responsive sensor mediates virulence and type III secretion during Pseudomonas aeruginosa-host cell interactions. Microbiology 158, 1057–1070 (2012). [DOI] [PubMed] [Google Scholar]
- Gooderham W. J. & Hancock R. E. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev 33, 279–294 (2009). [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M. Nucleoside diphosphate kinase: role in bacterial growth, virulence, cell signalling and polysaccharide synthesis. Mol Microbiol 28, 875–882 (1998). [DOI] [PubMed] [Google Scholar]
- Wallet V. et al. Dictyostelium nucleoside diphosphate kinase highly homologous to Nm23 and Awd proteins involved in mammalian tumor metastasis and Drosophila development. Journal of the National Cancer Institute 82, 1199–1202 (1990). [DOI] [PubMed] [Google Scholar]
- Hogardt M. & Heesemann J. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int J Med Microbiol 300, 557–562 (2010). [DOI] [PubMed] [Google Scholar]
- Gilljam H., Ellin A., Fau - Strandvik B. & Strandvik B. Increased bronchial chloride concentration in cystic fibrosis. Scand J Clin Lab Invest 49, 121–124 (1989). [DOI] [PubMed] [Google Scholar]
- Frank D. W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol 26, 621–629 (1997). [DOI] [PubMed] [Google Scholar]
- Postel E. H., Berberich S. J., Rooney J. W. & Kaetzel D. M. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. Journal of bioenergetics and biomembranes 32, 277–284 (2000). [DOI] [PubMed] [Google Scholar]
- Waters C. M., Wu J. T., Ramsey M. E., Harris R. C. & Bassler B. L. Control of the type 3 secretion system in Vibrio harveyi by quorum sensing through repression of ExsA. Appl Environ Microbiol 76, 4996–5004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogardt M., Roeder M., Fau - Schreff A. M., Schreff A. m., Fau - Eberl L., Eberl L., Fau - Heesemann J. & Heesemann J. Expression of Pseudomonas aeruginosa exoS is controlled by quorum sensing and RpoS. Microbology 150, 843–851 (2004). [DOI] [PubMed] [Google Scholar]
- Kong W., Liang H., Shen L. & Duan K. [Regulation of type III secretion system by Rhl and PQS quorum sensing systems in Pseudomonas aeruginosa]. Wei Sheng Wu Xue Bao 49, 1158–1164 (2009). [PubMed] [Google Scholar]
- Wilder C. N., Diggle S. P. & Schuster M. Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J 5, 1332–1343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. P., Pesci E. C. & Iglewski B. H. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol 179, 5756–5767 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brint J. M. & Ohman D. E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol 177, 7155–7163 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Park H., Egger L. A. & Inouye M. Nucleoside-diphosphate kinase-mediated signal transduction via histidyl-aspartyl phosphorelay systems in Escherichia coli. J Biol Chem 271, 32886–32893 (1996). [DOI] [PubMed] [Google Scholar]
- Munoz-Dorado J., Almaula N., Inouye S. & Inouye M. Autophosphorylation of nucleoside diphosphate kinase from Myxococcus xanthus. J Bacteriol 175, 1176–1181 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M. & Ajitkumar P. Histidine 117 in the His-Gly-Ser-Asp motif is Required for the Biochemical Activities of Nucleoside Diphosphate Kinase of Mycobacterium smegmatis. Open Biochem J 6, 71–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R. & Liu J. Scarless and sequential gene modification in Pseudomonas using PCR product flanked by short homology regions. BMC Microbiol. 10, 209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. et al. Elastase LasB of Pseudomonas aeruginosa promotes biofilm formation partly through rhamnolipid-mediated regulation. Can. J. Microbiol. 60, 227–235 (2014). [DOI] [PubMed] [Google Scholar]
- Hashimoto Y. et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc Natl Acad Sci USA 98, 6336–6341 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisz M., Lee P. C. & Rietsch A. ExoS controls the cell contact-mediated switch to effector secretion in Pseudomonas aeruginosa. J Bacteriol 190, 2726–2738 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma S. L. et al. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189, 8165–8178 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsen S., Collin-Faure V., Gidrol X. & Lemercier C. The opportunistic pathogen Pseudomonas aeruginosa activates the DNA double-strand break signaling and repair pathway in infected cells. Cell Mol Life Sci 70, 4385–4397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essar D. W., Eberly L., Hadero A. & Crawford I. P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol 172, 884–900 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. et al. Elastase LasB of Pseudomonas aeruginosa promotes biofilm formation partly through rhamnolipid-mediated regulation. Can J Microbiol 60, 227–235 (2014). [DOI] [PubMed] [Google Scholar]
- Palmer G. C., Schertzer J. W., Mashburn-Warren L. & Whiteley M. Quantifying Pseudomonas aeruginosa quinolones and examining their interactions with lipids. Methods Mol Biol 692, 207–217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.