Abstract
Two screening methods to detect staphylococcal colonization in humans were compared. Direct plating to CHROMagar (BD Diagnostics) was compared to a broth preenrichment followed by plating to Baird-Parker agar. The broth-enrichment method was comparable to CHROMagar for methicillin-resistant Staphylococcus aureas (MRSA) detection, but the enrichment method was optimum for recovery of coagulase-positive Staphylococcus spp.
TEXT
Patients with community-associated methicillin-resistant Staphylococcus aureus (MRSA) and their household companions often develop recurrent episodes of MRSA infection. Cyclical reinfection within households, potentially driven by environmental or animal reservoirs, contributes to the burden of MRSA infection (1). Culture of specific organisms from the environment typically requires enrichment or other selection methods to address contamination with nontarget bacteria (2, 3). Methods that allow culture of Staphylococcus spp. other than S. aureus are critical to assess cocolonization outcomes and the presence of animal-associated staphylococci, given the potential role for these microbiota to modulate colonization by pathogens (4). Currently, the culture-based screening methods to identify S. aureus from human samples are generally different from methods for environmental or animal samples. Use of the same culture method for all types of samples (human, environmental, animal) would enhance the comparability of results. However, methods designed for environmental and animal specimens may not be optimal for use on human samples. The aim of this study was to compare a method to screen for MRSA and methicillin-susceptible S. aureus (MSSA) using CHROMagar media (5) with a broth-enrichment method, used on the same specimens, that was optimized to detect methicillin-susceptible and methicillin-resistant staphylococci from environmental and animal specimens (2, 6).
(Portions of this work were presented at the Consortium of Universities for Global Health conference [7] and the 2015 ASM-ESCMID Conference on Methicillin-Resistant Staphylococci in Animals.)
Index participants with MRSA skin or soft tissue infection (SSTI) and their household members were recruited as part of a three-arm nonblinded, randomized, controlled trial (NCT00966446), i.e., the Commonwealth Universal Research Enhancement (CURE) trial (8). This trial evaluated the effect of two similar householdwide decolonization protocols using nasal mupirocin ointment and chlorhexidine body wash versus education control on human MRSA colonization. A subset of these households participated in a nested evaluation of home environments and companion animals, i.e., the Pets and Environmental Transmission of Staphylococci (PETS) study (6, 9). Two home visits were conducted at a 3-month interval; randomization and treatment occurred between these visits. People sampled themselves using Copan ESwabs (Copan Diagnostics, Murrieta, CA) at (i) both nares and (ii) axillae and groin creases (pooled, referred to as the skin site). Index patients submitted a third ESwab from the site of the original MRSA SSTI lesion. Self-swabbing has been validated for use in this context (10).
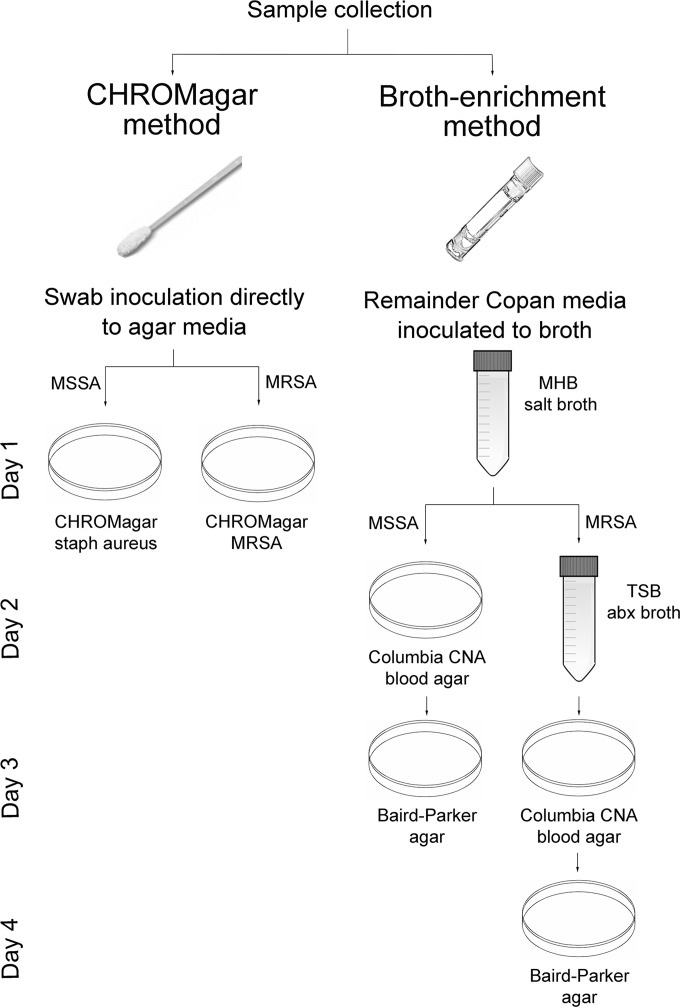
Figure 1 illustrates the protocols and timing of inoculation for the two methods. For the CHROMagar method, swabs transported in Copan Amies medium were cultured onto BBL CHROMagar MRSA and CHROMagar Staph aureus (BD Diagnostics, Sparks, MD) per manufacturer's guidelines (5). Isolates presumptively identified as MRSA based on colony morphology (mauve colonies) were confirmed as MRSA by mecA PCR. For the broth-enrichment method, to isolate S. pseudintermedius and other animal-associated staphylococci from the same human swabs, residual medium (typically at least 100 μl) from the Copan swabs was then subjected to broth-enrichment culture as previously described (2), modified to use Columbia CNA blood agar. This protocol was designed for environmental samples and included parallel arms for nonselective and methicillin-resistance-selective culture. Isolates that demonstrated tellurite reduction and lecithinase activity on Baird-Parker agar were presumptively identified as coagulase-positive staphylococci (CPS) (11). Personnel performing the CHROMagar and broth-enrichment protocols were blinded to results from the other protocol. For additional details, see the supplemental material.
FIG 1.
Protocols for the two culture-based methods (CHROMagar and broth enrichment) compared in this analysis. Additional incubation time (24 to 48 h) before reading of the inoculated agar media plates (CHROMagar or Baird-Parker) is not depicted but was comparable between the methods. For the broth-enrichment method, multiple colonies were selected from the blood agar plates based on distinct colony morphology, and these were subcultivated as needed before pure colony growth was inoculated onto Baird-Parker agar; this may result in an additional day or (rarely) days to complete the protocol.
All index participants who submitted swabs for the joint CURE/PETS study were selected for complete identification of all species of CPS isolated from the broth-enrichment culture protocol. A subset of their household members (25 households), identified a priori, were also selected for evaluation (6). All presumptive CPS isolates identified from these swabs were subjected to species identification by PCR using nuc (S. aureus, S. pseudintermedius, S. schleiferi) and mecA/C genes (12, 13). Any nasal CPS isolates that were not identified by PCR were identified biochemically to species using the BD Phoenix system (BD Diagnostics). Results of the CHROMagar-based and the broth-enrichment culture methods were compared using the kappa statistic (14) and chi-squared analysis in Stata 13 (College Station, TX).
The University of Pennsylvania and Johns Hopkins University Institutional Review Boards and Institutional Animal Care and Use Committees approved this study. Participants provided written informed consent.
Swabs were obtained from 79 (90%) of the 88 index participants evaluated in the broth-enrichment study and from an additional 68 household members in the subset of 25 households, for a total of 147 participants. Of these, 54 (68% of 79) index participants and 16 household members remained in the study and were evaluated at the subsequent 3-month visit. At enrollment, 9 households failed to provide swabs concurrently with the home visit and were excluded from this analysis. Dropout following enrollment was due to noncompliance with the protocol (n = 18), censoring for incomplete data (n = 10), or participant withdrawal (n = 5); these participants are included only for enrollment data.
Table 1 compares MRSA and MSSA results obtained by the CHROMagar method with the results obtained by the broth-enrichment method, demonstrating almost perfect strength of agreement for the kappa statistic for MRSA (14) but weaker agreement for MSSA results. Results were similar for the aggregated person-level analysis and the swab-level analysis considering nares, skin, and lesion sites separately (swab-level data not shown). The weaker agreement for MSSA results was driven by more frequent MSSA detection from nasal swabs using the broth-enrichment protocol. Broth enrichment has been shown to enhance test sensitivity for S. aureus (15, 16). Combined prevalence (designating a person as positive for MRSA or MSSA if either protocol yielded a confirmed isolate) was higher than prevalence for either study alone. Although these tests were completed sequentially, they were performed independently (i.e., simultaneous testing); hence, this increase in net sensitivity over individual test sensitivity was expected (17).
TABLE 1.
Prevalence of methicillin-resistant and methicillin-susceptible Staphylococcus aureus colonization and correlation of results according to the CURE CHROMagar and PETS broth-enrichment study protocols
| Study group |
S. aureus prevalence (no. positive [%]) |
Test sensitivity (%) |
Test agreement |
Sample size | ||||
|---|---|---|---|---|---|---|---|---|
| Jointa | CHROMagar | Broth enrichment | CHROMagar | Broth enrichment | Kappa (95% confidence interval)b | Kappa interpretationc (agreement) | ||
| MRSA | ||||||||
| Enrollment visit | ||||||||
| Subsetd,e | 37 (40.7) | 27 (29.7) | 29 (31.9) | 73.0 | 78.4 | 0.54 [0.35–0.72]*** | Moderate | 91 |
| Index only | 32 (40.5) | 26 (32.9) | 24 (30.4) | 81.3 | 75 | 0.59 [0.40–0.78]*** | Moderate | 79 |
| 3-mo visit | ||||||||
| Subsetd,e | 12 (48.0) | 12 (48.0) | 10 (40.0) | 100 | 83.3 | 0.84 [0.63–1.00]*** | Almost perfect | 25 |
| Index only | 29 (53.7) | 22 (40.7) | 26 (48.2) | 75.9 | 89.7 | 0.63 [0.42–0.83]*** | Substantial | 54 |
| MSSA | ||||||||
| Enrollment visit | ||||||||
| Subsetd,e | 20 (22.0) | 8 (8.8) | 15 (16.5) | 40 | 75 | 0.17 [-0.08–0.42]* | Slight | 91 |
| Index only | 12 (15.2) | 7 (8.9) | 8 (10.1) | 58.3 | 66.7 | 0.34 [0.00–0.67]** | Fair | 79 |
| 3-mo visit | ||||||||
| Subsetd,e | 1 (4.0) | 1 (4.0) | 1 (4.0) | 100 | 100 | 1.00 [1.00–1.00]*** | Almost perfect | 25 |
| Index only | 8 (14.8) | 3 (5.6) | 8 (14.8) | 37.5 | 100 | 0.50 [0.15–0.86]*** | Moderate | 54 |
Joint prevalence combines test results from the CHROMAgar and broth-enrichment protocols and is positive if either protocol yielded the target organism (gold standard), and the sensitivity is calculated by dividing the number positive for the individual tests by the number positive according to the gold standard (17).
*, P ≤0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
According to criteria by Landis and Koch (14).
Subset was all household members from the first 25 homes enrolled, of which 14 continued in the study at the 3-month visit.
No lesion swabs were included in the subset analysis; the subset includes index participants in order to mimic whole-household testing.
To demonstrate the utility of the data on Staphylococcus spp. from the broth-enrichment protocol, we next assessed cocolonization between MRSA and MSSA, methicillin-susceptible Staphylococcus pseudintermedius (MSSP), and other presumptive CPS. Table 2 provides these findings, demonstrating that only one participant colonized nasally with MRSA was also colonized nasally with Staphylococcus epidermidis and Staphylococcus lugdunensis. Identification of MRSA and other staphylococci together occurred more frequently with skin swabs; however, these swab samples were pooled from axillae and groin creases. Thus, cocolonization at the same skin site could not be determined, which is a limitation of this study. Table 3 lists the identified Staphylococcus spp.; S. lugdunensis and S. epidermidis were the most common. Many of these isolates, testing positive for lecithinase on Baird-Parker agar, were identified as coagulase-negative Staphylococcus (CNS) species instead of CPS. This finding was unexpected and may indicate poor correlation between lecithinase and coagulase activity in this context or may indicate coagulase and/or lecithinase positivity by another mechanism in the cultured strains (see further discussion in the supplemental material).
TABLE 2.
Methicillin-resistant Staphylococcus aureus cocolonization among index participants and household members from 25 PETS study homes and all PETS study index participants from whom swabs were obtained
| Group, sample location, and isolatea | Cocolonization (n [%]) in people: |
Chi-squared P valued | |
|---|---|---|---|
| Colonized with MRSA | Not colonized with MRSA | ||
| All participants from subset of 25 households | |||
| Enrollment (n = 91; 181 sites, nares and skin only) | |||
| Nares only | 19 (21) | 72 (79) | |
| MSSA | 0 | 13 | 0.04 |
| MSSP | 0 | 4 | 0.29 |
| Other CPS | 0 | 15 | 0.03 |
| Nares or skin | 37 (20) | 144 (80) | |
| MSSA | 3 | 15 | 0.68 |
| MSSP | 0 | 4 | 0.30 |
| Other CPS | 0 | 33 | 0.001 |
| 3mo visit (n = 25; 50 sites, nares and skin only) | |||
| Nares only | 7 (28) | 18 (72) | |
| MSSA | 0 | 1 | 0.52 |
| MSSP | 0 | 0 | NE |
| Other CPS | 0 | 6 | 0.08 |
| Nares or skinb | 11 (22) | 39 (78) | |
| MSSA | 0 | 1 | 0.59 |
| MSSP | 0 | 0 | NE |
| Other CPS | 1 | 17 | 0.04 |
| All index participants | |||
| Enrollment (n = 79; 236 sites, nares, skin, lesion site) | |||
| Nares only | 10 (13) | 69 (87) | |
| MSSA | 0 | 7 | 0.29 |
| MSSP | 0 | 2 | 0.58 |
| Other CPS | 0 | 9 | 0.23 |
| Nares, skin or lesionb | 41 (17) | 195 (83) | |
| MSSA | 1 | 10 | 0.45 |
| MSSP | 0 | 3 | 0.42 |
| Other CPS | 1 | 33 | 0.02 |
| 3-mo visit (n = 54; 162 sites, nares, skin, lesion site) | |||
| Nares only | 16 (30) | 38 (70) | |
| MSSA | 0 | 6 | 0.09 |
| MSSP | 0 | 0 | NE |
| Other CPS | 1c | 8 | 0.18 |
| Nares, skin or lesionb | 40 (25) | 122 (75) | |
| MSSA | 1 | 9 | 0.27 |
| MSSP | 0 | 0 | NE |
| Other CPS | 6 | 36 | 0.07 |
MSSA, methicillin-susceptible S. aureus; MSSP, methicillin-susceptible S. pseudintermedius; CPS, coagulase-positive Staphylococcus.
Anatomical-site-level analysis.
S. epidermidis and S. lugdunensis cocolonization.
NE, not estimable.
TABLE 3.
Staphylococcus spp., identified as lecithinase positive on Baird-Parker agar, other than S. aureus that were cultured from the nares of 147 participants in the PETS study
| Participants | Species (no. positive isolates) identified at: |
|
|---|---|---|
| Enrollmenta | 3-mo visitb | |
| Index groupc | S. epidermidis (2) | S. lugdunensis (4)i |
| S. pseudintermedius (2) | S. epidermidis (2)i | |
| S. lugdunensis (1) | S. kloosii (2)e | |
| S. kloosii (1)e | S. haemolyticus (1)f | |
| S. haemolyticus (1)f | S. sciuri (1) | |
| S. warneri (1) | ||
| S. chromogenes (1) | ||
| Unidentified (2)j | ||
| Household membersd | S. epidermidis (3) | S. lugdunensis (2) |
| S. lugdunensis (3)g | S. haemolyticus (2) | |
| S. haemolyticus (2)h | ||
| S. pseudintermedius (2)g | ||
| S. warneri (2)h | ||
| S. equorum (1) | ||
| Unidentified (2)j | ||
n = 11 (12%) for index group and 13 (19%) for household members.
n = 9 (17%) for index group and 4 (25%) for household members.
n = 91 at enrollment and 54 at 3 mo.
n = 68 at enrollment and 16 at 3 mo. While all index participants in the PETS study were evaluated fully, only household members from the first 25 homes were included in this analysis.
One index participant S. kloosii positive at both visits.
One index participant S. haemolyticus positive at both visits.
One household member cocolonized with S. pseudintermedius and S. lugdunensis.
One household member cocolonized with S. haemolyticus and S. warneri.
One index participant cocolonized with S. epidermidis, S. lugdunensis, and MRSA.
Unidentified isolates were cultured and were confirmed not to be S. aureus, S. pseudintermedius, or S. schleiferi using PCR but were not further identified to the species level using the BD Phoenix instrument, due to isolate loss.
This study demonstrates similar recovery of MRSA from a CHROMagar method and a broth-enrichment method optimized for the culture of staphylococci from environmental samples. Despite being more resource and time intensive, the broth-enrichment method provided optimum recovery of all coagulase-positive staphylococci, including MSSA. Given its benefits, researchers should consider the use of the broth-enrichment method to reduce bias in comparisons among human, animal, and environmental samples.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the study participants and to study personnel and students, particularly Kathleen Julian, Julie Vallati, Amy Shelly, Jacqueleen Wise, Robyn Smith, Grace Ndicu, John Ndicu, Aimee Vasse, Danielle Searson, Elana Youssef, Haley Keller, and Krista Reynolds. John Groopman, David Sack, and Ellen Silbergeld provided laboratory and other resources.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00132-16.
REFERENCES
- 1.Davis MF, Iverson SA, Baron P, Vasse A, Silbergeld EK, Lautenbach E, Morris DO. 2012. Household transmission of meticillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect Dis 12:703–716. doi: 10.1016/S1473-3099(12)70156-1. [DOI] [PubMed] [Google Scholar]
- 2.Davis MF, Baron P, Price LB, Williams DL, Jeyaseelan S, Hambleton IR, Diette GB, Breysse PN, McCormack MC. 2012. Dry collection and culture methods for recovery of methicillin-susceptible and methicillin-resistant Staphylococcus aureus strains from indoor home environments. Appl Environ Microbiol 78:2474–2476. doi: 10.1128/AEM.06886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weese JS. 2007. Environmental surveillance for MRSA. Methods Mol Biol 391:201–208. doi: 10.1007/978-1-59745-468-1_15. [DOI] [PubMed] [Google Scholar]
- 4.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. 2010. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One 5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Z, Lautenbach E, Fishman N, Nachamkin I. 2007. Evaluation of mannitol salt agar, CHROMagar Staph aureus and CHROMagar MRSA for detection of meticillin-resistant Staphylococcus aureus from nasal swab specimens. J Med Microbiol 56:43–46. doi: 10.1099/jmm.0.46777-0. [DOI] [PubMed] [Google Scholar]
- 6.Iverson SA, Brazil AM, Ferguson JM, Nelson K, Lautenbach E, Rankin SC, Morris DO, Davis MF. 2015. Anatomical patterns of colonization of pets with staphylococcal species in homes of people with methicillin-resistant Staphylococcus aureus (MRSA) skin or soft tissue infection (SSTI). Vet Microbiol 176:202–208. doi: 10.1016/j.vetmic.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Davis MF, Morris D, Bilker WB, Tolomeo P, Julian K, Baron P, Brazil A, Ferguson J, Iverson S, Hu B, Rankin S, Nachamkin I, Lautenbach E. 2015. Companion animals and home surface contamination in community-associated methicillin-resistant Staphylococcus aureus colonization of people. Ann Global Health 81:126. doi: 10.1016/j.aogh.2015.02.790. [DOI] [Google Scholar]
- 8.Cluzet VC, Gerber JS, Nachamkin I, Metlay J, Zaoutis T, Julian KG, Royer D, Linkin DR, Coffin SE, Margolis DJ, Hollander JE, Mistry RD, Gavin LJ, Tolomeo P, Wise JA, Wheeler MK, Bilker W, Han X, Hu B, Fishman NO, Lautenbach E, CDC Prevention Epicenters Program. 2014. A randomized controlled trial of the effect of total household decolonization on termination of colonization with methicillin-resistant Staphylococcus aureus, abstr 1336 Abstr IDWeek Sci Conf. [Google Scholar]
- 9.Misic A, Davis M, Tyldsley A, Hodkinson B, Bugayev J, Nachamkin I, Lautenbach E, Morris D, Grice E. 2015. The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome 3:2. doi: 10.1186/s40168-014-0052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lautenbach E, Nachamkin I, Hu B, Fishman NO, Tolomeo P, Prasad P, Bilker WB, Zaoutis TE. 2009. Surveillance cultures for detection of methicillin-resistant Staphylococcus aureus: diagnostic yield of anatomic sites and comparison of provider- and patient-collected samples. Infect Control Hosp Epidemiol 30:380–382. doi: 10.1086/596045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baird RM, Lee WH. 1995. Media used in the detection and enumeration of Staphylococcus aureus. Int J Food Microbiol 26:15–24. doi: 10.1016/0168-1605(93)E0028-P. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Alvarez L, Holden MTG, Lindsay H, Webb CR, Brown DFJ, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RLR, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. 2011. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, Kawakami T, Fukata T, Hiramatsu K. 2010. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol 48:765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landis JR, Koch GG. 1977. An application of hierarchical kappa-type statistics in the assessment of majority agreement among multiple observers. Biometrics 33:363–374. doi: 10.2307/2529786. [DOI] [PubMed] [Google Scholar]
- 15.Cookson BD, Webster M, Phillips I. 1987. Control of epidemic methicillin-resistant Staphylococcus aureus. Lancet 1:696. [DOI] [PubMed] [Google Scholar]
- 16.van Ogtrop ML. 1995. Effect of broth enrichment cultures on ability to detect carriage of Staphylococcus aureus. Antimicrob Agents Chemother 39:2169. doi: 10.1128/AAC.39.9.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordis L. 2014. Epidemiology, 5th ed Elsevier, Philadelphia, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.