Abstract
Eumycetoma is a debilitating, chronic, fungal infection that is endemic in India, Indonesia, and parts of Africa and South and Central America. It remains a neglected tropical disease in need of international recognition. Infections follow traumatic implantation of saprophytic fungi and frequently require radical surgery or amputation in the absence of appropriate treatment. Several fungal species can cause black-grain mycetomas, including Madurella spp. (Sordariales), Falciformispora spp., Trematosphaeria grisea, Biatriospora mackinnonii, Pseudochaetosphaeronema larense, and Medicopsis romeroi (all Pleosporales). We performed phylogenetic analyses based on five loci on 31 isolates from two international culture collections to establish the taxonomic affiliations of fungi that had been isolated from cases of black-grain mycetoma and historically classified as Madurella grisea. Although most strains were well resolved to species level and corresponded to known agents of eumycetoma, six independent isolates, which failed to produce conidia under any conditions tested, were only distantly related to existing members of the Pleosporales. Five of the six isolates shared >99% identity with each other and are described as Emarellia grisea gen. nov. and sp. nov; the sixth isolate represents a sister species in this novel genus and is described as Emarellia paragrisea. Several E. grisea isolates were present in both United Kingdom and French culture collections and had been isolated independently over 6 decades from cases of imported eumycetoma. Four of the six isolates involved patients that had originated on the Indian subcontinent. All isolates were all susceptible in vitro to the azole antifungals, but had elevated MICs with caspofungin.
INTRODUCTION
Described over 3,000 years ago in the Atharva Veda, and present in Mesoamerica during the pre-Hispanic period (1), mycetoma has preponderance for men of working age and children (2). Eumycetoma (fungal mycetoma), which is acquired after traumatic inoculation usually on exposed body sites, is characterized by a chronic progressive destruction of soft tissue and adjoining structures with associated tumefaction and purosanguineous sinuses that drain fungal grains (2–5). Without adequate treatment, the infection eventually invades the skeletal system, and clinical and mycological cure is impossible without extensive debridement/amputation (2–5). The extruded grains are either pale or dark (black), depending on the etiological agent (4, 5). Historically, the most common agents of black-grain eumycetoma were classified within the genus Madurella (Sordariales), which is characterized by organisms that produce dark gray, brown or black colonies, and dematiaceous hyphae which either remain sterile or sporulate only very reluctantly (4). The absence of conidiogenesis in isolates recovered from most black-grain mycetoma infections has historically hindered accurate species identification, and it was only recently that molecular approaches have demonstrated that Madurella mycetomatis (6) (Sordariales) and “Madurella grisea” (7) (Pleosporales) are genetically unrelated (8). More recently, genuine sister species have been discovered in Madurella, and M. pseudomycetomatis (9), M. fahalii, and M. tropicana (10) have been described. In addition, the identities and taxonomic affiliations of a number of other agents of black-grain mycetoma have been better elucidated. Classified within Pleosporales, they include Medicopsis romeroi (formerly Pyrenochaeta), Falciformispora senegalensis, and F. tompkinsii (both formerly Leptosphaeria) (11), Biatriospora mackinnonii (formerly Pyrenochaeta and Nigrograna), Trematosphaeria grisea (including some former M. grisea isolates), Pseudochaetosphaeronema larense, and Roussoella spp. (12). Two further poorly or nonsporulating dematiaceous fungi, Rhytidhysteron rufulum (Patellariales) and Roussoella percutanea (Arthopyreniaceae/Roussoellaceae), have also recently been described from subcutaneous mycetoma-like infections in humans, although grain and sinus tract formation has not been reported to date (13–15).
Since Madurella grisea was distinguished historically by gray velvety colonies, and lack of production of both conidia and diffusing pigment (7), the species was likely to be polyphyletic. Indeed, many of the isolates originally identified as M. grisea have now been reclassified and placed in Medicopsis, Biatriospora, and Trematosphaeria (12). However, it is highly probable that additional historical “M. grisea” isolates might still include more taxa, including undescribed species. The present study sought to address this hypothesis, using isolates of “M. grisea” archived in two international culture collections, the National Collection of Pathogenic Fungi (NCPF) housed at the UK National Mycology Reference Laboratory, Bristol, United Kingdom, and the Institut Pasteur Culture Collection (UMIP), housed at the Institut Pasteur, Paris, France. Isolates from proven cases of black-grain mycetoma which fulfilled the historical characteristics of “M. grisea,” and other known pleosporalean fungi implicated in mycetoma were subjected to a phylogenetic analysis using up to five loci: the internal transcribed spacer region 1 (ITS1), the large ribosomal subunit (LSU), translation elongation factor 1α (TEF1), the second largest RNA polymerase subunit (RPB2), and actin (ACT). Using this approach, we have identified six isolates that constitute two sister species in a novel pleosporalean genus, which we describe here as Emarellia.
MATERIALS AND METHODS
Strains analyzed.
Clinical isolates obtained from confirmed cases of dark grain mycetoma were obtained from the NCPF or UMIP culture collections as detailed in Table 1. All isolates were subcultured twice on plates of Oxoid Sabouraud dextrose agar containing 0.5% (wt/vol) chloramphenicol (Unipath Limited, Basingstoke, England).
TABLE 1.
Isolates and sequences used in the present studya
| Organism | Culture no. | Source, origin, yr | GenBank accession no. |
||||
|---|---|---|---|---|---|---|---|
| LSU | ITS | TEF1 | RPB2 | Other | |||
| Biatriospora mackinnonii | CBS674.75T | Mycetoma, Venezuela, 1975 | KF015612 | KF015654 | KF407986 | KF015703 | |
| CBS110022 | Mycetoma, Mexico, 1999 | KF015609 | KF015653 | KF407985 | KF015704 | ||
| IP1034.71* | Mycetoma, Costa Rica, 1971 | LT160875 | LT160874 | ||||
| IP72.65* | Mycetoma, Venezuela, 1965 | LT160877 | LT160876 | ||||
| IP67.52* | Mycetoma, Venezuela, 1952 | LT160879 | LT160878 | ||||
| NCPF 2290* | Mycetoma, Venezuela, 1981 | LT160881 | LT160880 | ||||
| NCPF 2292* | Mycetoma, Venezuela, 1981 | LT160883 | LT160882 | ||||
| NCPF 2294* | Mycetoma, Venezuela, 1981 | LT160885 | LT160884 | ||||
| NCPF 2297* | Mycetoma, Venezuela, 1981 | LT160887 | LT160886 | ||||
| NCPF 2323* | Mycetoma, Venezuela, 1982 | LT160889 | LT160888 | ||||
| Falciformispora senegalensis | NCPF 7023 | Mycetoma, Senegal, 1960 | LT160891 | LT160890 | |||
| CBS196.79T | Mycetoma, Senegal, 1979 | KF015631 | KF015673 | KF015687 | KF015717 | ||
| CBS197.79 | Mycetoma, Senegal, 1979 | KF015626 | KF015677 | KF015690 | KF015712 | ||
| IP612.60 | Mycetoma, Senegal, 1960 | LT160892 | |||||
| IP610.60 | Mycetoma, Senegal, 1960 | DQ836776 | |||||
| IP611.60 | Mycetoma, Senegal, 1960 | DQ836777 | |||||
| Falciformispora tompkinsii | CBS200.79 | Man, Senegal | KF015625 | KF015670 | KF015685 | KF015719 | |
| CBS201.79 | Man, Senegal | KF015624 | KF015671 | KF015686 | KF015718 | ||
| IP1151.76 | Mycetoma, Senegal, 1976 | DQ836778 | |||||
| IP1156.77 | Mycetoma, 1977 | LT160873 | |||||
| Medicopsis romeroi | CBS132878 | Mycetoma, India | KF015622 | KF015658 | KF015682 | KF015709 | |
| CBS252.60T | Mycetoma, Venezuela, 1960 | EU754207 | KF366446 | KF015678 | KF015708 | ||
| NCPF 2325 | Mycetoma, Venezuela, 1951 | AJ842341 | AJ842342 | ||||
| NCPF 2615* | Mycetoma, Pakistan, 1986 | LT160894 | LT160893 | ||||
| NCPF 2687* | Mycetoma, India, 1986 | LT160896 | LT160895 | ||||
| NCPF 2688* | Mycetoma, India, 1981 | LT160898 | LT160897 | ||||
| NCPF 2723* | Mycetoma, Pakistan, 1989 | LT160900 | LT160899 | ||||
| NCPF 2755* | Mycetoma, Unknown, 1990 | LT160902 | LT160901 | ||||
| NCPF 2859* | Mycetoma, India, 1992 | LT160904 | LT160903 | ||||
| NCPF 7251* | Mycetoma, Pakistan, 1996 | LT160906 | LT160905 | ||||
| NCPF 7521* | Mycetoma, India, 2000 | LT160908 | LT160907 | ||||
| NCPF 7525 | Mycetoma, Saudi Arabia, 2000 | LT160913 | |||||
| NCPF 7653* | Mycetoma, Pakistan, 2004 | LT160910 | LT160909 | ||||
| NCPF 7668* | Mycetoma, India, 2005 | LT160912 | LT160911 | ||||
| Pseudochaetosphaeronema larense | CBS640.73T | Mycetoma, Venezuela, 1973 | KF015611 | KF015656 | KF015684 | KF015706 | |
| CBS639.94 | Man, Venezuela, 1994 | KF015610 | KF015655 | KF015683 | KF015705 | ||
| Rhytidhysteron rufulum | IP234.61* | Mycetoma, Senegal, 1961 | LT160915 | LT160914 | |||
| IP696* | Mycetoma, Unknown | LT160917 | LT160916 | ||||
| IP1134.76* | Mycetoma, South Africa, 1976 | LT160919 | LT160918 | ||||
| NCPF 7709* | Mycetoma, Zimbabwe, 2005 | LT160921 | LT160920 | ||||
| Roussoella percutanea | CBS128203 | Subcutaneous, Aruba | KF366448 | KF322117 | KF407988 | KF366453 | |
| Trematosphaeria grisea | CBS332.50 (AUT) | Mycetoma, Chile, 1950 | KF015614 | KF015662 | KF015698 | KF015720 | |
| CBS246.66 | Abscess, India, 1966 | KF015615 | KF015661 | KF015697 | KF366454 | ||
| Trematosphaeria pertusa | CBS122371 | Plant, France | FJ201992 | KF015669 | KF015702 | GU371801 | |
| CBS122368 | Plant, France | FJ201990 | KF015668 | KF015701 | FJ795476 | ||
| Emarellia grisea gen. nov., sp. nov. | NCPF 7066T* | Mycetoma, India, 1976 | LT160923 | LT160922 | LT160934 | LT160933 | LT160932 |
| NCPF 7384* | Mycetoma, Sri Lanka, 1998 | LT160925 | LT160924 | LT160937 | LT160936 | LT160935 | |
| NCPF 7666* | Mycetoma, India, 2004 | LT160927 | LT160926 | LT160940 | LT160939 | LT160938 | |
| NCPF 7761* | Mycetoma, Sri Lanka, 2009 | FN600643 | FN600644 | LT160943 | LT160942 | FN600645; LT160941 | |
| IP66.52* | Mycetoma, unknown, 1952 | LT160929 | LT160928 | ||||
| Emarellia paragrisea gen. nov., sp. nov. | NCPF 7611T* | Mycetoma, unknown, 2002 | LT160931 | LT160930 | LT160946 | LT160945 | LT160944 |
Isolates indicated in boldface are novel to the present study; those denoted by an asterisk represent hitherto-unidentified organisms with colonial morphologies consistent with “Madurella grisea.” Accession numbers for the sequences of CBS isolates and NCPF 7761 were taken from references 12 and 34, respectively.
Morphological examination.
The clinical isolates included in this study were all subjected to morphological examination at the MRL. Isolates were cultured on slopes of Oxoid Sabouraud dextrose agar containing 0.5% (wt/vol) chloramphenicol, potato dextrose agar (PDA; 20 g of glucose, 4 g of potato extract, and 15 g of agar per liter), potato sucrose agar (PSA; 20 g of sucrose, 4 g of potato extract, and 15 g of agar per liter), Borelli's Lactrimel medium (honey, 10 g; wheat flour, 10 g; milk, 200 ml; agar, 18 g/liter), cornmeal agar (CMA; 2 g of cornmeal extract and 15 g of agar per liter), or CMA supplemented with sterilized, twice-autoclaved straw. Cultures were incubated at 30°C and at room temperature both in the dark and with sunlight/diurnal lighting for up to 6 months. Cultures were examined for conidiogenesis under a Kyowa Optical SDZ-PL dissecting microscope, and using a Nikon Eclipse E400 light microscope to examine microscopic mounts prepared in lactophenol mounting fluid. The growth rates of Emarellia species were determined on Sabouraud dextrose agar incubated for 14 days in the dark at 25, 30, 35, 37, and 40°C.
Molecular methods.
Genomic DNA was prepared from cultures after 2 weeks incubation on Sabouraud agar using Whatman FTA filter paper technology exactly as described previously (16). Amplification of a region of the large subunit gene (LSU) and the internal transcribed spacer 1 (ITS1) region was performed using the primers described in (17). Amplification of a fragment of the actin (ACT), RNA polymerase second largest subunit (RPB2) and translation elongation factor 1α (TEF) genes used the primer pairs ACT-512F/ACT-738R (18), fRPB2-5F/fRPB2-7cR (19), and EF-3983F/EF-2218R (http://www.aftol.org/pdfs/EF1primer.pdf), respectively, and the specific cycling conditions specified by the authors (after an initial enzyme activation step at 94°C for 15 min). In all cases, PCR amplification (100-μl reaction volumes) was performed in the presence of 200 μM concentrations of each deoxynucleoside triphosphate, 250 nM concentrations of the appropriate primers, 2 U of HotStar Taq polymerase (Qiagen, Valencia, CA), and a single FTA filter punch. The amplification success was evaluated by electrophoresis of a fraction of total amplification products in 1.2% (wt/vol) agarose gels run for 45 min at 120 V in Tris-borate buffer. Amplification products were sequenced in both directions using the forward and reverse primers described above for each gene target.
Sequence alignment and phylogenetic analyses.
Preliminary organism identification was achieved using BLASTN searches against fungal sequences in existing EMBL/GenBank DNA databases (20). For phylogenetic analyses, sequence alignments included reference sequences from the type strains of key members of the Pleosporales obtained from GenBank (see Table 1) (12). Sequences from this study were deposited in GenBank; the accession numbers are given in Table 1. Initial alignments of each gene were performed separately in MAFFT version 7 (21) and then combined for phylogenetic analyses. The MEGA program, version 6 (22), was used to construct phylogenetic trees based on the maximum-likelihood algorithm with a Tamura-Nei model and gaps treated as complete deletions. A 100-bootstrap replicate was used, and bootstrap values >80% were considered significant. The topological equivalence of individual trees was assessed using the TOPD/FMTS software described by Puigbo et al. (23). Pairwise comparisons of trees were made by using the nodal distance and compared to unguided random reiterations.
Antifungal agents and CLSI broth microdilution determination of MICs.
Antifungal drugs were obtained from their respective manufacturers as standard powders. Amphotericin B (AMB; Sigma Chemical, St. Louis, MO), and voriconazole (VRC; Pfizer Central Research, Sandwich, United Kingdom) were dissolved in dimethyl sulfoxide. Itraconazole (ITC; Janssen Research Foundation, Beerse, Belgium) was prepared in PEG400 with heating to 70°C. Caspofungin (CAS; Merck, Sharp, and Dhome, Hoddlesdon, United Kingdom) was resuspended in sterile water. Serial 2-fold dilutions of the various drugs were prepared in RPMI 1640 medium (with l-glutamine, without bicarbonate; Sigma) and buffered to pH 7.0 using a 0.165 M solution of morpholinepropanesulfonic acid (Sigma).
MICs were determined according to Clinical and Laboratory Standards Institute (CLSI) methodologies (24) in round-bottom 96-well plates with hyphal fragment suspensions. Suspensions were prepared under saline by gentle scraping of aerial mycelium and left to settle for 2 min, and the supernatant containing short hyphal fragments was diluted into RPMI 1640 and adjusted to final concentrations equivalent to 2.5 × 103 CFU/ml (spectrophotometric transmission at 530 nm of 70 to 80% compared to inoculum-free controls). Viability of hyphal fragments was verified by plating of dilutions of resulting suspensions on Sabouraud agar. All MIC assays included the quality control Aspergillus fumigatus strains NCPF 7097 and NCPF 7100 and were performed at least in duplicate. Inoculated plates were incubated at 35°C. MICs were read at 48 or 96 h (depending on growth rates for each organism) as the concentration of drug that elicited 100% inhibition of growth (AMB) and significant (ca. 80%) inhibition of growth compared with a drug-free control (ITC and VRC). For caspofungin, the minimum effective concentration was measured by microscopic examination as described previously (24) and scored as the lowest concentration that resulted in restricted compact microcolonies compared to hyphal growth in the absence of drug.
Accession numbers.
Sequences were deposited in GenBank under accession numbers LT160873 to LT160946, and data were deposited in MycoBank under accession numbers MB815958 to 815960.
RESULTS
In the present study, 31 fungal isolates from two international fungal culture collections (i.e., the NCPF and UMIP collections), including many that had been previously identified phenotypically as Madurella grisea on the basis of colonial morphology and absence of conidiation, were revived and subjected to molecular analyses. Isolates were predominantly from imported cases of eumycetoma in patients who had previously resided in central and South America, Africa, or the Indian subcontinent, and dated from 1951 through 2009 (Table 1).
Molecular and morphological analyses of isolates.
All isolates produced floccose, felt-like or velvety gray to dark brown, domed colonies on Sabouraud media, with dematiaceous (melanized) septate sterile hyphae and, in many cases, infrequent lateral, terminal, and intercalary chlamydospores (Fig. 1). Repeated attempts to induce conidiation were unsuccessful for all of the isolates included in the present study (data not shown). The media used included Sabouraud agar, PSA, PDA, Borelli's Lactrimel, CMA, and CMA supplemented with sterilized straw, along with incubation at room temperature in the dark, with diurnal lighting and UV/sunlight, or in the dark at 30, 35, and 37°C (see Materials and Methods). Incubation was continued for up to 6 months, with repeated subcultures.
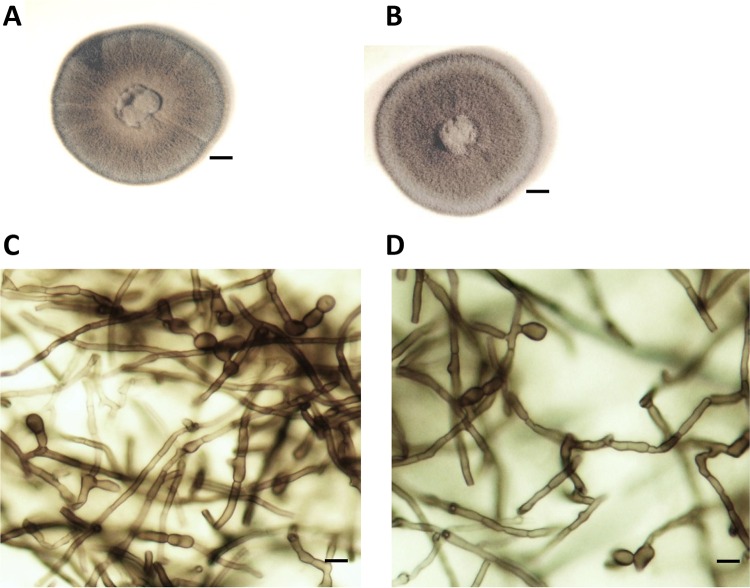
FIG 1.
Colonial and microscopic appearance of Emarellia grisea (NCPF 7066) (A and C) and E. paragrisea (NCPF 7611) (B and D). The colonial appearance after 2 weeks of culture on Sabouraud dextrose agar at 30°C (A and B) and the microscopic appearance showing lateral, terminal, and intercalary chlamydospores (prepared in lactophenol, ×400 magnification) are depicted (C and D). No discernible differences in colonial morphology or conidiation were noted on any media. Scale bar: 4 mm (A and C) or 5 μm (B and D).
In agreement with previous reports (12), all isolates grew readily in the temperature range 20 to 37°C, with modest species-specific differences in optimal growth temperature within this range (data not shown). PCR amplification and sequencing of the LSU and ITS1 regions of the various isolates, and comparison of the sequences of the resulting amplicons to those available in public synchronized databases in most cases allowed robust identification of the organisms to species level (>99% identity over both regions to known agents of eumycetoma; see Table 1).
Of 28 isolates historically considered to constitute M. grisea (denoted by asterisks in Table 1), 22 were in fact other, established pleosporalean agents of eumycetoma and included B. mackinnonii (8 isolates), M. romeroi (10 isolates), and R. rufulum (4 isolates). Thus, at least among the isolates in the two culture collections examined, B. mackinnonii and M. romeroi appear to be more common causes of eumycetoma than previously thought. Interestingly, a further six isolates (NCPF 7066, 7384, 7666, 7761, and 7611 and IP66.52), which were predominantly from patients originating in the Indian subcontinent, returned much poorer quality matches with existing sequences in the public databases; the closest matches with reliably identified organisms were to Trematosphaeria grisea isolates (ca. 80% identity over both LSU and ITS1 regions; data not shown).
Phylogenetic analyses.
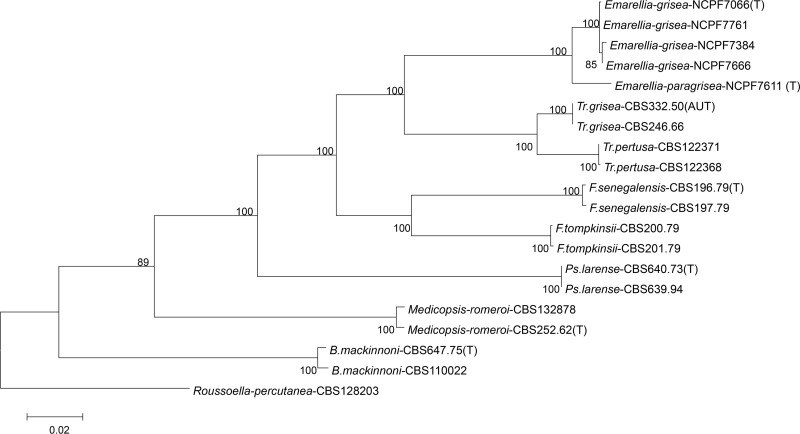
To further characterize these six novel isolates, PCR amplification and sequencing was extended to include portions of the RPB2 and TEF1 genes and, together with LSU and ITS1, these additional loci were used for phylogenetic comparisons with the established pleosporalean agents of eumycetoma. A total of 2,202 alignment characters were used, of which 355 were derived from LSU, 257 were derived from ITS1, 705 were derived from RPB2, and 882 were derived from TEF1. Maximum-likelihood phylogenetic trees were established for each of the four individual loci and for the combined four locus data set (Fig. 2 and see Fig. S1 in the supplemental material). Since attempts to amplify and sequence the RPB2 locus from strains of R. rufulum were unsuccessful, trees were rooted either with R. rufulum (LSU, ITS, and TEF1) or R. percutanea (RPB2 and combined data set). For clarity of the resulting trees, phylogenetic analyses were performed with sequences corresponding to key (type strains where available) examples of the other pleosporalean agents of eumycetoma rather than the whole sequence data set. Initial comparisons performed with 100 bootstrap replicates suggested that larger replicate numbers would not provide additional useful phylogenetic information (data not shown). The topological equivalence of individual trees was shown by low nodal distance values for all pairwise combinations of the four-gene, ITS, RPB, and TEF trees, as assessed through the TOPD/FMTS software (23). Slightly less equivalence was seen with comparisons for the LSU derived tree. This tree had less bootstrap support than the others and this, together with the lower levels of heterogeneity seen in these sequences suggests that this data set may have less resolution for these genera than the others. Lower levels of resolution with one gene are common in fungi as very few gene regions have been found to be equivalent across all groups (25). Importantly, all five phylogenetic trees strongly supported the existence of a new well-supported clade within the Pleosporales (Trematosphaeriaceae), encompassing isolates NCPF 7066, 7611, 7761, 7666, and 7384 and IP66.52. (Fig. 2 and see Fig. S1 in the supplemental material). Given the phylogenetic distance of this clade from all other known members of the Trematosphaeriaceae, we propose the erection of a new genus, Emarellia, and Emarellia grisea sp. nov. for isolates NCPF 7066, 7384, 7666, and 7761 and IP66.52, with NCPF 7066 as the type species.
FIG 2.
Consensus phylogenetic tree for representative members of the Pleosporales inferred from the combined data set of ITS1, LSU, RPB2, and TEF1. Trees were constructed in MEGA version 6.0 using maximum likelihood and rooted with R. percutanea. Bootstrap support values of >50 are indicated above branches.
In all phylogenetic trees, NCPF 7611 appeared consistently as a sibling outgroup to this clade. We further analyzed inter- and intraspecific variation among the isolates of Emarellia by including a fifth locus, ACT (see Table 1 for additional EMBL accession numbers). Sequence divergence within the isolates of E. grisea (NCPF 7066, 7384, 7666, 7761 and IP66.52) was extremely low with 0, 0, 2, 2, and 3 polymorphic nucleotides identified between the various organisms in the LSU (386 nucleotides [nt]), ACT (195 nt), ITS1 (222 nt), TEF1 (930 nt), and RPB2 (705 nt) regions examined, respectively. Conversely, isolate NCPF 7611 differed from isolates of E. grisea with substitutions at 2, 6, 9, 33, and 13 nt positions, respectively, over the same loci. Representative portions of the aligned sequences are depicted in Fig. S2 in the supplemental material. Given this degree of divergence (% nucleotide identity with sequences of E. grisea strains of 99.5 [LSU], 96.0 [ITS1], 96.9 [ACT], 98.1 [RPB2], and 96.5 [TEF1]), we propose NCPF 7611 as the type of a sister species within Emarellia, Emarellia paragrisea.
Antifungal susceptibility.
Antifungal susceptibility testing results for the previously described pleosporalean agents of eumycetoma were broadly concordant with previous reports (26–28). With the exception of R. rufulum, all of the species tested (including E. grisea and E. paragrisea) exhibited high MICs with caspofungin (Table 2). All isolates tested appeared susceptible to voriconazole, at least in vitro, with mean MICs of 0.59 mg/liter (B. mackinnonii), 0.25 mg/liter (M. romeroi), 0.18 mg/liter (R. rufulum), and 0.19 mg/liter (E. grisea). The majority of isolates of B. mackinnonii, R. rufulum, and Emarellia grisea/E. paragrisea also appeared susceptible in vitro to amphotericin B and itraconazole, with mean MIC ranges of 0.25 to 0.76 mg/liter and 0.25 to 0.71 mg/liter, respectively (Table 2). In contrast to the other Pleosporales included in the present study, isolates of M. romeroi exhibited significantly higher MICs to amphotericin B and itraconazole (mean MICs of 1.48 and 2 mg/liter, respectively, and MIC ranges 0.125 to 4 mg/liter and 1 to 4 mg/liter, respectively; Table 2), in agreement with results from previous case reports concerning single isolates of this organism (26, 28). Similarly, Ahmed et al. (27) previously reported elevated MICs for itraconazole with 4 of 5 isolates of M. romeroi (range, 0.5 to >16 mg/liter). However, in that study, MICs with amphotericin were consistently lower than those seen with all other pleosporalean eumycetomal agents (mean MIC, 0.13 mg/liter). Since at least one isolate (CBS123975) was common to both the studies by Ahmed et al. (27) and Badali et al. (28), it is likely that these minor discrepancies in MICs observed with amphotericin B and M. romeroi are a result of variations in assay conditions. In agreement with our current clinical opinion regarding the treatment of dark-grain eumycetoma, of the antifungal agents included in the present study, voriconazole appeared to have the broadest spectrum of activity, as judged by lower MICs with all of the eumycetomal agents than the other antifungal agents tested.
TABLE 2.
Antifungal MICs for B. mackinnonii, M. romeroi, R. rufulum, and Emarellia isolates
| Species (no. of isolates) | MIC (mg/liter)a |
Source or reference | |||
|---|---|---|---|---|---|
| AMB | ITC | VRC | CAS | ||
| Biatriospora mackinnoni (8) | 0.77 (0.25–2) | 0.65 (0.25–2) | 0.59 (0.5–1) | 8.72 (2–16) | This study |
| Biatriospora mackinnoni (2) | 0.71 (0.5–1) | 0.5 (0.5) | 0.18 (0.13–0.25) | >16 (>16) | 29 |
| Medicopsis romeroi (9) | 1.47 (0.125–4) | 2 (1–4) | 0.25 (0.125–0.5) | 5.04 (4–8) | This study |
| Medicopsis romeroi (5) | 0.13 (0.13) | 12.1 (0.5–>16) | 0.17 (0.13–0.25) | 16 (4–>16) | 29 |
| Medicopsis romeroi (1) | 4 (4) | 0.5 (0.5) | 4 (4) | 8 (8) | 30 |
| Medicopsis romeroi (1) | 8 (8) | 3 (3) | 0.008 (0.008) | 6 (6) | 28 |
| Rhytidhysteron rufulum (2) | 0.25 (0.125–0.5) | 0.71 (0.5–1) | 0.18 (0.125–0.25) | 0.25 (0.125–0.5) | This study |
| Emarellia grisea (5) | 0.76 (0.25–4) | 0.25 (0.06–0.5) | 0.19 (0.125–0.5) | 9.19 (4–16) | This study |
| Emarellia paragrisea (1) | 1 (1) | 0.125 (0.125) | 0.125 (0.125) | 8 (8) | This study |
Amphotericin B (AMB), itraconazole (ITC), voriconazole (VRC), and caspofungin (CAS) MICs were determined. Data represent geometric mean MICs (MIC ranges are indicated in parentheses). MIC values of >X were taken as the next highest doubling dilution for geometric mean MIC calculations.
TAXONOMY
Emarellia Borman, Desnos-Ollivier, Campbell, Bridge, Dannaoui et Johnson, gen. nov.
MycoBank accession number: MB815958. Type species: Emarellia grisea. Etymology: Named after the clinical syndromes and country of origin; Emarellia = EuMycetoma And RElated Localised Lesions Indian Agent. Colonies gray-brown, felt-like, slow growing. Hyphae moderately thick-walled, brown, smooth, septate, branching with acute angles. Chlamydospores occasionally formed. Conidiophores, conidia and other fruiting bodies are absent. No known teleomorph.
Emarellia grisea Borman, Desnos-Ollivier, Campbell, Bridge, Dannaoui et Johnson, sp. nov.
MycoBank accession number: MB815959. Etymology: grisea is retained as species epithet since these organisms were classically consistent with Madurella grisea. Colonies on Sabouraud dextrose agar slowly expanding, centrally slightly domed, felt-like, grayish brown to brown, attaining approximately 2 cm in diameter after 14 days. Optimum growth temperature is 30°C, growth is observed up to 37°C. Hyphae are moderately thick-walled, smooth, septate, branching and conspicuously brown. Conidiophores and conidia are absent, chlamydospores are occasionally formed. No known teleomorph.
Holotype dried culture NCPF-H 7066; isolated from a case of human black-grain mycetoma in an Indian male by A. Thammayya, School of Tropical Medicine, Kolkata, 1976. Living Type material is preserved in sterile water in metabolically inert form under the unique identifier NCPF 7066, at the NCPF, Bristol, United Kingdom.
Emarellia paragrisea Borman, Desnos-Ollivier, Campbell, Bridge, Dannaoui et Johnson, sp. nov.
MycoBank accession number: MB815960. Etymology: to denote the sibling, paralogous relationship with E. grisea. Colonies on Sabouraud dextrose agar slowly expanding, centrally slightly domed, felt-like, grayish brown, attaining approximately 2 cm in diameter after 14 days. Optimum growth temperature is 30°C, growth is observed up to 37°C. Hyphae are moderately thick-walled, smooth, septate, branching and conspicuously brown. Conidiophores and conidia are absent, chlamydospores are occasionally formed. No known teleomorph.
Holotype dried culture NCPF-H 7611; isolated from a case of human black-grain mycetoma at the Royal Victoria Hospital, Belfast, 2002. Country of origin of patient unknown. Living Type material is preserved in sterile water in metabolically inert form under the unique identifier NCPF 7611, at the NCPF, Bristol, United Kingdom.
DISCUSSION
Historically, numerous nonsporulating fungal species have been implicated as the etiological agents in cases of black-grain mycetoma. However, formal identification of the responsible organisms and full taxonomic characterizations were hindered by inability to induce conidiation. The advent of molecular phylogenetic approaches, including ITS barcoding and multilocus sequence approaches have allowed these issues to begin to be addressed (9–14, 29, 30). Within the Sordariales, Madurella mycetomatis, M. fahalii, M. pseudomycetomatis, and M. tropicana have been formally described and studied in detail (8–10). Many of the other species included in the historical literature as Madurella are now considered to be doubtful, with the exception of Madurella grisea (7), which was still considered to be a valid species (31). M. grisea was distinguished by its gray-brown colonies, an endemicity principally restricted to South America and India, and the absence of conidiation or soluble pigment production in culture (5, 7). However, phylogenetic analyses have formally shown that many of the known agents of black-grain eumycetoma are phylogenetically very distant from the genera in which they were originally placed, with many isolates considered to represent M. grisea actually being members of the Pleosporales (8). The novel genera Medicopsis and Nigrograna were introduced to accommodate the former species Pyrenochaeta romeroi and Pyrenochaeta mackinnonii, respectively (11). A further recent revision of the pleosporalean agents of black-grain eumycetoma (12) made considerable further progress in clarifying the phylogeny and taxonomic affiliations of these organisms: Nigrograna mackinnonii was shown to be taxonomically very close to Biatriospora marina and was thus reclassified as B. mackinnonii; Leptosphaeria senegalensis and L. tompkinsii and several historical isolates of M. grisea were shown to belong to the Trematosphaeriaceae and were accordingly reclassified as Falciformispora senegalensis, F. tompkinsii, and Trematosphaeria grisea, respectively, the latter due to its close phylogenetic proximity to T. pertusa (12).
Despite this considerable progress, the present study has shown that the list of pleosporalean fungi capable of causing black-grain mycetoma, and a full understanding of the epidemiology of these organisms, is not complete. The present study subjected 28 fungi that had been isolated from black-grain eumycetoma and had failed to produce any structures by which they could be conventionally identified to comprehensive molecular analyses (isolates denoted by asterisks; Table 1). Although the existing literature suggests that M. romeroi, B. mackinnonii, and R. rufulum are relatively rare causes of eumycetoma (8, 27), phylogenetic analyses identified 10 of 28 strains as M. romeroi, 8 of 28 strains as B. mackinnonii, and 4 of 28 strains as R. rufulum (Table 1). Importantly, 6 of 28 isolates from clinically proven cases of black-grain eumycetoma from two international culture collections could not be accommodated in any of the current species or genera within the Trematosphaeriaceae. On the basis of phylogenetic analyses of 4 different loci, and of the combined data set of over 2,200 alignment characters, all six isolates formed a well-defined outgroup within the Trematosphaeriaceae (Fig. 2). The six isolates all exhibited sufficient nucleotide sequence divergence from other known genera to merit the erection of a new pleosporalean genus, Emarellia to encompass them. Sequence variation among five of the six isolates was extremely low, and these isolates are described as Emarellia grisea with NCPF 7066 as the type of the genus. The sixth isolate (NCPF 7611) formed a sister taxon to Emarellia grisea in analyses both of the individual loci and the combined data set (Fig. 2 and see Fig. S1 in the supplemental material). Sequence divergence of NCPF 7611 from isolates of E. grisea was significant (Fig. 2) and consistent with variation seen in other sister species in related genera within the Trematosphaeriaceae (see Table S1 in the supplemental material). On the basis of this variation, we propose that NCPF 7611 represents a sibling species within Emarellia, E. paragrisea.
Given the historical practice of grouping nonsporulating fungal isolates from cases of black-grain mycetoma under the generic umbrella “Madurella grisea,” it is perhaps not surprising that those organisms encompass both known pleosporalean agents of eumycetoma that had failed to sporulate in vitro and also a variety of novel mycetomal agents. For organisms that can be induced to produce characteristic features in culture, the rates of misidentification in our experience are much lower. For instance, all six isolates in the NCPF that had been previously identified phenotypically as M. romeroi were confirmed genetically to be this organism. Similarly, 11 of 13 isolates of M. mycetomatis had been correctly identified, with the remaining 2 isolates identified as the related M. fahalii (10) using molecular approaches (data not shown). Since different culture collections receive culture deposits from different contributors and countries of origin, it is difficult to accurately assess the relative prevalence of the different agents of black-grain mycetoma from culture collection data alone. However, an extended molecular analysis of the 54 viable eumycetomal agent isolates present in the NCPF revealed M. romeroi (15 isolates), B. mackinnonii (13 isolates), M. mycetomatis (11 isolates), E. grisea (5 isolates), F. senegalensis (3 isolates), M. fahalii (2 isolates), E. paragrisea, R. rufulum, and R. percutanea (1 isolate each) and 2 different, currently undescribed Pleosporales (the present study and unpublished data).
The six strains of Emarellia spp. described here exhibited remarkably similar antifungal susceptibility profiles, showing low amphotericin B, itraconazole, and voriconazole MICs, but elevated caspofungin MICs. From the present study, and previously published reports, in vitro echinocandin resistance appears to be a consistent feature of many of the pleosporalean agents of black-grain mycetoma (26–28), which is also shared by the sordarialean Madurella spp. (10, 32). The present data agree with previous reports suggesting that isolates of B. mackinnonii are broadly susceptible to amphotericin B, itraconazole, and voriconazole (27). However, most in vitro data indicate that amphotericin B and itraconazole are less likely to be efficacious in infections with M. romeroi, since MICs with these antifungals are generally above the ranges seen with the other members of the Pleosporales (Table 2). In the absence of a definitive molecular identification, in the light of these data, and based on anecdotal clinical successes in treating infections caused by several of the different species described here, we would recommend the use of voriconazole for the empirical treatment of confirmed cases of black-grain eumycetoma.
Studies on other species with Trematosphaeriaceae have highlighted a propensity for halotolerance and an association with natural marine habitats (12, 33). While the natural habitat(s) of Emarellia spp. remain to be investigated, it is noteworthy that four of the six strains described here were isolated from cases of eumycetoma from patients who had originated in India/Sri Lanka, suggesting that these fungi are perhaps geographically constrained to the Indian subcontinent. Certainly, Emarellia species are not recently “emerging” human pathogens. The six strains described here were isolated from independent infections over 6 decades, further adding weight to the argument that Emarellia spp. should be considered bona fide agents of eumycetoma. Finally, it is also perhaps noteworthy that the 28 previously unidentified isolates examined in the present study, which had been collected over 6 decades from infections originating in three different continents, could all be accommodated in only four genera within Pleosporales (after the creation of Emarellia), suggesting that perhaps only a limited number of fungal species possess the necessary shared features that make them so adept at provoking this devastating clinical entity.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Sarah-Jane Miles, whose earlier work on some of the isolates described here provided the impetus for the present study. We also thank the other members of our laboratories for their support and interest in this work.
During the past 5 years, E.D. has received grants from Gilead, Ferrer, and Bio-Rad, payment for lectures from Gilead, MSD, and Schering, and has been a consultant for Astellas and Innothera; E.M.J. has received research funding and/or been a consultant and/or invited speaker for Astellas, Gilead Sciences, Merck Sharp & Dohme, Pfizer, and Schering-Plough.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00477-16.
REFERENCES
- 1.Mansilla-Lory J, Contreras-Lopez EA. 2009. Mycetoma in prehispanic Mexico: review in the skeletical collection of Tlatilco culture. Rev Med Inst Mex Seguro Soc 47:237–242. [PubMed] [Google Scholar]
- 2.Fahal AH, Sabaa AH. 2010. Mycetoma in children in Sudan. Trans R Soc Trop Med Hyg 104:117–121. doi: 10.1016/j.trstmh.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed AA, van de Sande WWJ, Fahal A, Bakker-Woudenberg I, Verbrugh H, van Belkum A. 2007. Management of mycetoma: major challenge in tropical mycoses with limited international recognition. Curr Opin Infect Dis 20:146–151. doi: 10.1097/QCO.0b013e32803d38fe. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed AO, van Leeuwen W, Fahal A, van de Sande W, Verbrugh H, van Belkum A. 2004. Mycetoma caused by Madurella mycetomatis: a neglected infectious burden. Lancet Infect Dis 4:566–574. doi: 10.1016/S1473-3099(04)01131-4. [DOI] [PubMed] [Google Scholar]
- 5.McGinnis MR. 1996. Mycetoma. Dermatol Clin 41:97–104. [DOI] [PubMed] [Google Scholar]
- 6.Kwon-Chung KJ, Bennett JE. 1992. Medical mycology. Lea & Febiger, Philadelphia, PA. [Google Scholar]
- 7.Mackinnon JE, Ferrada-Urzua LV, Montemayor L. 1949. Madurella grisea n.sp. Mycopathologia 4:384–393. [Google Scholar]
- 8.de Hoog GS, Adelmann D, Ahmed AO, van Belkum A. 2004. Phylogeny and typification of Madurella mycetomatis, with a comparison of other agents of eumycetoma. Mycoses 47:121–130. doi: 10.1111/j.1439-0507.2004.00964.x. [DOI] [PubMed] [Google Scholar]
- 9.Yan J, Deng J, Zhou CJ, Zhong BY, Hao F. 2010. Phenotypic and molecular characterization of Madurella pseudomycetomatis sp. nov., a novel opportunistic fungus possibly causing black-grain mycetoma. J Clin Microbiol 48:251–257. doi: 10.1128/JCM.00018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Hoog GS, van Diepeningen AD, Mahgoub el-S, van de Sande WW. 2012. New species of Madurella, causative agents of black-grain mycetoma. J Clin Microbiol 50:988–994. doi: 10.1128/JCM.05477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Gruyter J, Woudenberg JH, Aveskamp MM, Verkley GJ, Groenewald JZ, Crous PW. 2013. Redisposition of phoma-like anamorphs in Pleosporales. Stud Mycol 75:1–36. doi: 10.3114/sim0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed SA, van de Sande WW, Stevens DA, Fahal A, van Diepeningen AD, Menken SB, de Hoog GS. 2014. Revision of agents of black-grain eumycetoma in the order Pleosporales. Persoonia 33:141–154. doi: 10.3767/003158514X684744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahajan VK, Sharma V, Prabha N, Thakur K, Sharma NL, Rudramurthy SM, Chauhan PS, Mehta KS, Abhinav C. 2014. A rare case of subcutaneous phaeohyphomycosis caused by a Rhytidhysteron species: a clinico-therapeutic experience. Int J Dermatol 53:1485–1489. doi: 10.1111/ijd.12529. [DOI] [PubMed] [Google Scholar]
- 14.Mishra K, Das S, Goyal S, Gupta C, Rai G, Ansari MA, Saha R, Singal A. 2014. Subcutaneous mycoses caused by Rhytidhysteron species in an immunocompetent patient. Med Mycol Case Rep 5:32–34. doi: 10.1016/j.mmcr.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed SA, Stevens DA, van de Sande WW, Meis JF, de Hoog GS. 2014. Roussoella percutanea, a novel opportunistic pathogen causing subcutaneous mycoses. Med Mycol 52:689–698. doi: 10.1093/mmy/myu035. [DOI] [PubMed] [Google Scholar]
- 16.Borman AM, Fraser M, Linton CJ, Palmer MD, Johnson EM. 2010. An improved protocol for the preparation of total genomic DNA from isolates of yeast and mould using Whatman FTA filter papers. Mycopathologia 169:445–449. doi: 10.1007/s11046-010-9284-7. [DOI] [PubMed] [Google Scholar]
- 17.Campbell CK, Borman AM, Linton CJ, Bridge PD, Johnson EM. 2006. Arthroderma olidum, sp. nov.: a new addition to the Trichophyton terrestre complex. Med Mycol 44:451–459. [DOI] [PubMed] [Google Scholar]
- 18.Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 19.Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol 16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 20.Pearson WR, Lipman DJ. 1988. Improved tools for biological science comparison. Proc Natl Acad Sci U S A 85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puigbo P, Garcia-Vallve S, McInerney JO. 2007. TOPD/FMTS: a new software to compare phylogenetic trees. Bioinformatics 23:1556–1558. doi: 10.1093/bioinformatics/btm135. [DOI] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, 2nd ed; approved standard CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Schoch CL, Keith A, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan K, Ahmad S, Kapilka K, Ranaswamy NV, Alath P, Joseph L, Chandy R. 2011. Pyrenochaeta romeroi: a causative agent of phaeohyphomycotic cyst. J Med Microbiol 60:842–846. doi: 10.1099/jmm.0.029520-0. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed SA, de Hoog GS, Stevens Da Fahal AH, van de Sande WWJ. 2015. In vitro antifungal susceptibility of coelomycete agents of black grain mycetoma to eight antifungals. Med Mycol 53:295–301. doi: 10.1093/mmy/myu098. [DOI] [PubMed] [Google Scholar]
- 28.Badali H, Chander J, Gulati N, Attri A, Chopra R, Najafzadeh MJ, Chhabra S, Meis JFGM, de Hoog GS. 2010. Subcutaneous phaeohyphomycotic cyst caused by Pyrenochaeta romeroi. Med Mycol 48:763–768. doi: 10.3109/13693780903440383. [DOI] [PubMed] [Google Scholar]
- 29.Desnos-Ollivier M, Bretagne S, Dromer F, Lortholary O, Dannaoui E. 2006. Molecular identification of black-grain mycetoma agents. J Clin Microbiol 44:3517–3523. doi: 10.1128/JCM.00862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed AO, Desplaces N, Leonard P, Goldstein F, De Hoog S, Verbrugh H, van Belkum A. 2003. Molecular detection and identification of agents of eumycetoma: detailed report of two cases. J Clin Microbiol 41:5813–5816. doi: 10.1128/JCM.41.12.5813-5816.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Hoog GS, Guarro J, Gené J. 2000. Atlas of clinical fungi, 2nd ed Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands. [Google Scholar]
- 32.van de Sande WW, Fahal AH, Bakker-Woudenberg IA, van Belkum A. 2010. Madurella mycetomatis is not susceptible to the echinocandin class of antifungal agents. Antimicrob Agents Chemother 54:2738–2740. doi: 10.1128/AAC.01546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Crous PW, Schoch CL, Hyde KD. 2012. Pleosporales. Fungal Divers 53:1–221. doi: 10.1007/s13225-011-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gulati V, Bakare S, Tibrewal S, Ismail N, Sayani J, Aali A, Kubba F, Lynn W, Borman A, Baghla DP. 2012. A rare presentation of concurrent Scedosporium apiospermum and Madurella grisea eumycetoma in an immunocompetent host. Case Rep Pathol 2012:154201. doi: 10.1155/2012/154201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.