Abstract
When using genome sequencing for molecular epidemiology, short sequence reads are aligned to an arbitrary reference strain to detect single nucleotide polymorphisms. We investigated whether reference genome selection influences epidemiological inferences of Mycobacterium tuberculosis transmission by aligning sequence reads from 162 closely related lineage 4 (Euro-American) isolates to 7 different genomes. Phylogenetic trees were consistent with use of all but the most divergent genomes, suggesting that reference choice can be based on considerations other than M. tuberculosis lineage.
TEXT
Whole-genome sequencing (WGS), which demonstrates higher resolution than classic molecular typing methods (see, e.g., references 1–5), has become the gold standard for molecular epidemiology of Mycobacterium tuberculosis. Epidemiological inferences depend on the detection of single nucleotide polymorphisms (SNPs) that distinguish isolates. Identification of SNPs using short-read data typically involves alignment (mapping) of reads to a single reference genome (e.g., M. tuberculosis H37Rv). As the difference between the genome of the reference strain and the clinical isolates increases (e.g., due to insertions/deletions/SNPs), fewer sequence reads are successfully mapped against the reference genome. As these data are essentially lost, the results are potentially biased, and true differences may go undetected. One solution in studies of other bacterial pathogens has been de novo assembly of a closely related isolate; this is then used in lieu of existing, more genetically distant reference genomes (6). However, this approach requires additional resources in terms of cost, technical expertise, and time; if short-read data are used for de novo assembly, a much greater sequencing depth is required (>100×) to ensure sufficient overlap of reads to facilitate accurate assembly(7), while alternative sequencing platforms are necessary to generate longer reads.
We asked whether the use of different reference genomes influences phylogenetic trees and epidemiological inferences of M. tuberculosis transmission, utilizing an existing data set of 163 lineage 4 (Euro-American) isolates from northern Quebec. DNA extraction and MiSeq-based WGS were performed as previously described (National Center for Biotechnology Information's Sequence Read Archive project under accession no. SRP039605, BioProject no. PRJNA240330) (8). Mixed infection with Mycobacterium avium was identified in 1 isolate using the Basic Local Alignment Search Tool (9); while this had no influence on previous phylogenies, it was excluded from the current analysis to avoid bias in coverage calculations (see below). Read quality was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were trimmed using Trimmomatic, v.0.32 (10), with a minimum length of 70 base pairs (bp), then aligned using the Burrows-Wheeler aligner (BWA) maximal exact matches (MEM) algorithm (11) to 7 different reference genomes (Table 1 [21–27]; for divergence in average nucleotide identity, see Table S1 in the supplemental material). PCR and optical duplicates were marked using Picard tools, v.1.118 (available at http://broadinstitute.github.io/picard/), and reads were locally realigned around insertions/deletions (indels). Reads aligning to >1 locus in the reference or with a mapping quality of <30 were excluded. The proportion of reads that aligned to each reference was calculated using SAMtools, v.1.2 (12). Genome coverage and average depth of coverage were calculated excluding duplicates in QualiMap, v.2 (13) and the Integrative Genomics Viewer (14) (see Tables S2 and S3 in the supplemental material).
TABLE 1.
Alignment and genome coverage across various reference genomes within the genus Mycobacteria
| Reference genome species | Reference genome name | Accession no. | Reference | Reference genome length (bp) | % of reads successfully aligned to referencea (median [IQR]) | Genome coverage (median [IQR])b at: |
||
|---|---|---|---|---|---|---|---|---|
| ≥1× depth | ≥10× depth | ≥20× depth | ||||||
| Mycobacterium tuberculosis | ||||||||
| Lineage 4 | H37Rv | NC_000962.3 | 21 | 4,411,532 | 98.0 (97.9–98.1) | 98.9 (98.8–98.9) | 98.1 (98.0–98.3) | 97.2 (96.8–97.5) |
| Lineage 4 | CDC1551 | NC_002755.2 | 22 | 4,403,837 | 98.2 (98.1–98.3) | 99.3 (99.2–99.3) | 98.5 (98.4–98.7) | 97.6 (97.2–97.9) |
| Lineage 2 | CCDC5079 | CP001641 | 23 | 4,398,812 | 97.8 (97.7–97.9) | 98.8 (98.7–98.8) | 98.1 (97.9–98.2) | 97.1 (96.8–97.4) |
| Mycobacterium africanum | GN041182 | FR878060.1 | 24 | 4,389,314 | 97.5 (97.4–97.5) | 98.9 (98.8–98.9) | 98.1 (98.0–98.3) | 97.2 (96.8–97.4) |
| Mycobacterium bovis | AF2122/97 | NC_002945.3 | 25 | 4,345,492 | 97.6 (97.5–97.7) | 99.3 (99.2–99.3) | 98.5 (98.4–98.7) | 97.6 (97.2–97.9) |
| Mycobacterium canettii | CIPT 140010059 | NC_015848.1 | 26 | 4,482,059 | 96.5 (96.3–96.6) | 95.1 (95.0–95.1) | 94.3 (94.2–94.4) | 93.4 (93.0–93.8) |
| Mycobacterium kansasiic | ATCC 12478 | NC_022663.1 | 27 | 6,432,277 | 52.7 (51.6–53.4) | 34.5 (34.2–35.5) | 28.4 (27.8–29.1) | 25.5 (24.6–26.4) |
Calculated using SAMtools (flagstat) as (total mapped − secondary alignments − duplicate reads)/(total reads surviving trimming − duplicate reads).
QualiMap includes secondary alignments marked by BWA-MEM (range, 1% to 3% of total mapped), double counted in coverage calculations. Duplicates excluded.
pMK plasmid sequence not used for alignment.
The highest proportion of reads were mapped to the CDC1551 (lineage 4) reference, followed by H37Rv (Table 1). As both are lineage 4, this is unsurprising. The median proportions of the CDC1551 and H37Rv references that had at least 1 read aligned (genome coverage) were also the highest across all analyses. As the reference strain became more genetically divergent from the sequenced isolates (lineage 2 M. tuberculosis, Mycobacterium africanum, Mycobacterium bovis, and Mycobacterium canettii), the percentage of total reads aligned and genome coverage declined slightly. When aligning against Mycobacterium kansasii, these values decreased by 45.5% and 64.8%, respectively, compared to those for CDC1551.
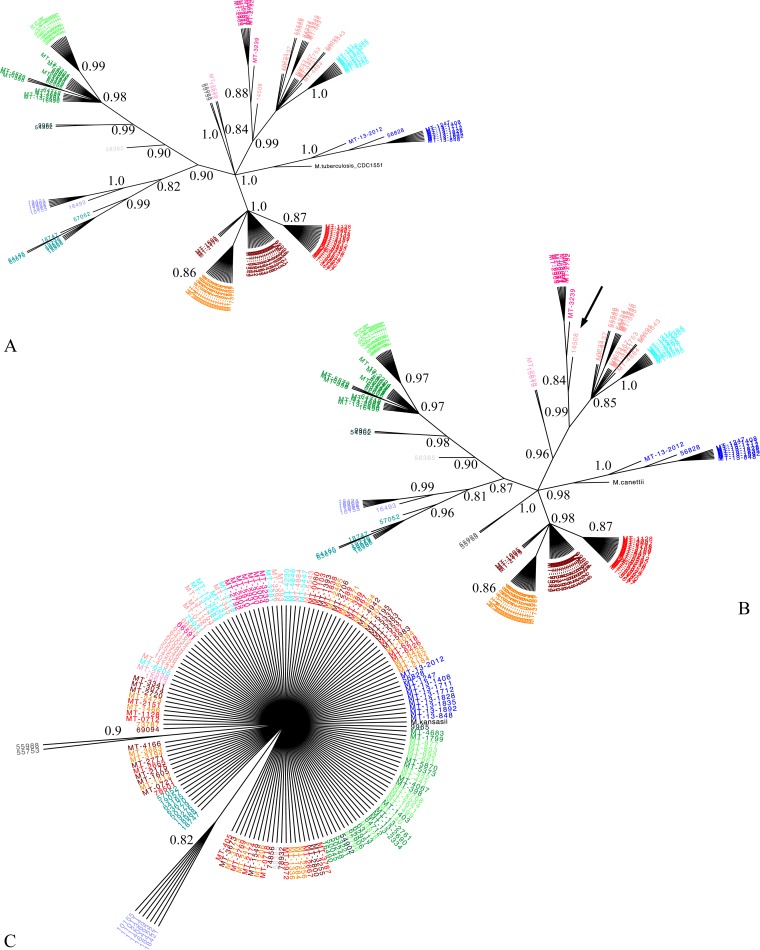
SNPs and indels were then identified (called) for each reference analysis using the Genome Analysis Toolkit (GATK), v.3.3 (15). SNPs were filtered for quality based on GATK recommendations, including assessment of strand bias. In addition, we required a Phred score of ≥50 (where Phred = −10 · log Perror, corresponding to a 1/100,000 probability of error) for each SNP locus, a minimum depth of coverage (i.e., the number of reads that are aligned to that locus) of 8 bp, and individual Phred-scaled genotype quality of ≥15 to confidently call an SNP. SNPs within 12 bp of one another or indels and heterozygous calls were excluded. Concatenated SNPs from each alignment were then used to generate phylogenetic trees using the maximum-likelihood method (16) with 1,000 bootstrap replicates (17). The model of nucleotide substitution was chosen based on the Bayesian information criterion. Because repetitive PE_PGRS genes, PPE genes, and mobile elements were not consistently annotated across all reference genomes, SNPs in these regions were included; however, any bias due to these SNPs should be nondifferential across references. Trees from each analysis were compared qualitatively and were largely consistent with a previously reported deletion-based phylogeny (8). As illustrated in Fig. 1 and Fig. S1 in the supplemental material, small changes in clustering became evident at the level of M. canettii, while resolution was almost entirely lost with M. kansasii.
FIG 1.
Impact of reference genome choice on phylogeny. Maximum likelihood trees with 1,000 bootstrap replicates. Branches of <80% bootstrap threshold are collapsed (branch lengths are therefore not to scale). For clarity, bootstrap P values are indicated up to the most proximal node defining each cluster. Isolates were colored for their respective clusters, identified according to CDC1551 (and H37Rv) (8). Isolates were then kept the same color across all panels to facilitate quick comparison between the new reference analysis and CDC1551 (see Table S4 in the supplemental material for cluster names). (A) Reference M. tuberculosis lineage 4 CDC1551, using the Tamura 3 parameter model of nucleotide substitution with 1,522 SNP loci (19). (B) Reference M. canettii, using the general time-reversible (GTR) model of nucleotide substitution with 17,406 SNP loci (20). Using M. canettii as a reference, a single isolate changed clusters (arrow). (C) Reference M. kansasii, using the GTR model of nucleotide substitution with 34,127 SNP loci.
To examine whether reference choices influenced our interpretation of direct patient-to-patient transmission, we restricted our analysis to 49 isolates from a well-defined epidemiological outbreak in a single Quebec community. All cases were diagnosed within a 1-year period, and previous work suggested a threshold for recent direct transmission of 0 to 1 SNP (5). Matrices of pairwise SNPs between isolates were generated. Using classifications with CDC1551 as the gold standard, because its genetic similarity to our isolates was the closest, we calculated the sensitivity and specificity for classifying each pair as probable recent transmission or not. As shown in Table 2, the sensitivity and specificity for detecting recent transmission was 100% across all reference genomes, except M. kansasii. In the latter, nearly all of the SNPs that formerly ruled out transmission between some pairs were missed because of low mapping to the reference, yielding an unacceptably high number of false positives.
TABLE 2.
Comparing pairwise single nucleotide polymorphisms and probable recent transmission by reference genome, using CDC1551 as gold standard
| Reference genome species | Reference genome name | Median pairwise SNP compared to reference (IQR)a | Median pairwise SNP between isolates (IQR)b | Sensitivity for recent transmission (95% CI)c | Specificity for recent transmission (95% CI) |
|---|---|---|---|---|---|
| Mycobacterium tuberculosis | |||||
| Lineage 4 | H37Rv | 781 (780–781) | 3 (2–6) | 100 (98.7–100) | 100 (99.6–100) |
| Lineage 4 | CDC1551 | 619 (618–619) | 3 (2–6) | ||
| Lineage 2 | CCDC5079 | 1,247 (1,246–1,247) | 3 (2–6) | 100 (98.7–100) | 100 (99.6–100) |
| Mycobacterium africanum | GN041182 | 1,908 (1,907–1,908) | 3 (2–6) | 100 (98.7–100) | 100 (99.6–100) |
| Mycobacterium bovis | AF2122/97 | 2,000 (1,999–2,000) | 3 (2–6) | 100 (98.7–100) | 100 (99.6–100) |
| Mycobacterium canettii | CIPT 140010059 | 16,637 (16,636–16,637) | 3 (2–6) | 100 (98.7–100) | 100 (99.6–100) |
| Mycobacterium kansasii | ATCC 12478 | 34,081 (34,081–34,081) | 0 (0–0) | 100 (98.7–100) | 0.1 (0.0–0.06) |
49 pairwise comparisons with reference genome. IQR, interquartile range.
1,176 pairwise comparisons.
95% CI, 95% confidence interval.
Overall, we have shown that that the choice of reference genome, within the M. tuberculosis complex, has negligible influence on phylogeny and epidemiological studies of M. tuberculosis transmission. Our ability to demonstrate the robustness of these analyses using a data set with very limited strain diversity (153/163 isolates were separated by a maximum distance of 72 SNPs, and clusters were distinguished by as few as 2 SNPs) (5, 8) indicates that our findings are generalizable to settings with greater genetic diversity and robust to differences in M. tuberculosis lineage. Therefore, epidemiological studies of tuberculosis can base reference choices on aspects such as quality of annotation rather than matching strain lineage.
Our findings also indicate that there is a threshold of genome coverage beyond which transmission can no longer be accurately discriminated. This can have implications particularly for nonclonal pathogens, which have greater genetic diversity than M. tuberculosis. One approach with such organisms restricts short-read alignment to the core genome region (e.g., in Escherichia coli, this represents only 40% of all possible genes [18]), while another approach restricts it to variation within preselected genes (e.g., housekeeping genes, used for multilocus sequence typing). These subsets are then used to build phylogenetic trees and delineate clusters of transmission. When limited to only a subset of the genome, epidemiologically relevant genetic diversity can be overlooked, as demonstrated when aligning to M. kansasii. A more optimal approach might involve aligning to both core and accessory genes and >1 reference from the same species, to capture a more complete portrait of bacterial diversity. To facilitate this, efforts must be made to further sequence, close, and annotate such genomes.
Supplementary Material
Funding Statement
The funding agency had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00364-16.
REFERENCES
- 1.Niemann S, Köser CU, Gagneux S, Plinke C, Homolka S, Bignell H, Carter RJ, Cheetham RK, Cox A, Gormley NA, Kokko-Gonzales P, Murray LJ, Rigatti R, Smith VP, Arends FPM, Cox HS, Smith G, Archer JAC. 2009. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS One 4:e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJM, Brinkman FSL, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 3.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TE. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee RS, Radomski N, Proulx JF, Manry J, McIntosh F, Desjardins F, Soualhine H, Domenech P, Reed MB, Menzies D, Behr MA. 2015. Reemergence and amplification of tuberculosis in the Canadian arctic. J Infect Dis 211:1905–1914. doi: 10.1093/infdis/jiv011. [DOI] [PubMed] [Google Scholar]
- 5.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ekblom R, Wolf JBW. 2014. A field guide to whole-genome sequencing, assembly and annotation. Evol Appl 7:1026–1042. doi: 10.1111/eva.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee RS, Radomski N, Proulx J-F, Levade I, Shapiro BJ, McIntosh F, Soualhine H, Menzies D, Behr MA. 2015. Population genomics of Mycobacterium tuberculosis in the Inuit. Proc Natl Acad Sci U S A 112:13609–13614. doi: 10.1073/pnas.1507071112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 10.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997. http://arxiv.org/abs/1303.3997.
- 12.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okonechnikov K, Conesa A, García-Alcalde F. 2016. QualiMap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorvaldsdottir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 18.Mira A, Martín-Cuadrado AB, D'Auria G, Rodríguez-Valera F. 2010. The bacterial pan-genome: a new paradigm in microbiology. Int Microbiol 13:45–57. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K. 1992. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9:678–687. [DOI] [PubMed] [Google Scholar]
- 20.Waddell PJ, Steel MA. 1997. General time-reversible distances with unequal rates across sites: mixing Γ and inverse Gaussian distributions with invariant sites. Mol Phylogenet Evol 8:398–414. doi: 10.1006/mpev.1997.0452. [DOI] [PubMed] [Google Scholar]
- 21.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 22.Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, Hickey E, Kolonay JF, Nelson WC, Umayam LA, Ermolaeva M, Salzberg SL, Delcher A, Utterback T, Weidman J, Khouri H, Gill J, Mikula A, Bishai W, Jacobs WR, Venter JC, Fraser CM. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol 184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Chen C, Liu J, Deng H, Pan A, Zhang L, Zhao X, Huang M, Lu B, Dong H, Du P, Chen W, Wan K. 2011. Complete genome sequences of Mycobacterium tuberculosis strains CCDC5079 and CCDC5080, which belong to the Beijing family. J Bacteriol 193:5591–5592. doi: 10.1128/JB.05452-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bentley SD, Comas I, Bryant JM, Walker D, Smith NH, Harris SR, Thurston S, Gagneux S, Wood J, Antonio M, Quail MA, Gehre F, Adegbola RA, Parkhill J, de Jong BC. 2012. The genome of Mycobacterium africanum West African 2 reveals a lineage-specific locus and genome erosion common to the M. tuberculosis complex. PLoS Negl Trop Dis 6:e1552. doi: 10.1371/journal.pntd.0001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garnier T, Eiglmeier K, Camus J-C, Medina N, Mansoor H, Pryor M, Duthoy S, Grondin S, Lacroix C, Monsempe C, Simon S, Harris B, Atkin R, Doggett J, Mayes R, Keating L, Wheeler PR, Parkhill J, Barrell BG, Cole ST, Gordon SV, Hewinson RG. 2003. The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci U S A 100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Supply P, Marceau M, Mangenot S, Roche D, Rouanet C, Khanna V, Majlessi L, Criscuolo A, Tap J, Pawlik A, Fiette L, Orgeur M, Fabre M, Parmentier C, Frigui W, Simeone R, Boritsch EC, Debrie A-S, Willery E, Walker D, Quail MA, Ma L, Bouchier C, Salvignol G, Sayes F, Cascioferro A, Seemann T, Barbe V, Locht C, Gutierrez M-C, Leclerc C, Bentley S, Stinear TP, Brisse S, Medigue C, Parkhill J, Cruveiller S, Brosch R. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet 45:172–179. doi: 10.1038/ng.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, McIntosh F, Radomski N, Dewar K, Simeone R, Enninga J, Brosch R, Rocha EP, Veyrier FJ, Behr MA. 2015. Insights on the emergence of Mycobacterium tuberculosis from the analysis of Mycobacterium kansasii. Genome Biol Evol 7:856–870. doi: 10.1093/gbe/evv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.