Abstract
Accurate typing methods are required for efficient infection control. The emergence of whole-genome sequencing (WGS) technologies has enabled the development of genome-based methods applicable for routine typing and surveillance of bacterial pathogens. In this study, we developed the Pseudomonas aeruginosa serotyper (PAst) program, which enabled in silico serotyping of P. aeruginosa isolates using WGS data. PAst has been made publically available as a web service and aptly facilitates high-throughput serotyping analysis. The program overcomes critical issues such as the loss of in vitro typeability often associated with P. aeruginosa isolates from chronic infections and quickly determines the serogroup of an isolate based on the sequence of the O-specific antigen (OSA) gene cluster. Here, PAst analysis of 1,649 genomes resulted in successful serogroup assignments in 99.27% of the cases. This frequency is rarely achievable by conventional serotyping methods. The limited number of nontypeable isolates found using PAst was the result of either a complete absence of OSA genes in the genomes or the artifact of genomic misassembly. With PAst, P. aeruginosa serotype data can be obtained from WGS information alone. PAst is a highly efficient alternative to conventional serotyping methods in relation to outbreak surveillance of serotype O12 and other high-risk clones, while maintaining backward compatibility to historical serotype data.
INTRODUCTION
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen and a major cause of mortality and morbidity among hospitalized and compromised patients, including those with cystic fibrosis (CF). P. aeruginosa is well known for its ability to cause chronic and extensively drug-resistant infections (1). The outer membrane lipopolysaccharide (LPS) layer is a major virulence factor of P. aeruginosa (2). LPS has been linked to antibiotic resistance and immune evasion. Furthermore, LPS is one of the receptors that determines the susceptibility of the bacterium to bacteriophages and pyocins (2–4). Our ability to control P. aeruginosa infections depends on the availability of accurate typing methods. Previously, serotyping was a benchmark typing method for P. aeruginosa. In the 1980s, the International Antigenic Typing Scheme (IATS) was established to classify the species P. aeruginosa into 20 serotypes (O1 to O20) (5–7). Today, serotyping is infrequently used in the clinic for typing purposes, mainly because of the time-consuming protocol, the need for a continuous supply of serotype-specific antisera, and a high prevalence of polyagglutinating or nontypeable isolates.
The loss of P. aeruginosa typeability has been known for decades and has often been linked to bacteria isolated from chronic infections, where typeability is lost over time during the course of infection (8, 9). A study performed by Pirnay et al. (10) showed that 65% of all P. aeruginosa isolates examined were either non- or multitypeable and therefore assigning a particular serotype to these strains would be difficult. The occurrence of these non- or multitypeable isolates was higher when isolates sampled from CF infections exclusively were evaluated (10). Multitypeability has been associated with poor prognosis for CF patients and is a trait of persistent or chronic infection. This correlates with the observation that P. aeruginosa isolates from chronic CF infections are initially resistant to human serum but evolve to becoming serum sensitive over time, likely due to the loss of production of O antigen, which protects the bacterial cell from the human serum (8). The mechanism underlying the loss of typeability over time is not fully understood but is most likely due to modifications of LPS structures over extended periods of bacterium-host interactions as a means to improve fitness in the host and to evade the host immune system, bacteriophages, and antibiotic therapy.
The knowledge concerning the serotype of an isolate is important for monitoring outbreaks and for understanding the structures of the LPS expressed on the surface of these bacteria. O11 and O12 are more predominant than other serotypes in the clinic, and, intriguingly, these serotypes have been associated with multidrug resistance (MDR) (10–13). This implies that these particular LPS structures improve fitness within the hosts and the hospital environments in ways that we currently do not understand. Specifically for the O12 serotype, it has been shown that horizontal gene transfer of LPS genes has resulted in MDR isolates and the switching of a certain serotype to O12 (14). To continuously monitor LPS structure and evolution, serotyping can help to improve our understanding of the isolates that successfully infect patients. The continued collection of these data will also enable retrospective population analysis, as the serotype has also been recorded for decades prior to the emergence of other DNA-based typing methods such as multilocus sequence typing (MLST) and PCR.
P. aeruginosa LPS is composed of three domains: lipid A, core oligosaccharide, and O antigen (2). Most P. aeruginosa isolates produce two forms of O antigen simultaneously: common polysaccharide antigen (CPA) and O-specific antigen (OSA). While CPA is relatively conserved, OSA is variable and defines the serotype of an isolate (2, 15). OSA is encoded in a gene cluster varying in size from just <15 kb to >25 kb. The OSA gene cluster is flanked by the genes ihfB/himD and wbpM. The 20 serotypes harbor 11 distinct OSA gene clusters, each with a high number of unique genes (16). With the emergence of whole-genome sequencing (WGS) methods, it is now possible to assign an isolate to one of 11 serogroups based on the sequence and structure of the OSA gene cluster (11, 14, 17).
The present study presents a program that our group has developed for fast and reliable in silico serotyping of P. aeruginosa isolates using WGS data: the Pseudomonas aeruginosa serotyper (PAst). The program has been made publically available as a web service and can enable high-throughput serotyping analysis based on analysis of the OSA gene cluster. Using PAst, issues with typeability of clinical isolates can be overcome, and serotyping can be performed in a rapid and cost-effective way in the clinic as whole-genome sequencing of isolates become accessible.
MATERIALS AND METHODS
PAst verification and isolates included in the study.
To evaluate the efficiency of the in silico serotyping using PAst, all available P. aeruginosa genomes were acquired and analyzed. These P. aeruginosa genomes were downloaded from NCBI and included 1,120 genome assemblies (extracted 18 August, 2015; see Table S1 in the supplemental material). An exclusively CF-related P. aeruginosa data set was constructed, due mainly to the documented high level of nontypeability in persistent infecting clones. The isolates described by Marvig et al. (475 genomes) (18) were used as the initial data set. These were assembled using SPAdes (19) prior to analysis. Additional CF isolates were recovered by searching for P. aeruginosa genome assemblies related to CF in PATRIC (54 genomes) (20). It was verified that frequently observed CF-specific strains such as DK2 and LES were part of the data set. The final data set included 529 CF-related P. aeruginosa genome assemblies. In silico serotyping of both data sets was performed using PAst in order to evaluate the typeability of the program. Nontypeable isolates (i.e., isolates in which the percent coverage of reference OSA was <95%) were manually examined for either biological or technical explanations of the lack of typeability.
PAst specifications.
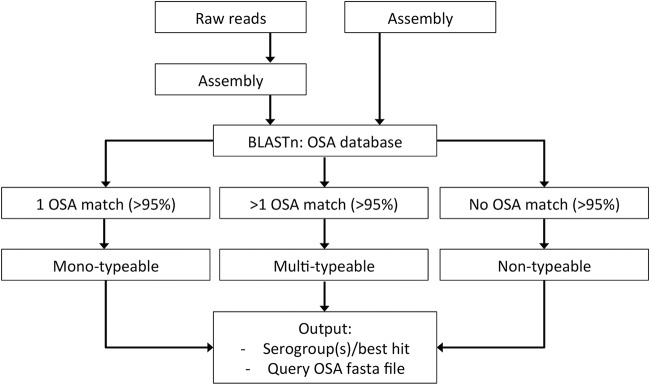
The PAst program was developed using the programming language Perl for in silico serotyping of P. aeruginosa isolates using WGS data. It is based on a BLASTn analysis of the assembled input genome, against an OSA cluster database. OSA clusters with >95% coverage in the query genome represent a positive hit for a serogroup. Since P. aeruginosa isolates which harbor either multiple OSA clusters or no clusters at all have been described, the program accommodates multi-, mono-, and nontypeability based on analysis of the number of positive OSA hits and coverage (Fig. 1). Compared to other studies (11, 14, 17), PAst optimizes in silico serotyping further by distinguishing members of the O2 serogroup through identification of the acquired phage-related wzyβ within serotypes O2 and O16 (21, 22). This enables typing into 12 serogroups as opposed to the 11 described by Raymond et al. (16). Together with a summary of the best hit(s) from the analysis and the BLAST report, the user receives a multi-FASTA file containing the sequence(s) of the OSA cluster from the isolate analyzed for use in future analysis.
FIG 1.
Workflow illustrating the in silico serotyping of the Pseudomonas aeruginosa serotyper (PAst).
The P. aeruginosa OSA cluster database.
The database was constructed using the WGS data of the 20 P. aeruginosa IATS serotype reference isolates (14). The genomes were assembled using SPAdes (19), and the OSA clusters were extracted via identification of the ihfB/himD gene flanking the cluster upstream and the wbpM gene flanking the cluster downstream. The clusters were aligned within their serotypes, described by Raymond et al., and their shared structure was confirmed (16). A representative cluster of each serotype was selected for the database (Table 1). Also included in the database was the wzyβ gene for distinguishing the O2 and O5 serotypes, as the two serogroups share the OSA cluster organization, but only the O2 and O16 serotypes harbor the wzyβ gene present on a prophage.
TABLE 1.
Serogroup definition in the PAst OSA database
| Serogroup | Reference OSA cluster | Reference gene | Size (bp) | Serotype(s) within serogroup |
|---|---|---|---|---|
| O1 | O1 | 18,368 | O1 | |
| O2 | O2 | wzyβ | 23,303 | O2, O16 |
| O3 | O3 | 20,210 | O3, O15 | |
| O5 | O2 | 23,303 | O5, O18, O20 | |
| O4 | O4 | 15,279 | O4 | |
| O6 | O6 | 15,649 | O6 | |
| O7 | O7 | 19,617 | O7, O8 | |
| O9 | O9 | 17,263 | O9 | |
| O10 | O10 | 17,635 | O10, O19 | |
| O11 | O11 | 13,868 | O11, O17 | |
| O12 | O12 | 25,864 | O12 | |
| O13 | O13 | 14,316 | O13, O14 |
In silico serotyping of P. aeruginosa isolates using PAst.
PAst has been implemented as a simple and user-friendly Web tool available on the Center for Genomic Epidemiology (CGE) service platform (https://cge.cbs.dtu.dk/services/PAst-1.0/). The tool accommodates raw reads, draft assemblies (contigs or scaffolds), and complete genomes from all WGS platforms. Raw read data are processed and assembled as previously described for other CGE tools (23). Following analysis of the input data, the Web tool outputs the predicted serogroup of the query genome, the percent coverage of the reference OSA cluster, and the OSA cluster sequence in multi-FASTA format, for the user to continue exploring the OSA genes (Fig. 1). If multiple positive hits are found (multitypeability), all of the identified OSA clusters are written for the user (Fig. 1). In the case of a nontypeable query genome (where no OSA cluster has >95% coverage), the best hit identified is written for the user together with the sequence of this hit (Fig. 1).
For batch analysis of larger data sets (only applicable for assembled genomes) the PAst Perl program has been made available on Github (https://github.com/Sandramses/PAst).
RESULTS
The PAst web server tool identifies and analyzes the nucleotide sequences of the O-specific antigen (OSA) gene clusters within the genomes provided and places them in 1 of 12 serogroups defined in Table 1. These serogroups are defined by sequence similarities between the 20 IATS serotypes (16) as well as by the absence/presence of the discriminatory wzyβ gene (21, 22) and are as such different from previous groupings of serotypes on the basis of in vitro serotyping data (11, 14, 17). All serogroups contained ≤3 of the 20 IATS serotypes (Table 1).
More than 97% of the genomes in the P. aeruginosa data set are typeable using PAst.
To evaluate the typeability efficiency of PAst, all P. aeruginosa genome assemblies available in NCBI (1,120 genomes on the date of extraction) were analyzed. A total of 97.68% (1,094) of the 1,120 genomes were typed unambiguously to a single serogroup by PAst (Fig. 2). This means that each genome assembly had a single BLAST hit of >95% OSA coverage to one sequence in our reference OSA database (Fig. 2). No isolates were found to be multitypeable and 2.32% (26 genomes) of the 1,120 genomes were found to be nontypeable (Fig. 2). In these cases, no significant BLAST hit of >95% OSA coverage to one of the sequence in the reference OSA database was identified. PAst correctly determined the serogroups of the 20 IATS strains as well as those of PAO1 (serotype O5), PA14 (serotype O10), and PAK (serotype O6).
FIG 2.
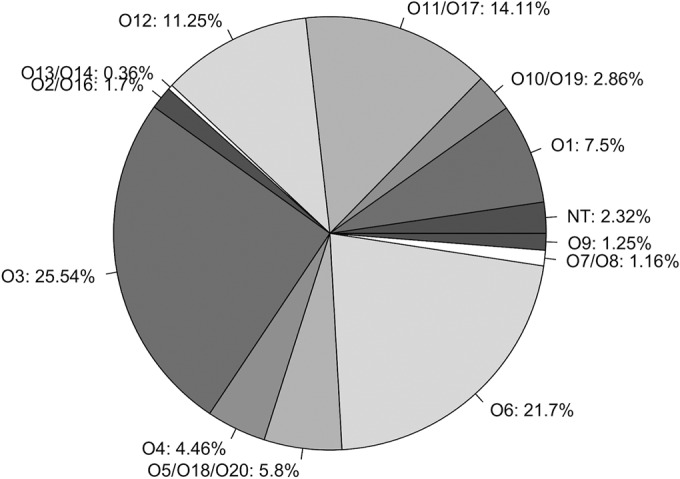
Distribution of the different serogroups (as percentages) identified via in silico serotyping of the P. aeruginosa data set using PAst. The analysis is based on all available P. aeruginosa genomes assemblies (n = 1,120). NT, nontypeable.
The analysis showed that all serogroups were represented in the 1,120 genomes (Fig. 2). Four of the 12 serogroups represented 70% of the genomes analyzed; these were O3, O6, O11, and O12 (Fig. 2). The smallest serogroup was O13, which contained only 4 genomes. We note that the same clone type might be present multiple times in the data set and that a substantial sampling bias would therefore be expected. The distribution of serotypes in our analysis thus describes what has been chosen for sequencing and does not necessarily match the distribution of serotypes in the actual P. aeruginosa population. This does not affect the high confidence of PAst, as it shows that unambiguous typing of multiple isolates from the same lineage is possible.
PAst overcomes nontypeability issues from in vitro typing of CF lineages.
P. aeruginosa isolates from CF infections are often nontypeable with conventional serotyping assays. To explore if our genome-based method enabled acquisition of serotype information in such isolates, we analyzed 529 genome assemblies of P. aeruginosa isolates sampled from CF infections. This data set contained multiple examples of isolates of the same lineage that had been sampled during the course of infection. This enabled us to investigate whether in silico typeability might be lost over time as has frequently been observed for in vitro serotyping of isolates from chronic CF infections. Interestingly, 99.81% of the genomes in the CF-specific data set were typed to single serogroups. More importantly, no multitypeable isolates were observed, and only one isolate was deemed nontypeable (Fig. 3). All serogroups were represented in the data set, except for O12. The absence of O12 serotypes among CF isolates has previously been reported (10). Serotypes O1, O6, and O7/O8 represented ∼65% of the CF-specific data set, and the smallest representation of serotypes was the O9 serogroup with only two isolates from these samples (Fig. 3).
FIG 3.
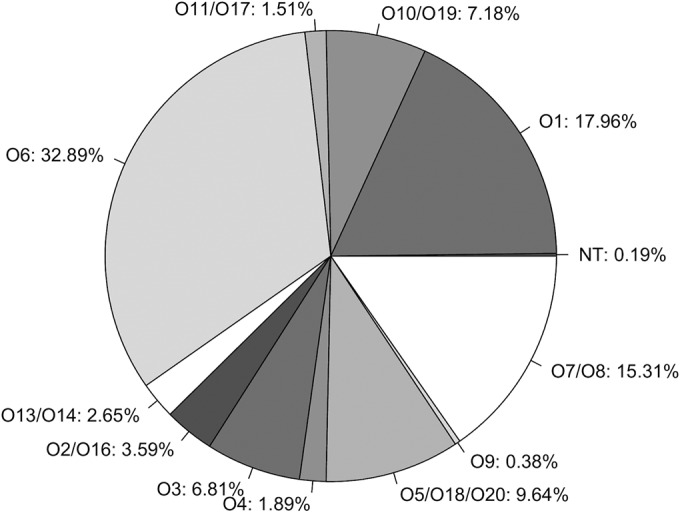
Distribution of the different serogroups (as percentages) identified via in silico serotyping of CF-specific P. aeruginosa isolates (n = 529) using PAst.
Well-known transmissible CF-specific clone types such as P. aeruginosa DK1 (24), DK2 (25), and LES (26) are represented in the data set due to multiple isolates being sampled from various patients over several decades. With our PAst tool, the typing problems documented from in vitro typing of such lineages were not observed, and the DK1, DK2, and LES isolates were consistently in silico serotyped with PAst. DK1 and DK2 were found to belong to the O3 serogroup, while the LES lineage belonged to the O6 serogroup.
Complete loss of O-specific antigen defining genes is a rare event.
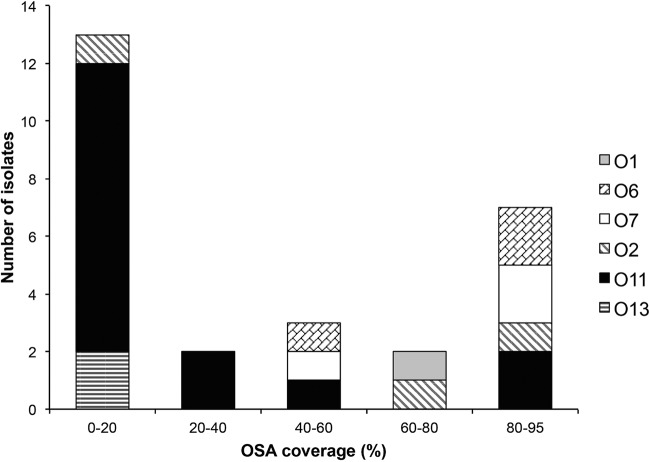
Out of two WGS-based data sets (n = 1649) that were in silico typed with PAst, our results yielded a total of 27 nontypeable isolates. The lack of typeability in these 27 genome assemblies was further investigated to resolve whether nontypeability in these cases was due to technical or biological reasons. We found that the percent OSA coverage of the nontypeable isolates ranged from a minimum of 1.91% to a maximum of 93.96% OSA coverage (see Table S2 in the supplemental material). Of the 27 isolates classified as nontypeable, 13 were found to have OSA coverage of 0 to 20%, whereas 7 had OSA coverage of 80 to 95% (Fig. 4). The best hit (serogroup) for each of the nontypeable isolates was then examined to evaluate if certain serogroups were more prone to be problematic in the PAst analysis and why. The 27 isolates were found to distribute across 6 serogroups (O1, O2, O6, O7, O11, and O13), while 15/27 isolates showed a best hit to be typed as the O11 serogroup (Fig. 4).
FIG 4.
Best-hit serotype distribution of the 27 nontypeable isolates as a function of the OSA coverage.
The group of nontypeable isolates with a best hit to the O11 serogroup were analyzed separately to identify the reason for the lack of typeability. Of the 15 O11 serogroup isolates, 9 had an OSA coverage of 14.94 to 15.84% (see Table S2 in the supplemental material); these corresponded to the presence of only the two flanking genes himD/ihfB and wbpM. This observation shows that a best hit of a nontypeable isolate to the O11 OSA cluster with a coverage of ∼15% is the result of a complete absence of an OSA cluster but the presence of the flanking genes. Two other isolates had an OSA coverage of <2% and corresponded to the absence of the entire OSA cluster as well as the flanking genes (see Table S2). In summary, a total of 11 of the 27 nontypeable isolates (or 11 of 1,649 isolates analyzed in total) were nontypeable due to a lack of the OSA cluster sequences.
Genome misassembly accounts for false nontypeability.
Since the seven nontypeable isolates with the highest OSA coverage (80 to 95%) in Fig. 4 were all candidates for harboring complete and functioning OSA clusters, we analyzed the cause of nontypeability in this group of isolates. For each of the isolates, we examined whether there were misassembly or assembly gaps within the OSA gene cluster; we also looked for the occurrence of insertion sequence (IS) elements, which often cause gaps in de novo assembly. Indeed, five of the seven isolates contained assembly gaps within their OSA cluster, which account for the observed lowered OSA coverage (Table 2). The remaining two isolates had no gaps within their OSA sequence (Table 2). However, both of these isolates had a best type hit to the O11 serogroup, which is known to contain OSA sequences of both the O11 and the O17 serotypes (16) (Table 1). Interestingly, the OSA clusters in these two serotypes differ only by the presence of two IS elements and a deletion in the O17 serotype OSA sequence (16). Alignment of the OSA sequence from the two nontypeable isolates to the O11 and O17 reference OSA sequences, respectively, contained an O17 OSA gene cluster, which had been misassembled into concatenated O11 serotype OSA clusters because of the O17 IS elements.
TABLE 2.
Nontypeable P. aeruginosa isolates with percent OSA coverage of 80 to 95% with specification of assemblies
| Strain | Size (Mb) | Scaffold | % GC | Best hit | % OSA | wbpM | himD | Gap |
|---|---|---|---|---|---|---|---|---|
| P. aeruginosa E2 | 6.36 | 196 | 66.4 | O7 | 83.31 | + | + | + |
| P. aeruginosa IGB83 | 6.48 | 249 | 66.4 | O2 | 84.46 | + | + | + |
| P. aeruginosa VRFPA04 | 6.82 | 1 | 66.5 | O11 | 86.96 | + | + | − |
| P. aeruginosa DR1 | 6.28 | 176 | 66.1 | O6 | 90.54 | + | + | + |
| P. aeruginosa 148 | 6.64 | 128 | 66.1 | O11 | 90.93 | + | + | − |
| P. aeruginosa ID4365 | 6.78 | 172 | 66.1 | O7 | 91.74 | + | + | + |
| P. aeruginosa C2773C | 6.72 | 200 | 65.9 | O6 | 93.96 | + | + | + |
DISCUSSION
The serotyping technique has been one of the standard tools for epidemiological studies and infection controls for many decades. The available historical records of P. aeruginosa serotypes offer a vast amount of information about P. aeruginosa epidemiology and population structures (27–30). Although problems with nontypeable isolates have been described since the implementation of the method, the serotype information is still applicable today for outbreak tracking, strain typing, and studies of LPS structure and evolution. The present study presents a newly developed web server tool called PAst, which is user friendly and reliable and has high throughput for in silico serotyping of P. aeruginosa isolates.
In contrast to conventional serology-based in vitro serotyping, PAst in silico serotyping has a very low occurrence of nontypeablility. Of the 1,649 analyzed genomes, only 27 nontypeable isolates were detected across two separate P. aeruginosa data sets. One data set represents all available whole-genome assemblies of P. aeruginosa, while the other specifically represents genomes from CF infections, which are known to contain high occurrences of nontypeability due to the adaptability of the bacteria into a biofilm lifestyle associated with chronicity of the infection (Fig. 1 and 2). Importantly, since the frequency of nontypeability of in vitro serotyped P. aeruginosa isolates may amount to >65% (10), analysis with PAst is clearly advantageous and superior compared to conventional in vitro serotyping. Importantly, the superiority of the PAst tool as a reliable and fast typing method is consistent with that of other published tools for in silico serotyping (31–35). Similar to SerotypeFinder (in silico serotyping of Escherichia coli) (31), LisSero (in silico serotyping of Listeria monocytogenes) (34, 35), and SeqSero (in silico serotyping of Salmonella) (32), PAst resolves the OSA cluster information to the most accurate typing possible as a serogroup representing 1 to 3 serotypes.
Interestingly, we observed a high level of conservation of the OSA gene cluster within the P. aeruginosa genome. In contrast to certain well-documented difficulties in serology-based in vitro serotyping, PAst identified complete OSA clusters (with >95% sequence being present) in 99.27% of the genomes analyzed. As such, only 12 of the 1,649 isolates examined were found to be devoid of the OSA cluster, and an additional 8 isolates were found to contain only a partial OSA cluster in their genomes (<80% OSA sequence compared to the reference). These findings indicate that the loss of typeability of P. aeruginosa isolates during the course of infection either is due to mutations (rather than larger deletions) or is linked to other parts of the LPS biosynthesis, such as regulatory genes or transport of the structure to the cell surface. A study by Bélanger et al. reported that mutation in any of the four wbp genes (wbpO, wbpP, wbpV, and wbpM) in the OSA gene cluster might disrupt the P. aeruginosa O6 OSA biosynthesis (36). Furthermore, key genes involved in the OSA assembly and translocation through the Wzx/Wzy-dependent pathway not localized within the OSA cluster, for instance, waaL, are essential for O-antigen expression (37, 38). It is possible that other OSA-related genes, which have not been discovered yet, might be present in the P. aeruginosa genomes. Overall, our study demonstrates that a complete lack of an OSA gene cluster is a rarely observed phenomenon in P. aeruginosa.
PAst will enable further investigations of the diversity, evolution, and variability of the OSA clusters. For example, the sequence of the cluster is part of the output material from the in silico serotyping which can then be readily analyzed for sequence variations to provide new knowledge on the mechanisms behind loss of typeability in vitro and in silico. Furthermore, PAst will enable systematic analysis of serotype switching by horizontal gene transfer and genetic recombination of the OSA gene cluster among different clone types. This recently described phenomenon has contributed to the evolution of the multidrug-resistant P. aeruginosa serotype O12 population that has successfully disseminated across hospitals worldwide (14). It is currently unknown if there are additional cases of such serotype switching by recombination.
The new PAst web server tool makes in silico serotyping of P. aeruginosa using WGS data a fast and reliable method. The use of PAst can play an important role in future surveillance of LPS evolution and possible outbreak detection. With the emergence of rapidly disseminating, high-risk clones of P. aeruginosa, such as the O12 ST111 clone, new and reliable typing techniques for improved monitoring and tracking of such outbreaks are becoming increasingly important (13). With the lowered cost of sequencing and the increased focus on WGS of pathogens in clinics and hospital settings, genome-based tools can assist in designing future treatments and containment of outbreaks.
Supplementary Material
ACKNOWLEDGMENTS
Funding for this study was provided by operating grants from the Villum Foundation to L.J. (VKR023113) and from the Canadian Institutes of Health Research (CIHR) to J.S.L. (MOP-14687). Additional support was provided by the A. N. Neergaard og Hustrus Foundation to L.J. and by a travel grant from the Knud Højgaards Foundation to S.W.T. V.L.T. was a recipient of a Cystic Fibrosis Canada Doctoral Studentship and a Queen Elizabeth II Graduate Scholarships in Science and Technology (QEII-GSST), and J.S.L. holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology.
We thank the Center for Genomic Epidemiology (CGE) at the Center for Biological Sequence analysis (CBS) at DTU, especially Johanne Ahrenfeldt and Rosa Allesøre, for expert assistance in setting up the PAst web service and hosting it on their web servers.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00349-16.
REFERENCES
- 1.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 2.Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D. 2011. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2:118. doi: 10.3389/fmicb.2011.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köhler T, Donner V, van Delden C. 2010. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J Bacteriol 192:1921–1928. doi: 10.1128/JB.01459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu PV, Wang S. 1990. Three new major somatic antigens of Pseudomonas aeruginosa. J Clin Microbiol 28:922–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanislavsky E, Lam J. 1997. Pseudomonas aeruginosa antigens as potential vaccines. FEMS Microbiol Rev 21:243–277. doi: 10.1111/j.1574-6976.1997.tb00353.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu PV, Matsumoto H, Kusama H, Bergan T. 1983. Survey of heat-stable, major somatic antigens of Pseudomonas aeruginosa. Int J Syst Bacteriol 33:256–264. doi: 10.1099/00207713-33-2-256. [DOI] [Google Scholar]
- 8.Penketh A, Pitt T, Roberts D, Hodson M, Batten J. 1983. The relationship of phenotype changes in Pseudomonas aeruginosa to the clinical condition of patients with cystic fibrosis. Am Rev Respir Dis 127:605–608. doi: 10.1164/arrd.1983.127.5.605. [DOI] [PubMed] [Google Scholar]
- 9.Ojeniyi B. 1994. Polyagglutinable Pseudomonas aeruginosa from cystic fibrosis patients—a survey. APMIS Suppl 46:1−44. [PubMed] [Google Scholar]
- 10.Pirnay J-P, Bilocq F, Pot B, Cornelis P, Zizi M, Van Eldere J, Deschaght P, Vaneechoutte M, Jennes S, Pitt T, De Vos D. 2009. Pseudomonas aeruginosa population structure revisited. PLoS One 4:e7740. doi: 10.1371/journal.pone.0007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witney AA, Gould KA, Pope CF, Bolt F, Stoker NG, Cubbon MD, Bradley CR, Fraise A, Breathnach AS, Butcher PD, Planche TD, Hinds J. 2014. Genome sequencing and characterization of an extensively drug-resistant sequence type 111 serotype O12 hospital outbreak strain of Pseudomonas aeruginosa. Clin Microbiol Infect 20:O609–O618. doi: 10.1111/1469-0691.12528. [DOI] [PubMed] [Google Scholar]
- 12.Cholley P, Thouverez M, Hocquet D, Van Der Mee-Marquet N, Talon D, Bertrand X. 2011. Most multidrug-resistant Pseudomonas aeruginosa isolates from hospitals in eastern France belong to a few clonal types. J Clin Microbiol 49:2578–2583. doi: 10.1128/JCM.00102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver A, Mulet X, López-Causapé C, Juan C. 2015. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 22:41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Thrane SW, Taylor VL, Freschi L, Kukavica-Ibrulj I, Boyle B, Laroche J, Pirnay J-P, Lévesque RC, Lam JS, Jelsbak L. 2015. The widespread multidrug-resistant serotype O12 Pseudomonas aeruginosa clone emerged through concomitant horizontal transfer of serotype antigen and antibiotic resistance gene clusters. mBio 6:e01396-15. doi: 10.1128/mBio.01396-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King JD, Kocíncová D, Westman EL, Lam JS. 2009. Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun 15:261–312. doi: 10.1177/1753425909106436. [DOI] [PubMed] [Google Scholar]
- 16.Raymond CK, Sims EH, Kas A, Spencer DH, Kutyavin TV, Ivey RG, Zhou Y, Kaul R, Clendenning JB, Olson MV. 2002. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J Bacteriol 184:3614–3622. doi: 10.1128/JB.184.13.3614-3622.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kos VN, Déraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm R a, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. doi: 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marvig RL, Sommer LM, Molin S, Johansen HK. 2015. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet 47:57–64. doi: 10.1038/ng.3148. [DOI] [PubMed] [Google Scholar]
- 19.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJC, Yoo HS, Zhang C, Zhang Y, Sobral BW. 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42:D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaluzny K, Abeyrathne PD, Lam JS. 2007. Coexistence of two distinct versions of O-antigen polymerase, Wzy-Alpha and Wzy-Beta, in Pseudomonas aeruginosa serogroup O2 and their contributions to cell surface diversity. J Bacteriol 189:4141–4152. doi: 10.1128/JB.00237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newton GJ, Daniels C, Burrows LL, Kropinski AM, Clarke AJ, Lam JS. 2001. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol Microbiol 39:1237–1247. doi: 10.1111/j.1365-2958.2001.02311.x. [DOI] [PubMed] [Google Scholar]
- 23.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markussen T, Marvig RL, Gómez-Lozano M, Aanæs K, Burleigh AE, Høiby N. 2014. Environmental heterogeneity drives within-host diversification and evolution of Pseudomonas aeruginosa. mBio 5:e01592-14. doi: 10.1128/mBio.01592-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marvig RL, Johansen HK, Molin S, Jelsbak L. 2013. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet 9:e1003741. doi: 10.1371/journal.pgen.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeukens J, Boyle B, Kukavica-Ibrulj I, Ouellet MM, Aaron SD, Charette SJ, Fothergill JL, Tucker NP, Winstanley C, Levesque RC. 2014. Comparative genomics of isolates of a Pseudomonas aeruginosa epidemic strain associated with chronic lung infections of cystic fibrosis patients. PLoS One 9:e87611. doi: 10.1371/journal.pone.0087611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam JS, MacDonald LA, Kropinski AM, Speert DP. 1988. Characterization of nontypeable strains of Pseudomonas aeruginosa from cystic fibrosis patients by means of monoclonal antibodies and SDS-polyacrylamide gel electrophoresis. Serodiag Immunother Infect Dis 2:365–374. doi: 10.1016/0888-0786(88)90064-9. [DOI] [Google Scholar]
- 28.Ojeniyi B, Wolz C, Doring G, Lam JS, Rosdahl VT, Høiby N. 1990. Typing of polyagglutinable Pseudomonas aeruginosa isolates from cystic fibrosis patients. Acta Pathol Microbiol Immunol Scand 98:423–431. doi: 10.1111/j.1699-0463.1990.tb01053.x. [DOI] [PubMed] [Google Scholar]
- 29.Ojeniyi B, Lam JS, Høiby N, Rosdahl VT. 1989. A comparison of the efficiency in serotyping of Pseudomonas aeruginosa from cystic fibrosis patients using monoclonal and polyclonal antibodies. Acta Pathol Microbiol Immunol Scand 97:631–636. doi: 10.1111/j.1699-0463.1989.tb00454.x. [DOI] [PubMed] [Google Scholar]
- 30.Speert DP, Campbell M, Puterman ML, Govan J, Doherty C, Høiby N, Ojeniyi B, Lam JS, Ogle JW, Johnson Z, Paranchych W, Sastry PA, Pitt TL, Lawrence L. 1994. A multicenter comparison of methods for typing strains of Pseudomonas aeruginosa predominantly from patients with cystic fibrosis. J Infect Dis 169:134–142. doi: 10.1093/infdis/169.1.134. [DOI] [PubMed] [Google Scholar]
- 31.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J Clin Microbiol 53:2410−2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685−1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Victor P. 2016. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, Bulach DM, Stinear TP, Seemann T, Howden BP. 2016. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bélanger M, Burrows LL, Lam JS. 1999. Functional analysis of genes responsible for the synthesis of the B-band O-antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145:3505–3521. doi: 10.1099/00221287-145-12-3505. [DOI] [PubMed] [Google Scholar]
- 37.Berry MC, McGhee GC, Zhao Y, Sundin GW. 2009. Effect of a waaL mutation on lipopolysaccharide composition, oxidative stress survival, and virulence in Erwinia amylovora. FEMS Microbiol Lett 291:80–87. doi: 10.1111/j.1574-6968.2008.01438.x. [DOI] [PubMed] [Google Scholar]
- 38.Abeyrathne PD, Daniels C, Poon KKH, Matewish MJ, Lam JS. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J Bacteriol 187:3002–3012. doi: 10.1128/JB.187.9.3002-3012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.