Abstract
Objective Adverse drug events (ADEs) are undesired harmful effects resulting from use of a medication, and occur in 30% of hospitalized patients. The authors have developed a data-mining method for systematic, automated detection of ADEs from electronic medical records.
Materials and Methods This method uses the text from 9.5 million clinical notes, along with prior knowledge of drug usages and known ADEs, as inputs. These inputs are further processed into statistics used by a discriminative classifier which outputs the probability that a given drug–disorder pair represents a valid ADE association. Putative ADEs identified by the classifier are further filtered for positive support in 2 independent, complementary data sources. The authors evaluate this method by assessing support for the predictions in other curated data sources, including a manually curated, time-indexed reference standard of label change events.
Results This method uses a classifier that achieves an area under the curve of 0.94 on a held out test set. The classifier is used on 2 362 950 possible drug–disorder pairs comprised of 1602 unique drugs and 1475 unique disorders for which we had data, resulting in 240 high-confidence, well-supported drug-AE associations. Eighty-seven of them (36%) are supported in at least one of the resources that have information that was not available to the classifier.
Conclusion This method demonstrates the feasibility of systematic post-marketing surveillance for ADEs using electronic medical records, a key component of the learning healthcare system.
Keywords: post market drug safety surveillance, pharmacovigilance, machine learning, EMR mining
BACKGROUND AND SIGNIFICANCE
Adverse drug events (ADEs) are undesired harmful effects resulting from use of a medication. It is estimated that ADEs occur in 30% of hospital stays, causing 2 million injuries, hospitalizations, and deaths each year in the United States at a cost of $75 billion.1–3 Preapproval clinical trials are the first line of defense for identifying ADEs but they are limited both in their power to detect rare events and their generalizability to patient populations with many co-morbidities and poly-pharmacy. These limits have driven efforts in postmarketing surveillance for ADEs using a variety of observational data sources as a key component of the learning healthcare system.4–10
These efforts have at their core the collection of counts of drugs being taken and adverse events occurring, derived from a variety of data sources. Most work has used spontaneous reporting system data such as the US Food and Drug Administration’s FDA Adverse Event Reporting System (FAERS), but other data sources such as claims data have also been used. Each of these data sources have well-documented biases. For instance, FAERS relies on voluntary reporting of suspected drug adverse event cases and suffers from reporting biases such that associations with adverse events with many possible causes are difficult to detect,11 while claims data suffers from biases arising from its primary use for billing instead of conveying clinical information.12
In contrast, electronic medical records (EMRs) and free text of clinical notes (CNs) provides arguably the most complete and unbiased picture of clinical events available.13 There has been much progress in using methods from Natural Language Processing (NLP) to extract structured information from the unstructured free text of CNs.5,14 These studies typically use NLP to generate counts of drug and disorder mentions in the clinical text, often subject to constraints that reflect, for instance, the intuition that the cause of an adverse event—for example, taking a drug—must precede the adverse event itself. Given such counts, there are 2 main approaches to identifying possible drug adverse event associations. One approach, disproportionality analysis (DPA), tackles the problem using the well-known framework of statistical hypothesis testing. These methods use counts of drugs, adverse events, and their co-occurrence to calculate a P-value for the association of the drug and adverse event relative to a null hypothesis of no association, with varying degrees of adjustment for confounding. LePendu et al.5 applied DPA to counts and co-mention counts to clinical text and achieved an area under the curve (AUC) of 0.79, matching the accuracy achieved by current state of the art DPA methods applied to FAERS in Harpaz et al.11 However, recent work has shown that using P-values to prioritize possible drug-adverse event associations is problematic, most notably because even with multiple testing corrections it is not possible to remove false positives.15 Furthermore, it has been noted that integration of other data sources is likely essential in effective postmarketing surveillance using observational data.12 However, it is not clear how to effectively incorporate some forms of relevant information, such as prior knowledge of known ADEs. Such prior knowledge may be especially helpful for detection of ADEs in which the drug or adverse event is rare.
An alternative to statistical hypothesis testing is using discriminative classifiers such as a logistic regression model. These methods differ from DPA in that they attempt to learn a function that guesses the validity of drug adverse event associations given inputs, or features, such as counts of the drug and the adverse event in the data. Importantly, the input can include features that reflect prior knowledge such as similarity to known ADEs. Such discriminative classifiers can be applied to spontaneous reports or to EMR derived counts. Harpaz et al.11 conducted a systematic review of drug adverse event algorithms using FAERS data that found logistic regression methods outperforming even the state of the art DPA methods across a variety of adverse events. Recent work by Noren et al. has shown that building such a discriminative classifier that uses additional information such as the geographic origin of an adverse event report and the timing of the submission achieves much better performance than existing methods.16–18 Cami et al.19 went even further and developed a logistic regression model using only features encoding prior knowledge about known ADEs that achieved high performance.
Liu et al.20 used a discriminative classifier to distinguish ADEs (pairs of drugs and adverse events in which the drug causes the adverse event) from drug usages (pairs of drugs and disorders in which the drug is used to treat the disorder). Their classifier used features derived from the free text of millions of CNs from the Stanford Translational Research Integrated Database Environment (STRIDE)21 that encoded the frequency of drug and disorder mentions and co-mentions in the text, subject to constraints on the order of the mentions in time. We have built on this work, developing a method suited for systematic, automated detection of potential ADEs from EMRs. Our method also uses a computationally efficient text processing system to extract mentions of drugs and disorders from the text of CNs. These mentions are further processed into statistics that are used by a discriminative classifier that outputs the probability that a given drug–disorder pair represents a valid ADE association. However, we address a different problem from Liu et al.—we seek to discriminate ADEs from all other drug-AE pairs, rather than from drug-indication pairs. Further, Liu et al. limited their study to drug–disorder pairs in which there were at least 1000 co-occurrences in the clinical text. We relax this restriction to all possible drug–disorder pairs in which the drug and disorder each appear at least once in the data in order to detect rare ADEs. Thus, we address a much harder classification problem. Because we are now making predictions on drug–-disorder pairs that may be very rare in the data, we also include features similar to those used in Jung et al.22 that encode prior knowledge about known ADEs and drug usages.
The classifier achieved an AUC of 0.94 on hold out test data. This result is a significant improvement on both the current state of the art DPA method and the classifier-based analysis applied to FAERS (0.79 and 0.83, respectively) reported in Harpaz et al.5 It is also improvement on DPA applied to electronic health record (her) free text (AUC 0.79) reported in LePendu et al.11 Applying it to all possible drug-adverse event pairs and filtering the predicted ADEs support in independent and complementary data sources—FAERS and MEDLINE—resulted in 240 well-supported, high-confidence ADEs. Our goal is to develop a scalable method that can exploit the free text of EHRs for comprehensive, timely surveillance for drug-adverse event associations. In such applications, it is critical to estimate the positive predictive value (PPV) of the method. We therefore validate our method using ADEs that were either withheld from the training data or became known after the dataset was created. We find that 87 of the 240 well-supported, high confidence ADEs are thus validated (36%). Figure 1 summarizes our approach.
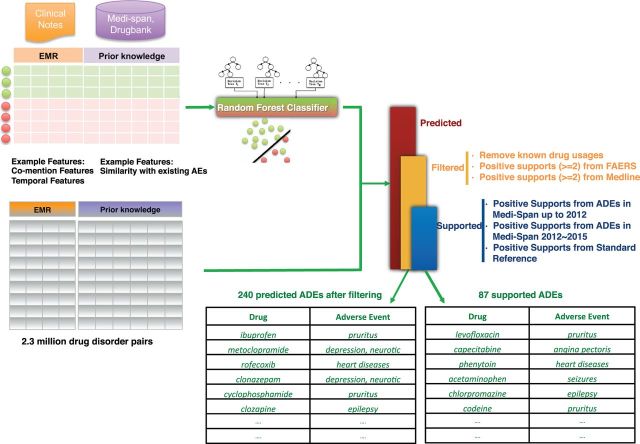
Figure 1:
Overview of methods and results. For each of the 2 362 950 possible drug–disorder pairs, we calculated 9 features from the free text of clinical notes in STRIDE, 8 features from known AEs in Medi-Span, and 12 features from known usages in Medi-Span and Drugbank. Based on these features, a Random Forest classifier was trained on the gold standard dataset to recognize the drug–AE relationships. Then, we applied the trained classifiers to the 2 362 950 possible drug–disorder pairs and filtered for support in FAERS and MEDLINE, yielding a set of 240 well supported, high confidence ADEs. Drug–AE pairs used in training are censored.
MATERIALS AND METHODS
Constructing training and test sets
Discriminative classifiers learn a function that maps input features to an output such as the probability that the inputs represent a true drug adverse event pair. In order to learn and evaluate the performance of this function, these methods require a set of examples of drug adverse event pairs whose status as true or false associations is known. We constructed such a set of positive and negative examples of drug-AE pairs using known ADEs from the Medi-Span® Adverse Drug Effects Database (from Wolters Kluwer Health, Indianapolis, IN, United States), a manually curated, commercial compendium of drug usages, side effects and pricing information, which was obtained under an academic license. Medi-Span up to 2012 contains 711 468 drug–disorder pairs comprising 13 000 unique drugs and 3403 unique disorders. Of these, 3550 pairs were assumed to be true ADE pairs because they occur in black box warnings with additional constraints (e.g., above moderate severity level), which are listed in Supplementary Materials Table S1. We used RxNorm to normalize drugs to their active ingredients and discarded pairs in which either the drug or adverse event did not occur in the EMR data. This resulted in 1898 positive examples for training and testing. To construct negative examples, we randomly sampled drugs and adverse events from among the drugs and adverse events in the positive set and ensured that the co-mention count distribution of the negative samples are roughly the same as those of the positive samples. Four thousand three hundred and thirty-six such negative samples remained after removing inadvertently generated positive examples. These drug adverse event pairs were then randomly split into 4358 training examples used to learn classifiers and 1877 test examples used to evaluate the performance of the classifiers.
Processing of clinical text-notes from STRIDE
An National Center for Biomedical Ontology (NCBO) Annotator–based text-processing pipeline23,24 was used to annotate 9.5 million CNs from STRIDE with mentions of drugs and disorders. Negated mentions (e.g., “MI was ruled out”) or those referring to other people (e.g., “father had a stroke”) were removed using NegEx25 and ConText,26 respectively. The notes spanned 18 years and 1.6 million patients. Drugs and disorders were mapped to Unified Medical Language System (UMLS) unique concept identifiers (CUIs). In this study we used the 2011AB version of the UMLS. Drugs were normalized to active ingredients using RxNorm27 as provided by UMLS2011AB—for example, Panocaps was normalized to lipase, protease, and amylase.
Feature construction
For each drug–disorder pair, we constructed 9 features from the CN-derived mentions—the drug and disorder frequency, co-mention frequency, drug first fraction (the fraction of patients in which the first mention of the drug precedes the first mention of the disorder), and association scores derived from these counts (e.g., chi squared statistic, odds ratio, and the conditional probability of drug mention given disorder mention).
In addition, we constructed 8 features encoding known drug-AEs and 12 features encoding known usages. Those features were motivated by the intuition that a drug may be more likely to cause a disorder if it is similar to other drugs known to cause that disorder. We calculate similarity as follows. First, we define different ways in which drugs may be similar to each other. For instance, we may calculate drug–drug similarity on the basis of known disorder associations. This prior knowledge is represented as a matrix in which the rows correspond to drugs and columns correspond to disorders. The (i, j)-th entry of the matrix is 1 if drug i is known to be associated with disorder j and 0 otherwise. Each drug is thus represented as a binary vector. We next define the set of other drugs to which we will compare drug i, the query drug, as the set of all other drugs that are known to be associated with disorder j. This corresponds to the set of drugs that have a 1 in column j. We then calculate cosine and Jacquard similarity between the query drug and each drug in this set using the binary vectors for the respective drugs. Finally, we pool the similarities over the set of drugs with max or mean operations over the sets of similarities. We also calculate drug-drug similarity using the same set of related drugs, on the basis of the known usage associations of the drugs, their targeted molecular pathways, and their drug classes. Disorder–disorder similarities are calculated in a similar manner, except that we calculate similarities between the columns of the matrix instead of the rows. This process is summarized in Figure 2. For usage-similarity, we adopted features described in Jung et al.22 In all, we used 29 features for each drug–disorder pair, listed in Supplementary Materials Table S2.
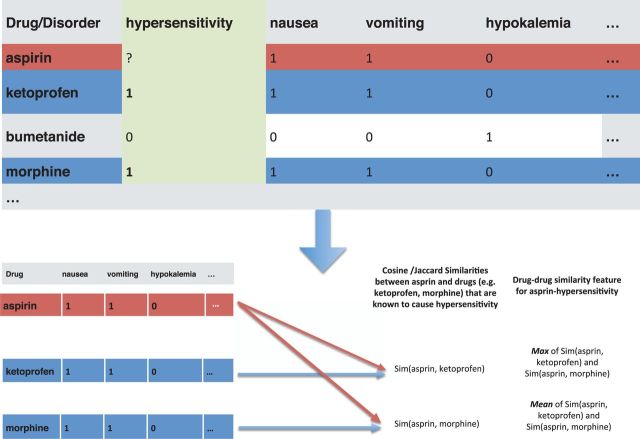
Figure 2:
Drug–drug and disorder–disorder similarity using known ADEs. We represent known drug–AEs as a matrix where the rows are drug names and columns are disorders, and the (i, j)-th entry is a binary indicator for whether or not the drug in the i-th row causes the disorder in the j-th column. In this way, each drug is represented as a binary vector. For a given query drug and adverse event (e.g., aspirin and hypersensitivity in panel a), we find other drugs that are known to be associated with hypersensitivity and calculate similarities between aspirin and those drugs. We summarize the similarities with 2 scalar values—the max and mean similarity.
Classifier development
We fit L1 regularized logistic regression,28 support vector machine with radial basis function kernels,29 and random forest classifiers30,31 to the training data. We used the R packages glmnet,32 e1071,33 and randomForest,34 respectively, and model hyper-parameters were tuned by cross validation on the training set. The classifiers were then evaluated on the hold out test set. The classifier development process is summarized in Figure 3. In order to investigate the contribution of different features, we performed an ablation analysis in which we evaluated classifiers trained on subsets of the features.
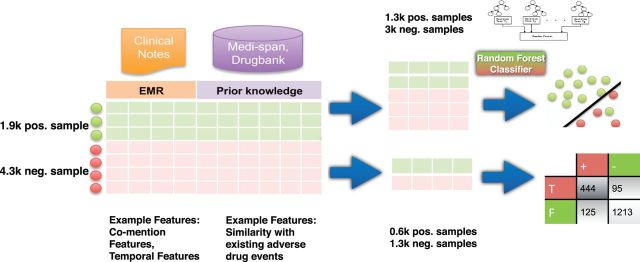
Figure 3:
Training a classifier to recognize drug–ADE relationships. Positive examples collected from known ADEs in Medi-Span and negative examples created through randomly sampling a drug and disorder with roughly the same co-mention distribution as the positive examples. For each drug–disorder pair in the gold standard, we used 9 features to characterize the pattern of drug and disorder mentions in 9.5 million clinical notes from STRIDE, 8 features to characterize the domain knowledge of drug, disorder, and known ADEs from Medi-Span, and 12 features to characterize the domain knowledge of drug, disorder, and known usages from Medi-Span and Drugbank. The gold standard dataset was randomly split into 70% for training and 30% for testing the classifier.
Identifying putative drug–AE associations
Performance on the test set indicated that the random forest classifier was superior to the other classifiers in all metrics (Supplementary Materials S3). We therefore focused on this model, and applied a classifier trained on the full gold standard dataset to all possible drug–AE combinations. One thousand six hundred and two unique drugs and 1475 unique disorders are each mentioned at least once in the clinical text of STRIDE, resulting in 2 362 950 possible drug–AE pairs. We focused on the most confident predicted associations by using a threshold of 0.7 for the posterior probability output by the classifier, yielding 41 248 predicted ADE associations.
In this study, the set of 3550 known ADEs were gathered from the Medi-Span® Adverse Drug Effects Database, where the documentation level is marked as “black box warning”; known usages were gathered from the Medi-Span Drug Indications Database (Wolters Kluwer Health, Indianapolis, IN, United States) and the National Drug File – Reference Terminology.35
Filtering in FAERS and MEDLINE
The classifier achieved an estimated specificity of 0.913 on hold out test data. However, even with such high specificity, we can expect on the order of 200 000 false positive results when we apply the classifier to 2.3 million possible ADEs because the prevalence of true ADE associations is likely very low. It is therefore critical to reduce the number of false positives in the candidate set for it to be practically useful. We note that this requirement is holds regardless of the method used to generate candidate ADEs; since DPA methods applied to both EHR data and FAERS report significantly lower accuracy in test sets, we can conclude that they will also suffer from a high false positive rate if applied to 2.3 million drug–disease pairs. We thus filter the predicted ADEs for positive support in FAERS36 and MEDLINE, two independent and complementary data sources that reflect clinical practice and published biomedical knowledge respectively. FAERS case reports contain explicit links between drugs and adverse events. We used all case reports from Q1 2005 through Q4 2013 to assess support for putative ADEs in terms of the number of case reports in which the drug was reported as the primary or secondary suspect drug for the adverse event. FAERS drugs and adverse events were mapped to UMLS CUIs, yielding a set of 4 508 892 drug-event reports covering 372 379 unique pairs. We also obtained reports of ADEs described in the biomedical literature from MEDLINE. Using an approach based on Avillach et al.,37 we obtained 118 552 unique ADEs from about 200 000 articles in MEDLINE that had been indexed with combinations of medical subject heading (MeSH) descriptors and qualifiers for both a drug involved in an adverse event (e.g., Ofloxacin/adverse effects) and its manifestation (e.g., Tendinopathy/chemically induced). We mapped the MeSH terms of drug– disease pairs (e.g., Ofloxacin–Tendinopathy) to their corresponding UMLS CUIs to make the findings compatible with our predictions. The query is provided in Supplementary Materials S5.
Validation in Medi-Span and time-indexed reference
We validate the drug–AE associations using drug–AE associations unknown to the system during training. The training examples used in our model were constructed from ADEs marked as “black box warning” in Medi-Span up to 2012; thus we only used well-established adverse events in training the classifier. Medi-Span also contains ADEs marked as “reported in multiple reports and uncontrolled studies” or “reported in few case reports and suggested links”; we refer to these ADEs with moderate support. We validated the drug–AE associations left after filtering based on FAERS and MEDLINE using ADEs with moderate support in both Medi-Span up to 2012 as well as with the additional Medi-Span data from 2012 to 2015. Medi-Span up to 2012 contains 95 115 ADEs with moderate support, and Medi-Span from 2012 to 2015 contains an additional 755 ADEs that were not reported in Medi-Span as of 2012. In addition, we used a time-indexed reference standard by Harpaz et al.38 to further assess the classifier’s ability to detect recent drug-AE associations. The reference standard was systematically curated from drug labeling revisions (e.g., new warnings issued and communicated by the US Food and Drug Administration in 2013), and included 62 positive test cases and 75 negative controls.
RESULTS
Performance of a classifier for drug–AE associations
The random forest classifier to detect drug–AE associations achieved an AUC of 0.94 in the hold out test set. We then fit a new random forest classifier using the entire gold standard and applied it to the 2 362 950 drug–AE pairs arising from all combinations of the 1602 unique drugs and 1475 unique disorders appearing in our data. After applying a cutoff on the classifier’s confidence (posterior probability of 0.7), we arrived at a set of 41 248 putative drug–AE associations. We refined these 41 248 drug–AE associations by filtering for support in FAERS and MEDLINE. Two hundred and forty associations were supported by at least 2 reports in both FAERS and MEDLINE. Table 1 lists the 10 associations with the highest level of support in FAERS; the complete table is available as Supplementary Materials Table S4.
Table 1:
List of 10 validated drug–AEs and their support in FAERS as well as MEDLINE.
| Drug_Name | Drug_CUI | Event_Name | Event_CUI | FAERS Support | MEDLINE Support |
|---|---|---|---|---|---|
| Etanercept | C0717758 | Pruritus | C0033774 | 11485 | 2 |
| Metoclopramide | C0025853 | Depression, neurotic | C0282126 | 1473 | 9 |
| Rofecoxib | C0762662 | Heart diseases | C0018799 | 861 | 3 |
| Clonazepam | C0009011 | Depression, neurotic | C0282126 | 691 | 2 |
| Diazepam | C0012010 | Depression, neurotic | C0282126 | 608 | 5 |
| Levofloxacin | C0282386 | Pruritus | C0033774 | 540 | 3 |
| Cyclosporine | C0010592 | Pruritus | C0033774 | 517 | 5 |
| Clozapine | C0009079 | Epilepsy | C0014544 | 410 | 13 |
| Lorazepam | C0024002 | Depression, neurotic | C0282126 | 403 | 3 |
| Ibuprofen | C0020740 | Pruritus | C0033774 | 374 | 4 |
Previous work by LePendu et al.5 demonstrated the use of the free text of EHRs for discovery of ADEs achieving an AUC of 0.79, while the current state of the art DPA method applied to FAERS in Harpaz et al.11 also achieved an AUC of 0.79. Our classifier thus achieves significantly higher performance than DPA methods using either EHR text of FAERS. We further note that the best performing method (logistic regression, a classifier based approach) using FAERS reported in11 achieved an AUC of 0.83. These results collectively suggest that the free text of EHRs is both a useful source of data about ADEs, and that classifier-based approaches outperform current DPA approaches in both EHR text and FAERS.
Assessing the quality of the predictions
Out of the 240 well-supported drug–AE associations, 76 occurred in the set of the ADEs with moderate support in Medi-Span up to 2012. We also found that 10 out of the 240 drug–AE associations occurred in the recent established ADEs included in the additional Medi-Span data from 2012 to 2015. Finally, 2 of the drug–ADE associations were also supported by a reference standard provided by Harpaz et al.38 Figure 4 shows the support from those independent and complementary data sources for the predicted drug–AE associations. Overall, from the 240 drug–AE associations, 87 of them (36%) are supported in at least one of the resources that have information not available to the classifier.
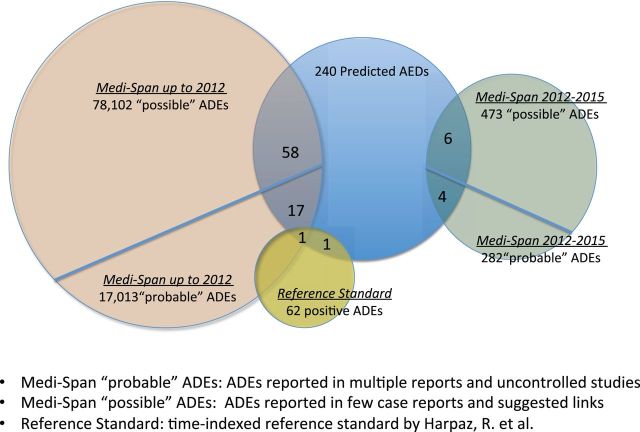
Figure 4:
Support from independent and complementary data sources. We validated the predicted drug–AE associations from three independent and complementary data sources. From the 240 drug–ADE associations, 76 occurred in the set of the ADEs with moderate support in Medi-Span up to 2012; 10 occurred in the recent established ADEs included in the additional Medi-Span data from 2012 to 2015; 2 occurred in the reference standard provided by Harpaz, R. et al. Overall, 87 of them (36%) were supported in at least one of the resources that have information that was not available to the classifier.
We also evaluated the PPV of the classifier alone, FAERS alone, and MEDLINE alone using the above methodology. We found that 823 of 41 248 novel ADEs predicted by the classifier were supported in the validation set, yielding a PPV of 2%. FAERS and MEDLINE alone each had PPVs of approximately 1% and 3.5%, respectively (Table 2). These results, together with the AUCs reported in various studies cited above, highlight the benefit of combining signals from multiple, independent data sources, with the PPV increasing from 1–3.5% to 36%. This result is consistent with the improvement in PPV reported for combining DPA signals from EHR text and FAERS in Harpaz et al.39
Table 2:
Performance of our method.
| Dataset | Method | AUC | PPV |
|---|---|---|---|
| EHR text alone | DPA [5] | 0.79 | N/A |
| EHR text alone | Our classifier | 0.94 | 0.020 |
| FAERS alone | DPA [11] | 0.79 | N/A |
| FAERS alone | Raw counts (≥2) | 0.72 | 0.010 |
| MEDLINE alone | Raw counts (≥2) | 0.69 | 0.035 |
| All | Classifier + raw counts in FAERS and MEDLINE | N/A | 0.36 |
Our method consists of a classifier based on EHR text in conjunction with filters based on counts in FAERS and MEDLINE. The classifier alone achieves an AUC of 0.94, but its PPV is only 0.020 because of the low prevalence of true ADE associations. Previous work shows that DPA applied to both EHR text and FAERS achieves an AUC of 0.79, significantly lower than that of our classifier. The PPV for these methods is not reported because these studies do not estimate PPV in a realistic setting; instead they calculate PPV in datasets highly enriched for true ADE associations. Using raw counts in FAERS and MEDLINE lead to similarly low PPV. In contrast, our method combining the classifier with counts in FAERS and MEDLINE achieved a PPV of 0.36.
Feature contribution
Feature ablation experiments were done to investigate the contribution of each group of features. We grouped the 29 features into 3 categories: features from CNs, features from known ADEs (KA), and features from known usages (KU). As shown in Table 3, classifiers using features from CNs, known ADEs, and known usages had AUCs of 0.92, 0.72, and 0.82, respectively. Classifiers that used both CN features and known ADE or usage information achieved an AUC of 0.93. Above the features derived from CNs, features from prior knowledge contributed little in our classifier. Thus, it would be helpful to incorporate the features used in Cami et al.’s work19 to encode the prior knowledge about known ADEs and usages.
Table 3:
Performance of Random Forest classifier on held-out test set with different feature sets.
| Subsets of Features | AUC | Precision/PPV | Specificity | Sensitivity/Recall |
|---|---|---|---|---|
| From clinical notes (CNs) | 0.920 | 0.763 | 0.803 | 0.702 |
| From known adverse-event (KA) | 0.723 | 0.526 | 0.624 | 0.550 |
| From known usage (KU) | 0.815 | 0.561 | 0.661 | 0.584 |
| CN + KA | 0.932 | 0.775 | 0.714 | 0.801 |
| CN + KU | 0.937 | 0.781 | 0.719 | 0.820 |
| All | 0.944 | 0.796 | 0.913 | 0.839 |
We performed feature ablation to investigate the contribution of different feature sets on the performance of the random forest classifier for detecting drug–AE relationships. The first column is the feature set used to train the classifier. The classifier performance was evaluated on the 1.9k withheld test examples. Individually, features from clinical notes (CNs) yielded higher performance than features from known ADEs (KA) and known usages (KU) in all metrics. Adding features from KA or KU to features from CNs significantly improved the classifier performance in terms of sensitivity, while all features together resulted in a sensitivity of 0.839 and an AUC of 0.944.
DISCUSSION
We have developed a method for systematic, automated detection of ADEs. The method achieves high specificity and sensitivity in a hold out test set using features derived from both CNs and prior knowledge about drug usages and ADEs. Our classifier does not make any assumptions on relationship between the inputs, allowing us to easily incorporate disparate types of knowledge into the predictions. In particular, features derived from CNs are empirical, and reflect clinical practice as is, while features encoding prior knowledge potentially allow us to make better predictions in cases where the empirical data is lacking.
The direct use of EMRs also opens the door to prioritizing potential novel ADEs using the observed frequency of co-occurrence of the drug and adverse event in the EMR, which may better reflect their true prevalence than data sources such as FAERS. We note that systematic detection of ADEs requires methods that are easily applicable to many healthcare institutions. Our method has several characteristics that make it especially suitable for such use. First, its input is primarily derived from clinical free text through a computationally efficient text processing system that can handle millions of notes in hours. Second, unlike other approaches to enabling cross institution analysis of EMR data, our method assumes only access to the text of CNs without assuming a common data model. Third, it is easy to adapt the classifier to different institutions because no components other than the classifier need site specific tuning. The latter is not an obstacle to adoption because the training and validation of the classifier can be entirely automated. These characteristics of our approach minimize the computational and organizational demands of implementing the method in a variety of settings.
A key contribution of our work is our quantification of the PPV of our system in a realistic manner. Direct comparison of this PPV with previously published results cannot be done because, to the best of our knowledge, no other study has evaluated signal detection methods on a large number of possible ADE associations. Instead, prior studies use evaluation datasets that are manually curated, contain “well-known” associations, and assume a high prevalence of true associations (close to 50% in Harpaz et al.11). The actual prevalence of true drug-adverse event associations is unknown, but is likely much lower than 50%. Therefore, PPVs calculated from such reference sets are hard to generalize.40–42.
We can compare our results with previous work using intrinsic measures of performance such as specificity, which does not depend on prevalence. Specifically, we can compare methods using the AUC, which summarizes sensitivity and specificity over the whole range of possible thresholds for signal detection. Our classifier has an AUC of 0.94, higher than the 0.79 AUC of the methods currently used for signal detection in FAERS. Note that these AUC estimates are calculated using different sets of manually curated ADE associations.
Our work has important limitations. First, we emphasize that the classifier generates hypotheses (i.e., “signals” of a putative association) instead of verified ADEs. Furthermore, our working definition of novelty in this study is novelty with respect to a set of well-established ADEs. We view this positively, as further validation of our method, because our primary aim is to demonstrate the feasibility of an approach to postmarketing surveillance which is suitable for large scale deployment across many healthcare institutions. We note that in practice, the set of “known ADEs” used in training could be tuned to change the sensitivity of the classifier. We also note that despite the classifier’s high specificity in test data, applying it to millions of hypothetical drug–AE pairs may result in a large absolute number of false positives. Second, as in previous studies that have used free text to discover relationships between drugs and disorders, the mismatch between terms as they are formally defined in formal ontologies versus how they are used in practice may lead to seemingly novel results that are in fact already known.22 Third, we note that while the use of CNs may provide an estimate of prevalence that can be used to prioritize findings for further study, it would be better still to combine prevalence with severity, which may require deeper NLP to ascertain severity. Fourth, we note that any biases that are present in the training set of known ADEs will likely carry over to predictions made on a wider range of drug adverse event pairs, potentially leading to missed associations. Furthermore, with respect to the use of this method for systematic, nation-wide postmarketing surveillance, we note that the problem of optimally integrating safety signals from multiple sites is an unsolved research problem despite recent progress.43 Meta-analytic approaches or weighted voting schemes may be necessary to combine the signals generated by such a classifier from multiple sites.
Despite these limitations, this method is an important first step toward automated, systematic, and comprehensive postmarketing surveillance for ADEs using EMRs as the primary source; such ability is an important use case envisioned for the learning healthcare system.10 We envision a future in which it is possible to generate hypotheses of ADEs automatically, in real time, and queue them up for potential review and submission to the Federal Adverse Event Reporting System.
CONCLUSION
We have developed and validated a data-mining method for identifying putative, new ADEs using clinical data and prior knowledge of known ADEs. Our classifier achieves high discrimination capability with an AUC of 0.94 on a held out test set. By applying the classifier to 2 362 950 drug–disorder pairs consisting of 1602 unique drugs and 1475 unique disorders we identified 240 high-confidence drug–AE associations. These high-confidence associations are well supported by multiple independent and complementary resources. Our method enables systematic post-marketing surveillance for new ADEs using EMRs.
CONTRIBUTORS
G.W. implemented the method and wrote the manuscript. K.J. contributed to the writing of the manuscript. N.H.S. conceived of the study, and edited the manuscript. R.W. performed the MEDLINE analysis and contributed to the writing.
FUNDING
This work was supported in part by NLM grant R01 LM011369, NIGMS grant R01 GM101430, and NHGRI grant U54 HG004028 for the National Center for Biomedical Ontology.
COMPETING INTERESTS
None.
ACKNOWLEDGEMENTS
We acknowledge the assistance of Juan M. Banda in accessing and processing data from FAERS and Medi-Span.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 1997;277(4):301–306. [PubMed] [Google Scholar]
- 2.Classen DC, Resar R, Griffin F, et al. ‘Global trigger tool' shows that adverse events in hospitals may be ten times greater than previously measured. Health Aff. 2011;30(4):581–589. [DOI] [PubMed] [Google Scholar]
- 3.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279(15):1200–1205. [DOI] [PubMed] [Google Scholar]
- 4.Tatonetti NP, Ye PP, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4(125):125ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lependu P, Iyer SV, Bauer-Mehren A, et al. Pharmacovigilance using clinical notes. Clin Pharmacol Therapeutics. 2013;93(6):547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harpaz R, Callahan A, Tamang S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Safety. 2014;37(10):777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman C. Discovering novel adverse drug events using natural language processing and mining of the electronic health record. AMIE 2009: Proceedings of the 12th Conference on Artificial Intelligence In Medicine. 2009:1–5. [Google Scholar]
- 8.Ryan PB, Madigan D, Stang PE, Overhage JM, Racoosin JA, Hartzema AG. Empirical assessment of methods for risk identification in healthcare data: results from the experiments of the Observational Medical Outcomes Partnership. Stat Med. 2012;31(30):4401–4415. [DOI] [PubMed] [Google Scholar]
- 9.Duke JD, Han X, Wang Z, et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. PLoS Comput Biol. 2012;8(8):e1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman CP, Wong AK, Blumenthal D. Achieving a nationwide learning health system. Sci Transl Med. 2010;2(57):57cm29. [DOI] [PubMed] [Google Scholar]
- 11.Harpaz R, DuMouchel W, LePendu P, Bauer-Mehren A, Ryan P, Shah NH. Performance of pharmacovigilance signal-detection algorithms for the FDA adverse event reporting system. Clin Pharmacol Therapeutics. 2013;93(6):539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan PB, Madigan D, Stang PE, Schuemie MJ, Hripcsak G. Medication-wide association studies. CPT: Pharmacometrics Syst Pharmacol. 2013;2:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poissant L, Taylor L, Huang A, Tamblyn R. Assessing the accuracy of an inter-institutional automated patient-specific health problem list. BMC Med Informat Dec Mak. 2010;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Hripcsak G, Markatou M, Friedman C. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. JAMIA. 2009;16(3):328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuemie MJ, Ryan PB, DuMouchel W, Suchard MA, Madigan D. Interpreting observational studies: why empirical calibration is needed to correct p-values. Stats Med. 2014;33(2):209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caster O, Juhlin K, Watson S, Noren GN. Improved statistical signal detection in pharmacovigilance by combining multiple strength-of-evidence aspects in vigiRank. Drug Safety. 2014;37(8):617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caster O, Noren GN, Madigan D, Bate A. Logistic regression in signal detection: another piece added to the puzzle. Clin Pharmacol Therapeutics. 2013;94(3):312. [DOI] [PubMed] [Google Scholar]
- 18.Harpaz R, DuMouchel W, LePendu P, Bauer-Mehren A, Ryan P, Shah NH. Response to “Logistic regression in signal detection: another piece added to the puzzle". Clin Pharmacol Therapeutics. 2013;94(3):313. [DOI] [PubMed] [Google Scholar]
- 19.Cami A, Arnold A, Manzi S, Reis B. Predicting adverse drug events using pharmacological network models. Sci Transl Med. 2011;3(114):114ra27. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Lependu P, Iyer S, Shah NH. Using temporal patterns in medical records to discern adverse drug events from indications. AMIA Summits on Translational Science proceedings AMIA Summit on Translational Science. 2012;2012:47–56. [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe HJ, Ferris TA, Hernandez PM, Weber SC. STRIDE–An integrated standards-based translational research informatics platform. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2009;2009:391–395. [PMC free article] [PubMed] [Google Scholar]
- 22.Jung KLP, Chen WS, Iyer SV, Readhead B, Dudley JT, Shah NH. Automated detection of off-label drug use. PloS ONE. 2014;9(2):e89324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lependu P, Iyer SV, Fairon C, Shah NH. Annotation analysis for testing drug safety signals using unstructured clinical notes. J Biomed Semantics. 2012;3 (Suppl 1):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung K, LePendu P, Iyer S, Bauer-Mehren A, Percha B, Shah NH. Functional evaluation of out-of-the-box text-mining tools for data-mining tasks. JAMIA. 2014;22(1):121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J BiomedI Inform. 2001;34:301–310. [DOI] [PubMed] [Google Scholar]
- 26.Chapman WW, Chu D, Downing JN. ConText: an algorithm for identifying contextual features from clinical text. Proceedings of the Workshop on BioNLP. 2007:81–88. [Google Scholar]
- 27.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. JAMIA. 2011;18(4):441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 29.Burges CJC. A tutorial on support vector machines for pattern recognition. Data Min KnowlDisc. 1998;2:121–167. [Google Scholar]
- 30.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 31.Liaw A, Wiener M. Classification and regression by randomforest. R News. 2002;2(3):18–22. [Google Scholar]
- 32.Friedman J, Hastie T, Simon N, Tibshirani R. http://cran.r-project.org/web/packages/glmnet/index.html. Accessed January 10, 2014. [Google Scholar]
- 33.Meyer D, Dimitriadou E, Hornik K, et al. http://cran.r-project.org/web/packages/e1071/index.html. Accessed January 10, 2014. [Google Scholar]
- 34.Liaw A. http://cran.r-project.org/web/packages/randomForest/index.html. Accessed March 25, 2014. [Google Scholar]
- 35.Brown SH, Elkin PL, Rosenbloom ST, et al. VA National Drug File Reference Terminology: a cross-institutional content coverage study. Stud Health Technol Inform. 2004;107(Pt 1):477–481. [PubMed] [Google Scholar]
- 36.FDA Adverse Event Reporting System (FAERS): Latest Quarterly Data Files, http://www.fda.gov/Drugs/GuidanceComplianceRegulatoryInformation/Surveillance/AdverseDrugEffects/ucm082193.htm. Accessed March 25, 2014.
- 37.Avillach P, Dufour JC, Diallo G, et al. Design and validation of an automated method to detect known adverse drug reactions in MEDLINE: a contribution from the EU-ADR project. JAMIA. 2013;20(3):446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harpaz R, Odgers D, Gaskin G, et al. A time-indexed reference standard of adverse drug reactions. Scientific Data. 2014;1:140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harpaz R, Vilar S, Dumouchel W, et al. Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. JAMIA. 2013;20(3):413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harpaz R, DuMouchel W, Shah NH. Comment on: “Zoo or savannah? Choice of training ground for evidence-based pharmacovigilance". Drug Safety. 2015;38(1):113–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noren GN, Caster O, Juhlin K, Lindquist M. Zoo or savannah? Choice of training ground for evidence-based pharmacovigilance. Drug Safety. 2014;37(9):655–659. [DOI] [PubMed] [Google Scholar]
- 42.Noren GN, Caster O, Juhlin K, Lindquist M. Authors' reply to Harpaz et al. comment on: “Zoo or savannah? Choice of training ground for evidence-based pharmacovigilance". Drug Safety. 2015;38(1):115–116. [DOI] [PubMed] [Google Scholar]
- 43.Harpaz R, DuMouchel W, LePendu P, Shah NH. Empirical bayes model to combine signals of adverse drug reactions. Knowledge Discovery and Data Mining'13. 2013:1339–1347. [Google Scholar]