Abstract
Dengue virus (DENV) and chikungunya virus (CHIKV) are important human pathogens with common transmission vectors and similar clinical presentations. Patient care may be impacted by the misdiagnosis of DENV and CHIKV in areas where both viruses cocirculate. In this study, we have developed and validated a one-step multiplex reverse transcriptase PCR (RT-PCR) to simultaneously detect, quantify, and differentiate between four DENV serotypes (pan-DENV) and chikungunya virus. The assay uses TaqMan technology, employing two forward primers, three reverse primers, and four fluorophore-labeled probes in a single-reaction format. Coextracted and coamplified RNA was used as an internal control (IC), and in vitro-transcribed DENV and CHIKV RNAs were used to generate standard curves for absolute quantification. The diagnostic 95% limits of detection (LOD) within the linear range were 50 and 60 RNA copies/reaction for DENV (serotypes 1 to 4) and CHIKV, respectively. Our assay was able to detect 53 different strains of DENV, representing four serotypes, and six strains of CHIKV. No cross-reactivity was observed with related flaviviruses and alphaviruses, To evaluate diagnostic sensitivity and specificity, 89 clinical samples positive or negative for DENV (serotypes 1 to 4) and CHIKV by the standard virus isolation method were tested in our assay. The multiplex RT-PCR assay showed 95% sensitivity and 100% specificity for DENV and 100% sensitivity and specificity for CHIKV. With an assay turnaround time of less than 2 h, including extraction of RNA, the multiplex quantitative RT-PCR assay provides rapid diagnosis for the differential detection of two clinically indistinguishable diseases, whose geographical occurrence is increasingly overlapping.
INTRODUCTION
Dengue is a mosquito-borne viral infection in humans that is of global importance. An estimated 390 million infections occur each year, of which 96 million result in clinically apparent infections, and in some epidemics the mortality rate may reach 5% (1, 2). Dengue viruses (DENVs) are members of the family Flaviviridae and consist of four antigenically distinct serotypes which exhibit 65% to 70% sequence homology (3). Disease manifestations range from mild undifferentiated acute dengue fever (DF) to severe dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (4, 5). After an incubation period of 2 to 10 days, a primary infection presents with the acute onset of high fever (≥40°C), accompanied by headache, retroorbital pain, generalized myalgias, arthralgias, and malaise. The early phase of DHF or DSS has similar clinical features; however, after defervescence, new abdominal pain, nausea, and vomiting, followed by thrombocytopenia and a rise in hematocrit, may progress to shock leading to multiorgan dysfunction, life-threatening hemorrhage, and death. DF symptoms may be clinically indistinguishable from acute febrile illness due to other infectious diseases such as influenza, malaria, measles, and chikungunya virus (CHIKV) infections. CHIKVs are alphaviruses belonging to the family Togaviridae. CHIKV causes an acute illness including fever, rash, severe incapacitating arthralgia, headache, fatigue, vomiting, and conjunctivitis (6). Although many patients infected with CHIKV recover within a few weeks of onset, neurologic disease and fatalities have been reported (7, 8). CHIKV was first isolated during the 1952-1953 epidemic of arthralgic disease in Tanzania and has been reported to have affected millions of people in Africa and Southeast and Central Asia (9–14). Since December 2013, CHIKV has been reported throughout the Americas, and local transmission has been observed on almost every island in the Caribbean, with more than 1,400,000 cases as of May 2015 (15–18). It is difficult to predict how long the outbreak in the Americas will last. Both DENV and CHIKV are transmitted to humans by Aedes aegypti and Aedes albopictus mosquitoes, which cocirculate in many of the tropical and subtropical areas of the world. As with dengue, chikungunya could become an endemic disease in this region. Since dengue is endemic throughout the Americas where there are ongoing chikungunya outbreaks and their illnesses are clinically similar, both DENV and CHIKV should be included in the differential diagnosis of people with acute febrile illness.
Accurate and rapid differential diagnosis of DENV and CHIKV infections is crucial for proper clinical management of patients presenting with an acute febrile illness. To distinguish dengue fever from chikungunya fever is critical in patients with DHF, which has the potential for a life-threatening outcome. The quantification of virus in clinical samples can be used as a marker of the degree of viral replication in patients at high risk of severe disease, as well as for monitoring the response to treatment. CHIKV has demonstrated the capacity to emerge and spread quickly, and infected people develop high viremia, which makes heightened surveillance a priority.
There are three main types of laboratory tests currently used for the diagnosis of DENV and CHIKV: virus propagation and isolation in susceptible cell lines, reverse transcriptase PCR (RT-PCR), and serology (immunoglobulin M [IgM] and IgG enzyme-linked immunosorbent assays [ELISAs]) (19). Serological methods are not appropriate in the first 5 to 6 days after the clinical onset. In early-stage infection, while the patient is viremic, nucleic acid amplification by PCR is the most rapid diagnostic tool. The use of molecular methods has increased, and several PCR-based assays have been developed for the diagnosis of DENV and CHIKV with human samples (20–25). The objective of this study was to develop a multiplex assay for the rapid, differential, quantitative diagnosis of DENV and CHIKV in a single-reaction format. This assay targets the 3′ untranslated region (UTRs) of CHIKV and DENV serotypes 1 through 4. The assay uses TaqMan technology employing two forward primers, three reverse primers, and four fluorophore-labeled probes in a single-reaction format. A real-time (closed) system was chosen because of increased speed, increased sensitivity, and lower risk of contamination (26). We used an exogenous internal control (IC) to increase the reliability of the RT-PCR results. In vitro-transcribed RNA was used for the standard curve for both DENV and CHIKV. The assay has been validated on material from spiked human sera and positive and negative field samples, and the results showed that the method is highly sensitive and specific for the early differential diagnosis of suspected DENV and CHIKV cases.
MATERIALS AND METHODS
DENV sequences and assay design.
The forward primer for the pan-DENV TaqMan assay was chosen from the last 200 nucleotides of the 3′ noncoding sequence of dengue virus (DENV) types 1 to 4 (DENV-1, GenBank accession number M87512.1; DENV-2, M20558.1; DENV-3, M93130.1; DENV-4, M14931) using Primer Express software (Applied Biosystems, Foster City, CA). Fluorogenic probes specific for DENV-2, DENV-3, and DENV-4 as well as two reverse primers detecting all four DENV serotypes have been described previously (27). A DENV-1-specific probe was designed and added to the multiplex assay. The CHIKV primers and probe were also designed based on its 3′-end genomic sequence (GenBank accession number AF369024.2, S27, African prototype) using Geneious software (Biomatters, Auckland, New Zealand). The DENV fluorogenic nucleotide probes were labeled with 5′ 6-carboxyfluorescein (FAM) and 3′ Black Hole Quencher (BHQ) dyes. The CHIKV nucleotide probe was labeled with 5′ cyanine 5 (Cy5) and 3′ BHQ (Integrated DNA Technologies, Coralville, IA). Sequences and genome positions for all oligonucleotide primers and fluorogenic probes are shown in Table 1.
TABLE 1.
Oligonucleotide primers and fluorogenic probes used in the quantitative multiplex RT-PCR assay
| Virus | Primer or probe | Sequence (5′→3′) | Genome position(s) |
|---|---|---|---|
| DENV-1,2,3,4 | F1 | GACTAGAGGTTAGAGGAGACCCCC | DENV-1, 10574–10597; DENV-2, 10581–10604; DENV-3, 10555–10578; DENV-4, 10508–10531 |
| DENV-1,2,3 | R1 | CATTCCATTTTCTGGCGTTCT | DENV-1, 10674–10694; DENV-2, 10680–10700; DENV-3, 10653–10673 |
| DENV-4 | R2 | CAATCCATCTTGCGGCGCTCT | 10605–10625 |
| DENV-1 | P1 | CTGTCTCTACAGCATCATTCCAGGCA | 10647–10672 |
| DENV-2,3 | P2 | CTGTCTCCTCAGCATCATTCCAGGCA | DENV-2, 10653–10678; DENV-3, 10626–10651 |
| DENV-4 | P3 | CTGTCTCTGCAACATCAATCCAGGCA | 10578–10603 |
| CHIKV | F1 | GTCACATACCACCCTCG | 11169–11185 |
| CHIKV | R1 | TGYCTCTTAGGGGACACATATACCT | 11332–11356 |
| CHIKV | P1 | GGGACTGGTTGTYGCTGTTGCCGC | 11244–11267 |
Viruses.
Cell culture supernatant harvested from Vero 81 (WHO) cells infected with DENV-1 (West Pac 74), DENV-2 (S16803), DENV-3 (CH53489), DENV-4 (341750), and CHIKV (181/25) (see Table 2) was used as virus stock for spiking negative human sera. The concentration of viral RNA or in vitro-transcribed RNA was measured as genome equivalents (GE)/gene copies. Genomic RNA preparations from 53 dengue viruses (including 10 strains of DENV-1, 24 strains of DENV-2, 10 strains of DENV-3, and 9 strains of DENV-4), 6 strains of CHIKV, Japanese encephalitis (JE) virus (SA 14-14-2, attenuated vaccine strain), yellow fever (YF) virus (17D, attenuated vaccine strain), Bebaru virus (BEBV) (ATCC VR-600), Ross River virus (RRV) (ATCC VR-373), and Sindbis virus (SBV) (ATCC VR-1248) (see Tables 2, 4, and 5) were obtained from BEI Resources-ATCC (Manassas, VA). DENV laboratory strains and JE and YF virus vaccine strains were received from the Walter Reed Army Institute of Research (WRAIR) (Silver Spring, MD), CHIKV R-91142, CHIKV S-27, CHIKV SL-15649, and all other DENV strains were received from BEI Resources-ATCC, the CHIKV proficiency panel was obtained from CDC (Fort Collins, CO), and CHIKV 181-25 (attenuated vaccine strain) was received from Shuenn-Jue Wu (NMRC, MD).
TABLE 2.
Detection and precision analysis of the DENV/CHIKV multiplex RT-PCR
| Virusa | GE/assay | No. of replicatesb |
CT value |
||
|---|---|---|---|---|---|
| Mean | SD | % CV | |||
| DENV-3 | 5 × 106 | 24 | 17.84 | 0.26 | 1.5 |
| 5 × 105 | 24 | 21.08 | 0.34 | 1.6 | |
| 5 × 104 | 24 | 24.43 | 0.35 | 1.4 | |
| 5 × 103 | 24 | 27.70 | 0.39 | 1.4 | |
| 5 × 102 | 24 | 31.17 | 0.50 | 1.6 | |
| 5 × 101 | 24 | 34.55 | 0.72 | 2.1 | |
| CHIKV | 6 × 106 | 24 | 18.08 | 0.28 | 1.5 |
| 6 × 105 | 24 | 21.30 | 0.31 | 1.5 | |
| 6 × 104 | 24 | 24.69 | 0.38 | 1.5 | |
| 6 × 103 | 24 | 28.14 | 0.38 | 1.3 | |
| 6 × 102 | 24 | 31.36 | 0.80 | 2.5 | |
| 6 × 101 | 24 | 34.59 | 1.10 | 3.2 | |
The multiplex RT-PCR was carried out on RNAs extracted from DENV and CHIKV in vitro transcripts.
Quadruplicate runs over 3 days were carried out by two different operators.
TABLE 4.
Viruses used to determine the multiplex RT-PCR specificity/inclusivity
| Virus | Strain | Resulta |
|---|---|---|
| DENV-1 | 228690 | + |
| TH-Sman | + | |
| Phi 003-89 | + | |
| 276RK1 | + | |
| BC89/94 | + | |
| 12150 | + | |
| 16607 | + | |
| West Pac 74b | + | |
| IQT 6152 | + | |
| OBS6152 | + | |
| DENV-2 | BC27/96 | + |
| S-14635 | + | |
| BC141/96 | + | |
| P8-1407MS | + | |
| 10674 | + | |
| S2 | + | |
| IQT2913 | + | |
| ArA6894 | + | |
| 429557 | + | |
| 328298 | + | |
| PM33974 | + | |
| DAKArA1247 | + | |
| BC171/96 | + | |
| 1349 | + | |
| K0049 | + | |
| BC100/98 | + | |
| BC102/94 | + | |
| IQT2124 | + | |
| NGC | + | |
| OBS8041 | + | |
| Phi 012-84 | + | |
| D80-100 | + | |
| S-16803b | + | |
| 16681 | + | |
| DENV-3 | BC14/97 | + |
| S-40580 | + | |
| 271242 | + | |
| BC188/97 | + | |
| S40921 | + | |
| MK594-87 | + | |
| 1187 | + | |
| VD8PR2517 | + | |
| Phi 075-84 | + | |
| CH53489b | + | |
| DENV-4 | D85-019 | + |
| BC13/97 | + | |
| BC258/97 | + | |
| BC123/97 | + | |
| BC287/97 | + | |
| Phi 011-90 | + | |
| H241 | + | |
| TVP360 | + | |
| 341750b | + | |
| JE virus | SA-14-14-2 | − |
| YF virus | 17D | − |
| CHIKV | 181/25b | + |
| India 2006 (CDC) | + | |
| R-91142 | + | |
| S-27 | + | |
| SL-15649 | + | |
| Grenada 2014 | + | |
| BEBV | MM2354 | − |
| RRV | T48 | − |
| SBV | Ar339 | − |
Positive or negative results are denoted + or −, respectively. Concentrations for DENV, CHIKV, JE virus, and YF virus were between 103 and 107 GE/reaction. Concentration for BEBV, RRV, and SBV were 100 ng per reaction.
Strain used to make tissue culture stocks.
TABLE 5.
Diagnostic sensitivity and specificity of the multiplex RT-PCR assay compared to virus isolation for detection of viruses in clinical serum samples
| Sample | No. of samples |
||
|---|---|---|---|
| Total | Positive by: |
||
| RT-PCRa | Virus isolation | ||
| DENV-1 positive | 4 | 4 | 4 |
| DENV-2 positive | 5 | 5 | 5 |
| DENV-3 positive | 5 | 4 | 5 |
| DENV-4 positive | 5 | 5 | 5 |
| DENV negative | 20 | 0 | 0 |
| CHIKV positive | 40 | 40 | 40 |
| CHIKV negative | 10 | 0 | 0 |
In comparison with the results of the reference method (virus isolation), the multiplex RT-PCR assay had 95% sensitivity and 100% specificity for DENV and 100% agreement for CHIKV.
RNA standards.
The target regions of DENV-3 (CH53489) and CHIKV (181/25) were amplified by RT-PCR (SuperScript III Platinum one-step qRT-PCR kit with ROX) (Invitrogen, Grand Island, NY) from supernatant of infected cells, generating a 119-bp fragment and a 188-bp fragment, respectively. The forward (DENV F and CHIKV F) and reverse (DENV R1 and CHIKV R1) primers are listed in Table 1. Amplicons were detected by 2% agarose gel electrophoresis, and 2 μl of the PCR product was used to clone amplicons into TOPO TA cloning kit with PCR 2.1 TOPO (Invitrogen). The presence of the cloned inserts was confirmed by restriction digestion (SpeI and NotI) followed by gel electrophoresis. Restriction digest plasmids were purified using the QIAprep spin miniprep kit (Qiagen Inc., Valencia, CA) and were CsCl purified for quantification by spectrophotometry. The copy number or genome equivalents (GE) of the plasmids were calculated based on their concentration and molecular weight. Purified linearized plasmids were used as templates for in vitro transcription (IVT) with the MEGAscript kit (Ambion, Thermo Fisher, Waltham, MA) according to the manufacturer's instructions. The IVT products were then treated with Turbo DNase for 15 min at 37°C, followed by inactivation of DNase at 70°C for 15 min. Tenfold dilution series of plasmids and IVT were prepared in duplicate to generate standard curves. The GE of the IVT preparations were calibrated using the calculated plasmid concentrations. For an exogenous internal control (IC) to be used as extraction and amplification control, DENV-1 Armored RNA was purchased from Asuragen Inc. (Austin, TX).
One-step multiplex real-time RT-PCR assay.
A real-time, one-step, multiplex RT-PCR assay was developed for the detection and quantitation of DENV and CHIKV RNAs in cell culture supernatants or human serum. Real-time PCR was performed under College of American Pathologists (CAP) restricted conditions in a CAP-accredited laboratory. Care was taken to perform RNA extraction, master mix preparation, template addition, and thermal cycling in different areas to avoid contamination. A total volume of 25 μl of reaction mixture consisted of 5 μl of extracted RNA, 12.5 μl of 2× RT buffer (Superscript III Platinum qRT-PCR kit with ROX), 0.5 μl of SuperScript III RT/Platinum Taq mix, 0.2 μl of RNase inhibitor (40 units/μl) (RNaseout; Invitrogen), 170 nM DENV.F1, 110 nM DENV.R1 and DENV.R2, 60 nM DENV1.P1, DENV.P2, and DENV.P3, and 150 nM CHIKV.F, CHIKV.R, and CHIKV.P. For the DENV probes, the 5′ and 3′ ends were labeled with 6-carboxyfluorescein (FAM) and Black Hole Quencher (BHQ), respectively, whereas cyanine 5 (Cy5) and BHQ were used for the CHIKV probe. The ABI 7500 DX gene detection system (Applied Biosystems, Carlsbad, CA) was used for PCR cycling, real-time data collection, and analysis. The amplification conditions were 50°C for 30 min, 95°C for 2 min, and then 40 cycles of 95°C for 15 s and 60°C for 40 s. The total run time for this protocol was 1 h 10 min. All reactions were carried out in MicroAmp Fast 96-well reaction plates (ABI). For clinical sample testing, all samples and controls were run in duplicate, and each plate contained a six-dilution standard curve of both DENV and CHIKV IVT product, an H2O nontemplate control (NTC), an H2O extraction control, an H2O extraction control containing an exogenous positive internal control (IC) (DENV-1 Armored RNA), extracted clinical sample, and extracted clinical sample containing the IC. The acceptance criteria for a successful run were a slope between −3.1 and −3.6 for both standard curves and correlation coefficient (r) values greater than 0.98. Samples were considered negative if the IC was positive but the patient sample without IC was negative.
Analysis of linearity, diagnostic LOD, and precision.
In vitro-transcribed RNA was used to establish standard curves for the quantitation of viral copy numbers and analysis of analytical sensitivity. Linearity studies were performed on serial 10-fold dilutions of both DENV and CHIKV IVT RNAs. Dilutions of IVT RNA ranged from 5 × 107 GE to 5 GE in triplicate. The IVT stock solutions for standard curves were diluted in nuclease-free water, prepared as 10-μl one-time-use aliquots, and stored at −80°C for future use. The linear range was established with 10-fold dilutions of IVT RNA and by fitting a best-fit line to the data by regression analysis, where the r2 value for this line was >0.98. Acceptance criteria for a successful run were established as a slope for the standard curve between −3.1 and −3.6 with an efficiency of >90%.
To establish the 95% limit of detection (LOD), the lowest concentrations of RNA from the linear-range study were used as the starting points for another five 2-fold dilutions. The LOD was defined as the lowest concentration of viral RNA that can be detected in ≥95% of 24 replicates with the amplification curves above the threshold of 0.04. The precision of the multiplex RT-PCR assay was evaluated using six 10-fold dilutions of IVT RNAs to generate standard curves, with one run per day in quadruplicate over 3 days by two different operators (see Table 2). Fresh dilutions were made for each run from one-time-use aliquots.
Specificity and sensitivity.
The specificity and sensitivity of the multiplex RT-PCR were determined using extracted RNA preparations from 53 dengue viruses, including 10 strains of DENV-1, 24 strains of DENV-2, 10 strains of DENV-3, and 9 strains of DENV-4, and 6 strains of CHIKV. To determine cross-reactivity of the dengue virus primers and probes, two related flaviviruses, JE virus and YF virus, at concentrations of 2 × 104 GE/reaction were evaluated in the assay. To determine the specificity of the CHIKV primers and probe, we included BEBV, RRV, and SBV at 100 ng/reaction (see Table 4). For the initial determination of sensitivity and cross-reactivity between DENV serotypes in the multiplex assay, serum from a healthy individual was spiked with 50, 500, or 5,000 GE of DENV-1 to -4 (see Table 3). A total of 12 spiked samples were run in duplicate in two separate runs.
TABLE 3.
Detection limit of the multiplex RT-PCR assay for the four DENV serotypes in spiked human serum
| Virus | Expected GE | Assay GEa |
||||||
|---|---|---|---|---|---|---|---|---|
| Run 1 |
Run 2 |
Mean | SD | % CV | ||||
| 1 | 2 | 1 | 2 | |||||
| DENV-1 | 5,000 | 4,700 | 4,000 | 2,800 | 2,700 | 3,550 | 968 | 27 |
| 500 | 525 | 430 | 237 | 325 | 379 | 125 | 33 | |
| 50 | 58 | 38 | 60 | 42 | 50 | 11 | 22 | |
| DENV-2 | 5,000 | 7,800 | 7,300 | 5,000 | 4,000 | 6,025 | 1,819 | 30 |
| 500 | 659 | 635 | 414 | 387 | 524 | 143 | 27 | |
| 50 | 75 | 43 | 33 | 57 | 52 | 18 | 35 | |
| DENV-3 | 5,000 | 2,800 | 2,700 | 1,600 | 2,000 | 2,275 | 574 | 25 |
| 500 | 245 | 381 | 125 | 46 | 199 | 146 | 73 | |
| 50 | 10 | 18 | 13 | 13 | 14 | 3 | 25 | |
| DENV-4 | 5,000 | 5,000 | 4,500 | 5,300 | 3,000 | 4,450 | 1,021 | 23 |
| 500 | 477 | 377 | 364 | 352 | 393 | 57 | 15 | |
| 50 | 58 | 37 | 41 | 40 | 44 | 9 | 22 | |
For each virus, a dilution series including the lowest LOD in two separate runs is shown.
Clinical samples.
A total of 89 human serum samples were used to evaluate the multiplex RT-PCR for its diagnostic potential. Fifty clinical samples provided by St. George's University (St. George, Grenada) were leftover specimen from serum samples collected in 2014 for clinically required testing for suspected dengue. Deidentified residual samples were shipped to the Naval Infectious Diseases Diagnostic Laboratory (NIDDL) (Silver Spring, MD) for use in the validation of our assay. The use of these deidentified coded samples for research purposes was covered under an Institutional Review Board (IRB) protocol (PJT-12-13, Collection of Human Samples for Use as Controls in Diagnostic Assays) allowing use of leftover samples collected for diagnostic purposes from febrile persons suspected of a possible arbovirus infection. Nineteen DENV isolation-confirmed and 20 location- and date-matched DENV-negative clinical samples from Honduras, Venezuela, and Peru were collected between 2010 and 2014 under study protocol NMRCD.2010.0010. This protocol was approved by the NAMRU-6 IRB in compliance with all U.S. federal regulations governing the protection of human subjects. The chikungunya virus RT-PCR proficiency panel was obtained from the CDC at Fort Collins, CO, and consisted of three samples representing chikungunya virus collected in India in 2006.
Isolation of viral RNA.
Viral RNA was extracted from 200 μl of serum samples or supernatant of infected cells using the EZ1 DSP virus kit (Qiagen) according to the protocol suggested by the manufacturer, using the EZ1 Advanced XL (Qiagen) automated extraction system. The RNA was eluted with 60 μl of elution buffer, aliquoted, and immediately stored at −80°C until further use.
Virus growth and isolation.
Vero cells were grown in Eagle's minimum essential medium (EMEM) (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS) (Sigma), 1% penicillin-streptomycin (Mediatech Inc., Manassas, VA), and 1% glutamine (Sigma, St. Louis, MO) at 37°C and 5% CO2. Isolation of viruses from acute-phase patient samples was carried out by adsorption of 0.3 ml of sample in 0.3 ml of EMEM–2% FBS to cells in a 12-cm2 flask for 1 h at 37°C. After adsorption, the cells were replenished with 7 ml of EMEM–2% FBS and monitored for cytopathic effect. Cells were harvested on appearance of cytopathic effect (compared to uninfected cells) or were kept for 14 days, with a medium change on day 7. The identification of the virus isolates obtained from the clinical samples was carried out by immunofluorescence using virus-specific antibodies.
Statistics.
Basic statistical analysis, including determination of means and standard deviations, was performed using Excel software (Microsoft, Bellevue, WA).
RESULTS
Primer design.
This assay targets the distinctive 3′ untranslated regions (UTRs) of CHIKV and four serotypes of DENV. The DENV/CHIK TaqMan assay consists of two forward primers, three reverse primers, and four fluorophore-labeled probes in a single-reaction format. The resultant real-time RT-PCR system can be used to differentially detect CHIKV as well as dengue virus serotypes 1 through 4. The oligonucleotide sequences for the reverse primers targeting DENV-1 through -4 and fluorescence-labeled probes specific to DENV-2, -3, and -4 have been previously described (27, 28). A universal forward primer was designed from a homologous region, and an additional DENV-1 probe was included in the multiplex assay to ensure the detection of all four DENV serotypes. The oligonucleotide sequences and genome positions of all primers and probes for the multiplex assay are listed in Table 1.
Assay performance and analytical sensitivity.
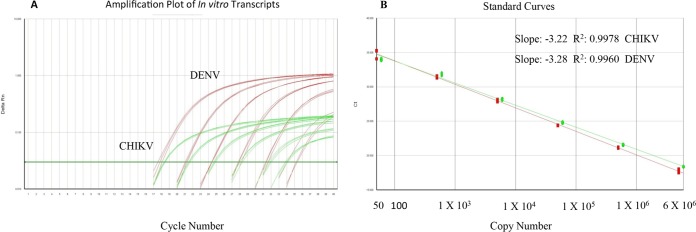
The multiplex assay was optimized as a single-well method incorporating a DENV group-specific forward primer, two DENV reverse primers which also serve at RT primers, a CHIKV primer pair, and four TaqMan probes for the detection of DENV serotypes 1 through 4 and CHIKV. The detection limits for both the DENV complex and CHIKV were determined through 10-fold serial dilutions of in vitro-transcribed RNA copies. The samples at each concentration were tested in quadruplicate in one run per day over 3 days by two different operators for a total of 24 runs per dilution (Table 2). The linear dynamic range for the multiplex RT-PCR assay was between 50 and 5 × 106 GE per reaction for DENV and between 60 and 6 × 106 GE for CHIKV (Fig. 1). The total imprecision (percent coefficient of variation [CV]) for the threshold cycle (CT) values for the six concentrations ranged from 1.4% to 2.1% for DENV and from 1.3% to 3.2% for CHIKV. The accepted amplification efficiency for each run was set at a slope of −3.1 and −3.6 with the correlation coefficient greater than 0.98. The 95% limit of detection (LOD) and limit of quantitation (LOQ) in which the signal was linear for the DENV complex (serotypes 1 to 4) and CHIKV RT-PCRs were 50 (3 × 103 GE/ml) and 60 (3.6 × 103 GE/ml) gene copies per reaction, respectively (Table 2). To ensure that the assay is not negatively impacted by components in human serum and to confirm the sensitivity and LOD for all DENV serotypes, dengue virus-negative human serum was spiked with three different dilutions of previously titrated virus stocks. Each dilution was extracted separately and assayed in two separate runs (Table 3). The results indicate equal sensitivity to all four DENV serotypes, and the assay is not impacted by components in human serum. The cutoff for positivity was defined by a CT value of ≤37, based on 50 independent runs where 100% of all replicates were below this value (data not shown).
FIG 1.
(A) Amplification plot for DENV/CHIKV multiplex assay (10-fold dilutions, triplicate), (B) The linear dynamic range of the multiplex RT-PCR is shown as gene copy number for DENV (5 × 101 to 5 × 106) and CHIKV (6 × 101 to 6 × 106).
Analytical specificity.
Analytical specificity distinguishes between the target analyte and other components in the sample and was expressed as inclusivity and exclusivity (29). Extracted RNAs from 53 dengue viruses, representing all four serotypes of DENV and six strains of CHIKV, were tested in duplicate to determine inclusivity of the multiplex RT-PCR. Two related flaviviruses (JE virus and YF virus) and three related alphaviruses (BEBV, RRV, and SBV) were tested to confirm exclusivity of the assay. The multiplex assay detected all DENV and CHIKV samples tested (Table 4). The five related flaviviruses and alphaviruses were not detected.
Diagnostic sensitivity and specificity.
To establish estimates of diagnostic sensitivity and specificity, clinical samples which had been defined positive or negative by virus isolation in Vero cells were used. Nucleic acid extracted from a total of 89 clinical samples positive or negative for DENV-1 to -4 and CHIKV were used for the calculation of diagnostic sensitivity and specificity (Table 5). Of the 19 samples positive for DENV by virus isolation, 18 samples (95%) tested positive by multiplex RT-PCR, and all 20 negative-control samples were negative in both assays. The one sample negative in the PCR but positive by virus isolation showed a CT value of 38.7 after a repeated run, which is below the validated LOD CT of 37 and is therefore recorded as negative. For the 50 positive and negative CHIKV clinical samples, 40 (100%) samples were positive by virus isolation and positive by RT-PCR. Ten (100%) negative-control sera were negative in both assays.
DISCUSSION
There are several studies describing the development of real-time PCR assays for the detection of DENV and CHIKV (20–25, 27, 28). Two previous studies reported on the development of an RT-PCR assay for the detection of dengue viruses using primers and probes designed in the 3′ UTR region (30, 31). These are assays either not quantitative or use a two-step method. Other real-time dengue virus RT-PCR assays use either a three- or four-step cycling method, which increases assay time, or use a SYBR green-based method, which is prone to nonspecific reaction products leading to increased background and false positives (20, 32–34). In 2010, the international external quality control assessment for the molecular diagnosis of dengue infections published their findings for 37 laboratories performing 46 tests and showed that 80% of these tests lacked sensitivity, specificity, or both (35). Similarly, several groups have developed singleplex real-time RT-PCR assays for the detection of CHIKV in clinical samples (24, 25, 36–39). Some of these assays are not quantitative or use plasmid DNA as a standard for quantitation (36, 37). Another study recently described the development of a multiplex real-time RT-PCR for the simultaneous detection of DENV and CHIKV with a detection limit of 1 to 50 PFU and an assay cutoff value CT of 32, due to nonspecific signal observed in the nontemplate control (40).
In this study, we describe the development and validation of a more sensitive, rapid, single-reaction, one-step, multiplex real-time RT-PCR for the detection, quantitation, and differentiation of DENV-1 to -4 and CHIK in patient samples. This assay targets the 3′ untranslated regions (UTRs) of all four DENV serotypes for species-specific detection but cannot be used for serotype determination. Serotype-specific determination is usually not required and does not affect clinical care and management, especially outside areas where dengue is endemic. In areas where DENV and CHIKV cocirculate, it is important to be able to differentiate between them due to different disease progression and outcome. Real-time PCR has brought true quantitation into the diagnostic laboratory. It allows monitoring of the progress of an infection and may be necessary when there is a lack of sequential specimens to indicate changes in virus levels or when viral load is used to differentiate active versus persistent infection. Our assay contains two differently labeled probes, and it uses external standard curves for quantitation that consist of serial dilutions of identically amplified templates within the same experimental run. The linear dynamic range in the assay covers six log10 copies of nucleic acid template, where the lower limit was set at the LOD CT of 37. To control for plate-to-plate variation, we adopted an acceptance criterion where the slope of the standard curves has to be within −3.1 to −3.6 with the correlation coefficient greater than 0.98. Due to the broad dynamic range of the assay, template quantitation from samples containing a large range of concentrations can be accomplished without the need to repeat the assay using a diluted sample. In addition to a nontemplate control and standard curves on every plate serving as negative and positive controls, both external and internal controls have been applied. Both patient sample and H2O controls were extracted with and without a spiked IC and served as false-negative and contamination/amplification controls. Inhibition of the IC amplification in the presence of high DENV or CHIKV RNA levels has been observed, but this was not detrimental since the IC is used to monitor false-negative results. Using this format, up to nine samples, including all controls, can be analyzed in about 2 h on a single plate, including RNA extraction. This assay fulfilled the validation criteria of specificity, linearity, and precision.
The sensitivity of the multiplex assay was evaluated by testing triplicate 10-fold serial dilutions of in vitro-transcribed RNAs for both DENV and CHIKV. The linear dynamic range of the assay was between 50 and 5 × 106 GE per reaction for DENV and between 60 and 6 × 106 GE for CHIKV. Sensitivity of 95% was achieved, with CT values in the range of 33 to 36, when samples included 50 or 60 GE for both targets, and these were therefore defined as the limit of detection (LOD). The limit of quantification (LOQ) corresponds to the last dilution in which the signal is linear and has an acceptable CV of <5%, which in this assay is equal to the LOD.
In this study, we demonstrated that the one-step multiplex RT-PCR assay was able to detect 53 different serotypes of DENV and 6 different strains of CHIKV at concentrations ranging from 103 to107 GE per reaction. The assay was found to have high analytical specificity, and no false-positive results were seen from testing of relatively high concentrations of related flavivirus and alphavirus RNAs. This study assessed the reproducibility of the assay by testing a range of concentrations from in vitro-transcribed RNA. The assay was highly reproducible for detecting virus at concentrations ranging from 50 to 6 × 106 gene copy numbers per reaction. Furthermore, low intra-assay and interassay variabilities were shown in measuring CT values over a wide range of concentrations, with different operators on different days, ranging from 1.4% to 3.2%. We have evaluated the diagnostic application of the multiplex RT-PCR assay using 89 archived serum samples representing all four serotypes of DENV and CHIKV. For the detection of DENV, the multiplex RT-PCR had a 95% diagnostic sensitivity and 100% specificity, whereas for CHIKV detection there was 100% positive agreement between the multiplex RT-PCR assay and the gold standard of virus isolation with both positive and negative samples tested.
In conclusion, the newly developed and validated one-step, multiplex quantitative RT-PCR enables simultaneous, sensitive, and specific detection and quantitation of DENV serotypes 1 to 4 or CHIKV in clinical specimens. The dynamic range of the new assay encompasses the copy numbers expected in diagnostic samples. The analytical sensitivity of the assay is 50 and 60 GE/reaction (or 10 and 12 copies/μl) for DENV-1 to -4 and CHIKV, respectively. The assay includes an exogenous internal control for the detection of false-negative results and two standard curves for quantitation, is technically robust, and has high-throughput potential. The screening assay described in the current study provides the means to rapidly detect and differentiate five distinct viruses in the same sample in less than 2 h, including extraction of RNA. The recent occurrence of both DENV and CHIKV in ever-closer geographical areas highlights the need to rapidly detect and differentiate these two diseases. This assay has been validated as an in-house diagnostic test in the CAP-accredited NIDDL and is used as a routine diagnostic tool for DENV and CHIKV infection.
ACKNOWLEDGMENTS
We thank Susana Widjaja for technical assistance and Kenneth Eckels and Shuenn-Jue Wu for providing virus stock. We thank Juan Perez, Instituto Nacional de Salud del Perú, DIRESA Loreto, Hospital La Merced (Junin), CS Pachitea (Piura), Stalin Vilcarromero, Crystyan Siles, Michel Valerio, and Victor Ocaña for their work in the field. We thank I. Lorenzana (Universidad Autonoma de Honduras) and G. Comach (LARDIDEV, Venezuela) for their help in collecting patient samples.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, or the U.S. Government.
We report no competing interests.
Funding Statement
This work was funded by a grant from the Armed Forces Health Surveillance Branch (AFHSB) Global Emerging Infections Surveillance and Response (GEIS) work unit number P0085-14-NM. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Ranjit S, Kissoon N. 2011. Dengue hemorrhagic fever and shock syndromes. Pediatr Crit Care Med 12:90–100. doi: 10.1097/PCC.0b013e3181e911a7. [DOI] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rico-Hesse R. 1990. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology 174:479–493. doi: 10.1016/0042-6822(90)90102-W. [DOI] [PubMed] [Google Scholar]
- 4.Halstead SB. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 5.Gubler DJ, Clark GG. 1995. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg Infect Dis 1:55–57. doi: 10.3201/eid0102.952004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson M. 1956. An epidemic of a dengue-like fever in the southern province of Tanganyika. Central Afr J Med 2:394. [PubMed] [Google Scholar]
- 7.Gérardin P, Fianu A, Malvy D, Mussard C, Boussaïd K, Rollot O, Michault A, Gaüzere B-A, Bréart G, Favier F. 2011. Perceived morbidity and community burden after a Chikungunya outbreak: the TELECHIK survey, a population-based cohort study. BMC Med 9:5. doi: 10.1186/1741-7015-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavalankar D, Shastri P, Bandyopadhyay T, Parmar J, Ramani KV. 2008. Increased mortality rate associated with chikungunya epidemic, Ahmedabad, India. Emerg Infect Dis 14:412. doi: 10.3201/eid1403.070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karabatsos N. (ed). 1985. International catalogue of arthropod-borne viruses, 3rd ed American Society for Tropical Medicine and Hygiene, San Antonio, TX. [Google Scholar]
- 10.Carey DE. 1971. Chikungunya and dengue: a case of mistaken identity? J Hist Med Allied Sci 26:243–262. [DOI] [PubMed] [Google Scholar]
- 11.Rao T. 1966. Recent epidemics caused by Chikungunya virus in India, 1963-1965. Sci Culture 32:215. [Google Scholar]
- 12.Nimmannitya S, Halstead SB, Cohen SN, Margiotta MR. 1969. Dengue and chikungunya virus infection in man in Thailand, 1962-1964. I. Observations on hospitalized patients with hemorrhagic fever. Am J Trop Med Hyg 18:954–971. [DOI] [PubMed] [Google Scholar]
- 13.Carey DE, Myers RM, DeRanitz C, Jadhav M, Reuben R. 1969. The 1964 chikungunya epidemic at Vellore, South India, including observations on concurrent dengue. Trans R Soc Trop Med Hyg 63:434–445. doi: 10.1016/0035-9203(69)90030-3. [DOI] [PubMed] [Google Scholar]
- 14.Jupp P, McIntosh B. 1988. Chikungunya virus disease, p 137–157. In Monath TP. (ed), The arboviruses: epidemiology and ecology, vol 2 CRC Press, Boca Raton, FL. [Google Scholar]
- 15.Powers AM. 2015. Risks to the Americas associated with the continued expansion of chikungunya virus. J Gen Virol 96:1–5. doi: 10.1099/vir.0.070136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodríguez-Morales AJ, Paniz-Mondolfi AE. 2015. Venezuela: far from the path to dengue and chikungunya control. J Clin Virol 66:60–61. doi: 10.1016/j.jcv.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Fischer M, Staples JE. 2014. Notes from the field: chikungunya virus spreads in the Americas-Caribbean and South America, 2013-2014. MMWR Morb Mortal Wkly Rep 63:500–501. [PMC free article] [PubMed] [Google Scholar]
- 18.Pan American Health Organization. 2015. Chikungunya: number of reported cases of chikungunya fever in the Americas. Pan American Health Organization, Washington, DC. [Google Scholar]
- 19.Blacksell SD, Jarman RG, Gibbons RV, Tanganuchitcharnchai A, Mammen MP, Nisalak A, Kalayanarooj S, Bailey MS, Premaratna R, de Silva HJ. 2012. Comparison of seven commercial antigen and antibody enzyme-linked immunosorbent assays for detection of acute dengue infection. Clin Vaccine Immunol 19:804–810. doi: 10.1128/CVI.05717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waggoner JJ, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, Gonzalez K, Ballesteros G, Balmaseda A, Karunaratne K, Harris E. 2013. Development of an internally controlled real-time reverse transcriptase PCR assay for pan-dengue virus detection and comparison of four molecular dengue virus detection assays. J Clin Microbiol 51:2172–2181. doi: 10.1128/JCM.00548-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waggoner JJ, Abeynayake J, Sahoo MK, Gresh L, Tellez Y, Gonzalez K, Ballesteros G, Guo FP, Balmaseda A, Karunaratne K. 2013. Comparison of the FDA-approved CDC DENV-1-4 real-time reverse transcription-PCR with a laboratory-developed assay for dengue virus detection and serotyping. J Clin Microbiol 51:3418–3420. doi: 10.1128/JCM.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito M, Takasaki T, Yamada K, Nerome R, Tajima S, Kurane I. 2004. Development and evaluation of fluorogenic TaqMan reverse transcriptase PCR assays for detection of dengue virus types 1 to 4. J Clin Microbiol 42:5935–5937. doi: 10.1128/JCM.42.12.5935-5937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol 30:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pastorino B, Bessaud M, Grandadam M, Murri S, Tolou HJ, Peyrefitte CN. 2005. Development of a TaqMan® RT-PCR assay without RNA extraction step for the detection and quantification of African Chikungunya viruses. J Virol Methods 124:65–71. doi: 10.1016/j.jviromet.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Laurent P, Le Roux K, Grivard P, Bertil G, Naze F, Picard M, Staikowsky F, Barau G, Schuffenecker I, Michault A. 2007. Development of a sensitive real-time reverse transcriptase PCR assay with an internal control to detect and quantify chikungunya virus. Clin Chem 53:1408–1414. doi: 10.1373/clinchem.2007.086595. [DOI] [PubMed] [Google Scholar]
- 26.Mackay IM, Arden KE, Nitsche A. 2002. Real-time PCR in virology. Nucleic Acids Res 30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houng HS, Chung-Ming Chen R, Vaughn DW, Kanesa-thasan N. 2001. Development of a fluorogenic RT-PCR system for quantitative identification of dengue virus serotypes 1-4 using conserved and serotype-specific 3′ noncoding sequences. J Virol Methods 95:19–32. doi: 10.1016/S0166-0934(01)00280-4. [DOI] [PubMed] [Google Scholar]
- 28.Houng HH, Hritz D, Kanesa-thasan N. 2000. Quantitative detection of dengue 2 virus using fluorogenic RT-PCR based on 3′-noncoding sequence. J Virol Methods 86:1–11. doi: 10.1016/S0166-0934(99)00166-4. [DOI] [PubMed] [Google Scholar]
- 29.Christensen DR, Hartman LJ, Loveless BM, Frye MS, Shipley MA, Bridge DL, Richards MJ, Kaplan RS, Garrison J, Baldwin CD. 2006. Detection of biological threat agents by real-time PCR: comparison of assay performance on the RAPID, the LightCycler, and the Smart Cycler platforms. Clin Chem 52:141–145. doi: 10.1373/clinchem.2005.052522. [DOI] [PubMed] [Google Scholar]
- 30.Gurukumar KR, Priyadarshini D, Patil JA, Bhagat A, Singh A, Shah PS, Cecilia D. 2009. Development of real time PCR for detection and quantitation of Dengue Viruses. Virol J 6:10. doi: 10.1186/1743-422X-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alm E, Lesko B, Lindegren G, Ahlm C, Söderholm S, Falk KI, Lagerqvist N. 2014. Universal single-probe RT-PCR assay for diagnosis of dengue virus infections. PLoS Negl Trop Dis 8:e3416. doi: 10.1371/journal.pntd.0003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dos Santos HW, Poloni TR, Souza KP, Muller VD, Tremeschin F, Nali LC, Fantinatti LR, Amarilla AA, Castro HL, Nunes MR, Casseb SM, Vasconcelos PF, Badra SJ, Figueiredo LT, Aquino VH. 2008. A simple one-step real-time RT-PCR for diagnosis of dengue virus infection. J Med Virol 80:1426–1433. doi: 10.1002/jmv.21203. [DOI] [PubMed] [Google Scholar]
- 33.Chien LJ, Liao TL, Shu PY, Huang JH, Gubler DJ, Chang GJ. 2006. Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. J Clin Microbiol 44:1295–1304. doi: 10.1128/JCM.44.4.1295-1304.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shu PY, Chang SF, Kuo YC, Yueh YY, Chien LJ, Sue CL, Lin TH, Huang JH. 2003. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J Clin Microbiol 41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Domingo C, Niedrig M, Teichmann A, Kaiser M, Rumer L, Jarman RG, Donoso-Mantke O. 2010. 2nd international external quality control assessment for the molecular diagnosis of dengue infections. PLoS Negl Trop Dis 4:e833. doi: 10.1371/journal.pntd.0000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edwards CJ, Welch SR, Chamberlain J, Hewson R, Tolley H, Cane PA, Lloyd G. 2007. Molecular diagnosis and analysis of Chikungunya virus. J Clin Virol 39:271–275. doi: 10.1016/j.jcv.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Carletti F, Bordi L, Chiappini R, Ippolito G, Sciarrone MR, Capobianchi MR, Di Caro A, Castilletti C. 2007. Rapid detection and quantification of Chikungunya virus by a one-step reverse transcription–polymerase chain reaction real-time assay. Am J Trop Med Hyg 77:521–524. [PubMed] [Google Scholar]
- 38.Grivard P, Le Roux K, Laurent P, Fianu A, Perrau J, Gigan J, Hoarau G, Grondin N, Staikowsky F, Favier F. 2007. Molecular and serological diagnosis of Chikungunya virus infection. Pathol Biol (Paris) 55:490–494. doi: 10.1016/j.patbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Kumar C, Johnson AA, Gopal D. 2007. Molecular characterization of chikungunya virus from Andhra Pradesh, India & phylogenetic relationship with Central African isolates. Indian J Med Res 126:534. [PubMed] [Google Scholar]
- 40.Cecilia D, Kakade M, Alagarasu K, Patil J, Salunke A, Parashar D, Shah P. 2015. Development of a multiplex real-time RT-PCR assay for simultaneous detection of dengue and chikungunya viruses. Arch Virol 160:323–327. doi: 10.1007/s00705-014-2217-x. [DOI] [PubMed] [Google Scholar]