Significance
One of the most enduring and widely debated questions in prehistoric archaeology concerns the origins of Europe’s earliest farmers: Were they the descendants of local hunter-gatherers, or did they migrate from southwestern Asia, where farming began? We recover genome-wide DNA sequences from early farmers on both the European and Asian sides of the Aegean to reveal an unbroken chain of ancestry leading from central and southwestern Europe back to Greece and northwestern Anatolia. Our study provides the coup de grâce to the notion that farming spread into and across Europe via the dissemination of ideas but without, or with only a limited, migration of people.
Keywords: paleogenomics, Neolithic, Mesolithic, Greece, Anatolia
Abstract
Farming and sedentism first appeared in southwestern Asia during the early Holocene and later spread to neighboring regions, including Europe, along multiple dispersal routes. Conspicuous uncertainties remain about the relative roles of migration, cultural diffusion, and admixture with local foragers in the early Neolithization of Europe. Here we present paleogenomic data for five Neolithic individuals from northern Greece and northwestern Turkey spanning the time and region of the earliest spread of farming into Europe. We use a novel approach to recalibrate raw reads and call genotypes from ancient DNA and observe striking genetic similarity both among Aegean early farmers and with those from across Europe. Our study demonstrates a direct genetic link between Mediterranean and Central European early farmers and those of Greece and Anatolia, extending the European Neolithic migratory chain all the way back to southwestern Asia.
It is well established that farming was introduced to Europe from Anatolia, but the extent to which its spread was mediated by demic expansion of Anatolian farmers, or by the transmission of farming technologies and lifeways to indigenous hunter-gatherers without a major concomitant migration of people, has been the subject of considerable debate. Paleogenetic studies (1–4) of late hunter-gatherers (HG) and early farmers indicate a dominant role for migration in the transition to farming in central and northern Europe, with evidence of only limited hunter-gatherer admixture into early Neolithic populations, but increasing toward the late Neolithic. However, the exact origin of central and western Europe’s early farmers in the Balkans, Greece, or Anatolia remains an open question.
Recent radiocarbon dating indicates that by 6,600–6,500 calibrated (cal) BCE sedentary farming communities were established in northwestern Anatolia at sites such as Barcın, Menteşe, and Aktopraklık C and in coastal western Anatolia at sites such as Çukuriçi and Ulucak, but did not expand north or west of the Aegean for another several hundred years (5). All these sites show material culture affinities with the central and southwestern Anatolian Neolithic (6).
Early Greek Neolithic sites, such as the Franchthi Cave in the Peloponnese, Knossos in Crete, and Mauropigi, Paliambela, and Revenia in northern Greece date to a similar period (7–9). The distribution of obsidian from the Cycladic islands, as well as similarities in material culture, suggest extensive interactions since the Mesolithic and a coeval Neolithic on both sides of the Aegean (8). Although it has been argued that in situ Aegean Mesolithic hunter-gatherers played a major role in the “Neolithization” of Greece (7), the presence of domesticated forms of plants and animals indicates nonlocal Neolithic dispersals into the area.
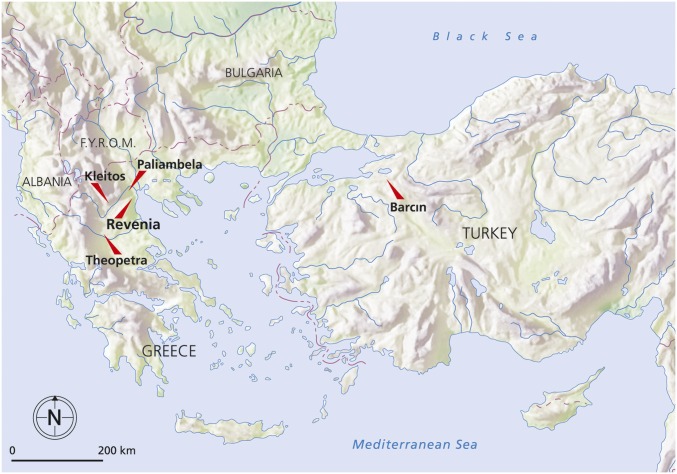
We present five ancient genomes from both, the European and Asian sides of the northern Aegean (Fig. 1); despite their origin from nontemperate regions, three of them were sequenced to relatively high coverage (∼2–7×), enabling diploid calls using a novel SNP calling method that accurately accounts for postmortem damage (SI Appendix, SI5. Genotype Calling for Ancient DNA). Two of the higher-coverage genomes are from Barcın, south of the Marmara Sea in Turkey, one of the earliest Neolithic sites in northwestern Anatolia (individuals Bar8 and Bar31). On the European side of the Aegean, one genome is from the early Neolithic site of Revenia (Rev5), and the remaining two are from the late and final Neolithic sites of Paliambela (Pal7) and Kleitos (Klei10), dating to ∼2,000 y later (Table 1). Estimates of mitochondrial contamination were low (0.006–1.772% for shotgun data) (Table 1; SI Appendix, SI4. Analysis of Uniparental Markers and X Chromosome Contamination Estimates.). We found unprecedented deamination rates of up to 56% in petrous bone samples, indicating a prehistoric origin for our sequence data from nontemperate environments (SI Appendix, Table S5).
Fig. 1.
North Aegean archaeological sites investigated in Turkey and Greece.
Table 1.
Neolithic and Mesolithic samples analyzed
| Site | Culture | Sample | Age (cal BCE, 95.4% calibrated range) | Genomic coverage (mean ± SD) | Contamination estimate (mt) | Sex | mtDNA haplogroup | Y haplogroup |
| Theopetra | Mesolithic | Theo5 | 7,605–7,529 | — | 1.84–6.71 | — | K1c | — |
| Theopetra | Mesolithic | Theo1 | 7,288–6,771 | — | 0.05–3.8 | — | K1c | — |
| Revenia | Early Neolithic | Rev5 | 6,438–6,264 | 1.16 ± 0.73 | 0.006–0.628 | XX | X2b | * |
| Barcın | Early Neolithic | Bar31 | 6,419–6,238 | 3.66 ± 2.04 | 0.006–0.628 | XY | X2m | G2a2b |
| Barcın | Early Neolithic | Bar8 | 6,212–6,030 | 7.13 ± 4.56 | 0.744–1.619 | XX | K1a2 | * |
| Paliambela | Late Neolithic | Pal7 | 4,452–4,350 | 1.28 ± 1.01 | 0.006–0.772 | XX | J1c1 | * |
| Kleitos | Final Neolithic | Klei10 | 4,230–3,995 | 2.01 ± 2.2 | 0.363–1.772 | XY | K1a2 | G2a2a1b |
Dates calibrated using Oxcal v4.2.2 and the Intcal13 calibration curve. For details on 14C dating and location of the sites (Fig. 1), see SI Appendix, SI1. Archaeological Background. Contamination was estimated on mitochondrial (mt) DNA. —, indicates no genomic data available; *, indicates not applicable.
Uniparental Genetic Systems
The mtDNA haplogroups of all five Neolithic individuals are typical of those found in central European Neolithic farmers and modern Europeans, but not in European Mesolithic hunter-gatherers (1). Likewise, the Y-chromosomes of the two male individuals belong to haplogroup G2a2, which has been observed in European Neolithic farmers (3, 10); in Ötzi, the Tyrolean Iceman (11); and in modern western and southwestern Eurasian populations, but not in any pre-Neolithic European hunter-gatherers (12). The mitochondrial haplogroups of two additional less well-preserved Greek Mesolithic individuals (Theo1, Theo5; SI Appendix, Table S6) belong to lineages observed in Neolithic farmers from across Europe; consistent with Aegean Neolithic populations, unlike central European Neolithic populations, being the direct descendants of the preceding Mesolithic peoples who inhabited broadly the same region. However, we caution against over-interpretation of the Aegean Mesolithic mtDNA data; additional genome-level data will be required to identify the Mesolithic source population(s) of the early Aegean farmers.
Functional Variation
Sequences in and around genes underlying the phenotypes hypothesized to have undergone positive selection in Europeans indicate that the Neolithic Aegeans were unlikely to have been lactase persistent but carried derived SLC24A5 rs1426654 and SLC45A2 rs16891982 alleles associated with reduced skin pigmentation. Because our Aegean samples predate the period when the rs4988235 T-allele associated with lactase persistence in Eurasia reached an appreciable frequency in Europe, around 4 kya (12–14), and because this allele remains at relatively low frequencies (<0.15) in modern Greek, Turkish, and Sardinian populations (15), this observation is unsurprising. However, despite their relatively low latitude, four of the Aegean individuals are homozygous for the derived rs1426654 T-allele in the SLC24A5 gene, and four carry at least one copy of the derived rs16891982 G-allele in the SLC45A2 gene. This suggests that these reduced-pigmentation–associated alleles were at appreciable frequency in Neolithic Aegeans and that skin depigmentation was not solely a high-latitude phenomenon (SI Appendix, SI12. Functional Markers). The derived rs12913832 G-allele in the HERC2 domain of the OCA2 gene was heterozygous in one individual (Klei10), but all other Aegeans for whom the allelic state at this locus could be determined were homozygous for the ancestral allele, indicating a lack of iris depigmentation in these individuals.
Examination of several SNPs in the TCF7L2 gene region indicates that the two Neolithic Anatolian individuals, Bar8 and Bar31, are likely to have carried at least one copy of a haplotype conferring reduced susceptibility to type 2 diabetes (T2D); the Klei10 and Rev5 individuals also carry a tag allele associated with this haplotype. Consistent with these observations, it has been previously estimated that this T2D-protective haplotype, which shows evidence for selection in Europeans, East Asians, and West Africans, originated ∼11,900 y ago in Europe (16).
A number of loci associated with inflammatory disease displayed the derived alleles, including rs2188962 C > T in the SLC22A5/IRF1 region, associated with Crohn’s disease; rs3184504 C > T in the SH2B3/ATXN2 region, associated with rheumatoid arthritis, celiac disease, and type 1 diabetes; and rs6822844 G > T in the IL2/IL21 region, associated with rheumatoid arthritis, celiac disease, and ulcerative colitis. Interestingly, we observe derived states for six of eight loci in a protein–protein interaction network inferred to have undergone concerted positive selection 2.6–1.2 kya in Europeans (17), suggesting that any recent selection on these loci acted on standing variation present at already appreciable frequency (SI Appendix, SI12. Functional Markers).
Principal Component Analysis, f-Statistics, and Mixture Modeling
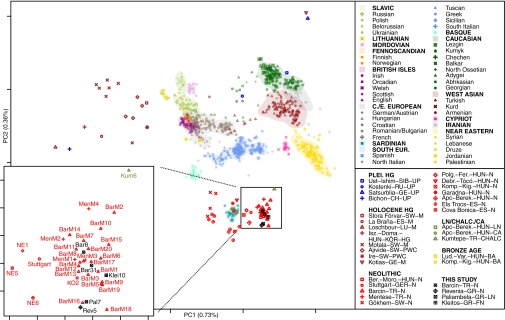
The first two dimensions of variation from principal component analysis (PCA) reveal a tight clustering of all five Aegean Neolithic genomes with Early Neolithic (EN) genomes from central and southern Europe (2, 3, 13) (Fig. 2). This cluster remains well-defined when the third dimension of variation is also considered (https://figshare.com/articles/Hofmanova_et_al_3D_figure_S4/3188767). Two recently published pre-Neolithic genomes from the Caucasus (20) appear to be highly differentiated from the genomes presented here and most likely represent a forager population distinct from the Epipaleolithic/Mesolithic precursors of the early Aegean farmers.
Fig. 2.
PCA of modern reference populations (18, 19) and projected ancient individuals. The Greek and Anatolian samples reported here cluster tightly with other European farmers close to modern-day Sardinians; however, they are clearly distinct from previously published Caucasian hunter-gatherers (20). This excludes the latter as a potential ancestral source population for early European farmers and suggests a strong genetic structure in hunter-gatherers of Southwest Asia. Central and East European (C./E. European), South European (South Eur.). Ancient DNA data: Pleistocene hunter-gatherer (Plei. HG) (20, 21, 22), Holocene hunter-gatherer (Holocene HG) (2, 4, 13, 20, 23), Neolithic (2, 4, 12, 13, 24), Late Neolithic/Chalcolithic/Copper Age (LN/Chalc./CA) (13, 25), and Bronze Age (13). Ancient samples are abbreviated consistently using the nomenclature “site-country code-culture”; see SI Appendix, Table S14 and Dataset S1 for more information. A 3D PCA plot can be viewed as a 3D figure (https://figshare.com/articles/Hofmanova_et_al_3D_figure_S4/3188767).
To examine this clustering of Early Neolithic farmers in more detail, we calculated outgroup f3 statistics (26) of the form f3 (‡Khomani; TEST, Greek/Anatolian), where TEST is one of the available ancient European genomes (SI Appendix, SI7. Using f-statistics to Infer Genetic Relatedness and Admixture Amongst Ancient andContemporary Populations and Figs. S8–S10; Dataset S2); ‡Khomani San were selected as an outgroup as they are considered to be the most genetically diverged extant human population. Consistent with their PCA clustering, the northern Aegean genomes share high levels of genetic drift with each other and with all other previously characterized European Neolithic genomes, including early Neolithic from northern Spain, Hungary, and central Europe. Given the archaeological context of the different samples, the most parsimonious explanation for this shared drift is migration of early European farmers from the northern Aegean into and across Europe (12).
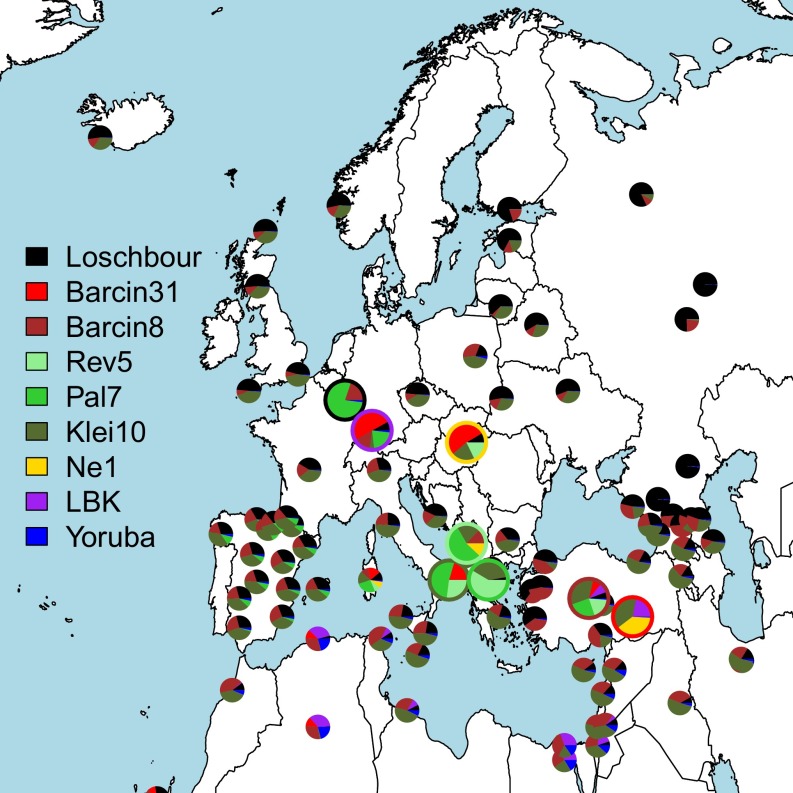
To better characterize this inferred migration, we modeled ancient and modern genomes as mixtures of DNA from other ancient and/or modern genomes, a flexible approach that characterizes the amount of ancestry sharing among multiple groups simultaneously (18, 27) (Fig. 3; SI Appendix, SI10. Comparing Allele Frequency Patterns Among Samples Using a Mixture Model). Briefly, we first represented each ancient or modern “target” group by the (weighted) number of alleles that they share in common with individuals from a fixed set of sampled populations (i.e., the “unlinked” approach described in ref. 27), which we refer to as the “allele-matching profile” for that target group. To cope with issues such as unequal sample sizes, we then used a linear model (28) to fit the allele-matching profile of the target group as a mixture of that of other sampled groups. Sampled groups that contribute most to this mixture indicate a high degree of shared ancestry with the target group relative to other groups. Under this framework the oldest Anatolian genome (Bar31) was inferred to contribute the highest amount of genetic ancestry (39–53%) to the Early Neolithic genomes from Hungary (13) and Germany (2) compared with any other ancient or modern samples, with the next highest contributors being other ancient Aegean genomes (Klei10, Pal7, Bar8) (SI Appendix, Figs. S23, S24, and S29). This pattern is not symmetric in that we infer smaller contributions from the German (<26%) and Hungarian (<43%) Neolithic genomes to any of the Anatolian or Greek ancient genomes. Furthermore, in this analysis modern samples from Europe and surrounding regions are inferred to be relatively more genetically related to the Aegean Neolithic genomes than to the Neolithic genomes from Germany and Hungary (Fig. 3; SI Appendix, SI10. Comparing Allele Frequency Patterns Among Samples Using a Mixture Model). These patterns are indicative of founder effects (29) in the German and possibly Hungarian Neolithic samples from a source that appears to be most genetically similar to the Aegean Neolithic samples (specifically, Bar31) and that distinguishes them from the ancestors of modern groups. Consistent with this, we found fewer short runs of homozygosity (ROH) (between 1 and 2 Mb) in our high-coverage Anatolian sample (Bar8) than in Early Neolithic genomes from Germany and Hungary (SI Appendix, SI11. Runs of Homozygosity and Fig. S31). However, it is not possible to infer a direction for dispersal within the Aegean with statistical confidence because both the Greek and Anatolian genomes copy from each other to a similar extent. We therefore see the origins of European farmers equally well represented by Early Neolithic Greek and northwestern Anatolian genomes.
Fig. 3.
Inferred mixture coefficients when forming each modern (small pies) and ancient (large pies, enclosed by borders matching key at left) group as a mixture of the modern-day Yoruba from Africa and the ancient samples shown in the key at left.
Ongoing gene flow into and across the Aegean is also indicated in the genome of a Chalcolithic individual from Kumtepe [Kum6 (25)], a site geographically close to Barcın but dating to ∼1,600 y later. Although archaeological evidence indicates a cultural break in many Aegean and West Anatolian settlements around 5,700/5,600 cal BCE [i.e., spanning this 1,600-y period (30)], Kum6 shows affinities to the Barcın genomes in “outgroup” f3-statistics in the form f3 (‡Khomani; TEST, Greek/Anatolian). The shared drift between Kum6 and both the early and late Neolithic Aegeans is similar in extent to the drift that Aegeans share with one another. However, f4 statistics of the form f4 (Aegean, Kum6, Early farmer, ‡Khomani) were often significantly positive (SI Appendix, Table S22; Dataset S2), suggesting that European Neolithic farmers [namely, Linearbandkeramik (LBK), Starcevo, and Early Hungarian Neolithic farmers] share some ancestry with early Neolithic Aegeans that is absent in Kum6. This is consistent with population structure in the Early Neolithic Aegean or with Kum6 being sampled from a population that differentiated from early Neolithic Aegeans after they expanded into the rest of Europe. Accordingly, compared with Barcın, Kum6 shares unique drift with the Late Neolithic genomes from Greece (Klei10 and Pal7), consistent with ongoing gene flow across the Aegean during the fifth millennium and with archaeological evidence demonstrating similarities in Kumtepe ceramic types with the Greek Late Neolithic (31). Finally, the Kum6, Klei10, and Pal7 genomes show signals of Caucasus hunter-gatherer (20) admixture that is absent in the Barcın genomes, suggesting post early Neolithic gene flow into the Aegean from the east.
It is widely believed that farming spread into Europe along both Mediterranean and central European routes, but the extent to which this process involved multiple dispersals from the Aegean has long been a matter of debate (32). We calculated f4 statistics to examine whether the Aegean Neolithic farmers shared drift with genomes from the Spanish Epicardial site Els Trocs in the Pyrenees (3, 12) that is distinct from that shared with Early Neolithic genomes from Germany and Hungary. In a test of the form f4 (Germany/Hungary EN, Spain EN, Aegean, ‡Khomani), we infer significant unique drift among Neolithic Aegeans (not significantly in Bar8) and Early Neolithic Spain to the exclusion of Hungarian and German Neolithic genomes (SI Appendix, Table S21). The best explanation for this observation is that migration to southwestern Europe started in the Aegean but was independent from the movement to Germany via Hungary. This is also supported by other genetic inferences (24) and archaeological evidence (33). An alternative scenario is a very rapid colonization along a single route with subsequent gene flow back to Greece from Spain. Potentially, preexisting hunter-gatherer networks along the western Mediterranean could have produced a similar pattern, but this is not supported by archaeological data. Interestingly, Ötzi the Tyrolean Iceman (11) shows unique shared drift with Aegeans to the exclusion of Hungarian Early Neolithic farmers and Late and Post Neolithic European genomes and feasibly represents a relict of Early Neolithic Aegeans (SI Appendix, SI7. Using f-statistics to Infer Genetic Relatedness and Admixture Amongst Ancient and Contemporary Populations and Table S18).
Hunter-Gatherer Admixture
Given that the Aegean is the likely origin of European Neolithic farmers, we used Bar8 and Bar31 as putative sources to assess the extent of hunter-gatherer admixture in European farmers through the Neolithic. f4 statistics of the form f4 (Neolithic farmer, Anatolian, HG, ‡Khomani) indicated small but significant amounts of hunter-gatherer admixture into both Spanish and Hungarian early farmer genomes, and interestingly, the Early Neolithic Greek genome. Our mixture modeling analysis also inferred a small genetic contribution from the Loschbour hunter-gatherer genome (3–9%) to each of the Early Neolithic Hungarian and German genomes, but evidence of a smaller contribution to any Aegean genomes (0–6%). These results suggest that mixing between migrating farmers and local hunter-gatherers occurred sporadically at low levels throughout the continent even in the earliest stages of the Neolithic. However, consistent with previous findings (3), both f4 statistics and ADMIXTURE analysis indicate a substantial increase in hunter-gatherer ancestry transitioning into the Middle Neolithic across Europe, whereas Late Neolithic farmers also demonstrate a considerable input of ancestry from steppe populations (SI Appendix, SI8. Proportions of Ancestral Clusters in Neolithic Populations of Europe and Fig. S32).
Relation to Modern Populations
Most of the modern Anatolian and Aegean populations do not appear to be the direct descendants of Neolithic peoples from the same region. Indeed, our mixture model comparison of the Aegean genomes to >200 modern groups (2) indicates low affinity between the two Anatolian Neolithic genomes and six of eight modern Turkish samples; the other two were sampled near the Aegean Sea at a location close to the site of the Neolithic genomes. Furthermore, when we form each Anatolian Neolithic genome as a mixture of all modern groups, we infer no contributions from groups in southeastern Anatolia and the Levant, where the earliest Neolithic sites are found (SI Appendix, Figs. S22 and S30 and Table S30; Dataset S3). Similarly, comparison of allele sharing between ancient and modern genomes to those expected under population continuity indicates Neolithic-to-modern discontinuity in Greece and western Anatolia, unless ancestral populations were unrealistically small (SI Appendix, SI9. Population Continuity). Instead, our mixing analysis shows that each Aegean Neolithic genome closely corresponds to modern Mediterraneans (>68% contributions from southern Europe) and in particular to Sardinians (>25%), as also seen in the PCA and outgroup f3 statistics with few substantial contributions from elsewhere. Modern groups matching the Neolithics—mostly from the Mediterranean and North Africa—strikingly match more to Bar8 from northwestern Anatolia than to the LBK genome from Stuttgart in Germany, indicating that the LBK genome experienced processes such as drift and admixture that were independent from the Mediterranean expansion route, consistent with the dual expansion model.
Concluding Remarks
Over the past 7 years, ancient DNA studies have transformed our understanding of the European Neolithic transition (1–4, 12, 13), demonstrating a crucial role for migration in central and southwestern Europe. Our results further advance this transformative understanding by extending the unbroken trail of ancestry and migration all of the way back to southwestern Asia.
The high levels of shared drift between Aegean and all available Early Neolithic genomes in Europe, together with the inferred unique drift between Neolithic Aegeans and Early Neolithic genomes from Northern Spain to the exclusion of Early Neolithic genomes from central Europe, indicate that Aegean Neolithic populations can be considered the root for all early European farmers and that at least two independent colonization routes were followed.
A key remaining question is whether this unbroken trail of ancestry and migration extends all the way back to southeastern Anatolia and the Fertile Crescent, where the earliest Neolithic sites in the world are found. Regardless of whether the Aegean early farmers ultimately descended from western or central Anatolian, or even Levantine hunter-gatherers, the differences between the ancient genomes presented here and those from the Caucasus (20) indicate that there was considerable structuring of forager populations in southwestern Asia before the transition to farming. The dissimilarity and lack of continuity of the Early Neolithic Aegean genomes to most modern Turkish and Levantine populations, in contrast to those of early central and southwestern European farmers and modern Mediterraneans, is best explained by subsequent gene flow into Anatolia from still unknown sources.
Methods
Ancient DNA Extraction and Sequencing.
Five Neolithic and two Mesolithic samples from both sides of the Aegean were selected for ancient DNA extraction and sequencing (Table 1). DNA was extracted, and Next Generation Sequencing libraries were constructed in dedicated ancient DNA facilities as previously described (1, 34) with slight modifications. DNA quality and quantity of all samples were derived from the combination of estimates of endogenous DNA content based on the percentage of reads mapping to the reference genome (GRCh37/hg19) after shallow Illumina Miseq sequencing and estimating the DNA copy number of extracts by quantitative PCR. The five Neolithic samples (Bar8, Bar31, Rev5, Klei10, and Pal7) showed endogenous DNA contents between 8.80 and 60.83% and underwent deep Illumina whole-genome resequencing. The two Mesolithic samples (Theo 1 and Theo 5) showed endogenous DNA content of only 0.05% and 0.62%, respectively, and were used to capture the full mitochondrial genome. SI Appendix, Fig. S1, displays the relationship between endogenous DNA content and copy number for each sample and DNA extraction. The enrichment of the mitochondrial genome was carried out with Agilent’s SureSelectXT in-solution target enrichment kit. The protocol for the preparation of further libraries for shotgun sequencing and capture was modified according to previously estimated sample quality, whereby some libraries from samples Bar8, Bar31, and Rev5 were prepared with USER treated DNA extract. Detailed information about the experimental setup is described in SI Appendix, SI2. Sample Preparation.
Bioinformatics.
All sequence reads underwent 3′ adapter trimming and were filtered for low-quality bases. For paired-end sequences only pairs with overlapping sequence were retained and merged into a single sequence. All sequences were aligned against the human reference build GRCh37/hg19 using BWA (35) and realigned using the Genome Analysis Toolkit (36) (SI Appendix, SI3. Read Processing). For genotyping, we developed a novel method to recalibrate quality scores and call genotypes that probabilistically accounts for postmortem damage patterns as estimated in mapDamage2.0 (37). For low-coverage genomes, we further developed a Bayesian haploid caller to reliably identify the most likely allele call for each site (code available on request from D.W.). For further details see SI Appendix, SI5. Genotype Calling for Ancient DNA.
Ancient DNA Authenticity.
The assessment of ancient DNA authenticity was performed using the sequence reads mapping to the mitochondrial genome following the likelihood approach described in Fu et al. (38) (SI Appendix, SI4. Analysis of Uniparental Markers and X Chromosome Contamination Estimates). Postmortem damage deamination rates were estimated using mapDamage 2.0 (37) and are displayed together with distribution of DNA fragment lengths of each sample (SI Appendix, Fig. S3). We used ANGSD (39) to determine X-chromosome contamination in male samples (SI Appendix, SI4. Analysis of Uniparental Markers and X Chromosome Contamination Estimates).
Analysis of Uniparental Markers.
Mitochondrial haplogroups were determined using HaploFind (40). Consensus sequences in FASTA format were created from alignments with SAMtools (41) (SI Appendix, SI4. Analysis of Uniparental Markers and X Chromosome Contamination Estimates).
To determine patrilineal lineages in ancient samples, we used clean_tree (42). This software requires BAM format files as input, and alleles are called with SAMtools mpileup at given SNP positions. These SNP positions were provided with the clean_tree software and contain 539 SNPs used for haplogroup determination (SI Appendix, SI4. Analysis of Uniparental Markers and X Chromosome Contamination Estimates).
PCA.
Principal component analysis was performed with LASER v2.02 (43). First, a reference space was generated on genotype data of modern individuals. For Fig. 2, we used European and Middle Eastern populations from a merged dataset published as part of Hellenthal et al. and Busby et al. (18, 19). In a second step, ancient samples provided as BAM files were projected into the reference space via a Procrustes analysis. See SI Appendix, SI6. PCA, for details.
D-Statistics and ADMIXTURE Analysis.
f3 and f4 statistics and the associated Z-scores (via block jackknife with default options) were calculated using the ADMIXTOOLS package (44) on haploid calls (SI Appendix, SI7. Using f-statistics to Infer Genetic Relatedness and Admixture Amongst Ancient and Contemporary Populations). Samples from this study were compared with the Haak et al. (3) dataset containing 2,076 contemporary and ancient individuals. Additionally, ADMIXTURE analysis (45) was performed on a subset of these data containing all Eurasian ancient samples that predate the Bronze Age (n = 77) and additionally with Caucasus hunter-gatherers (n = 79) and prehistoric individuals from the eastern European steppe (n = 89) (SI Appendix, SI8. Proportions of Ancestral Clusters in Neolithic Populations of Europe).
Mixture Modeling.
To compose a target group as mixtures of other sampled groups, we used the following two-step procedure. First, we used a previously described technique (27) to infer an allele-matching profile for each target group by comparing its allele frequencies independently at each SNP to that of a set of “donor” groups. In particular, at a given SNP for each chromosome in our target group, we identified all X nonmissing donor chromosomes that shared the same allele type as the target and assigned each of these donors a score of 1/X and all other donors a score of 0. We did this for each SNP and each target individual and then summed up these scores across SNPs and target individuals to give an allele-matching profile for the target group conditional on that set of donors. For each target group, the contributions from each donor group were rescaled to sum to 1. For analyses presented here, our donor groups consisted of modern individuals (2) (plus Neanderthal and Denisova). Our target groups included all modern and ancient groups. Next, analogous to ref. 18, we performed a multiple linear regression using the target group’s allele-matching profile as a response and a set of allele-matching profiles of different “surrogate” groups as predictors. In all analyses, we used three different sets of surrogate groups: (i) all (or a subset of) modern groups, (ii) all ancient groups and all (or a subset of) modern groups, and (iii) the modern Yoruba plus all (or a subset of) ancient groups. Mixture coefficients were inferred using nonnegative least squares in R with a slight modification to ensure that the coefficients sum to 1 (SI Appendix, SI10. Comparing Allele Frequency Patterns Among Samples Using a Mixture Model).
Population Continuity Test.
We used a forward-simulation approach to test for population continuity. For our purposes a continuous population is defined as “a single panmictic population without admixture from other populations.” Our approach is designed to test continuity using a single ancient genome and a set of modern genomes. We designate alleles as ancestral or derived by comparing them to the chimpanzee genome (panTro2) and consider only haploid calls for the ancient genome to avoid genotype calling biases. We examine the proportion of allele sharing between the ancient haploid and modern diploid genome calls that fall into each of the following six classes: A/AA; D/DD; A/DD; D/AA; A/AD; D/AD (where A = ancestral and D = derived alleles in the ancient haploid/modern diploid genomes, respectively). To generate expected proportions of these allele-sharing classes, we forward-simulate genetic drift by binomial sampling from a set of allele frequency vectors based on the modern site frequency spectrum. Finally, we use Fisher’s method to combine two-tailed P values for the observed sharing class fractions falling into the simulated ranges and compare the resultant χ2 values to those obtained by comparing each simulation against the set of all other simulations (46) to obtain a P value. We performed this test for a range of assumed ancient and modern effective population sizes (SI Appendix, SI9. Population Continuity).
Runs of Homozygosity.
The distribution of ROH for 5 ancient (2, 13) and 2,527 modern individuals (47) was determined with PLINK v1.90 (48) following the specifications used in ref. 13, with a set of 1,447,024 transversion SNPs called securely across all ancient samples (SI Appendix, SI11. Runs of Homozygosity).
Functional Markers.
Genotypes were determined using the diploid genotyping method described in SI Appendix, SI5. Genotype Calling for Ancient DNA, and further verified through direct observation of BAM files using samtools tview (htslib.org). We included sites having ≥2× coverage in the analysis (SI Appendix, SI12. Functional Markers).
Supplementary Material
Acknowledgments
We thank Songül Alpaslan for help with sampling in Barcın and Eleni Stravopodi for help with sampling in Theopetra. Z.H. and R.M. are supported by a Marie Curie Initial Training Network (BEAN/Bridging the European and Anatolian Neolithic, GA 289966) awarded to M.C., S.J.S., D.G.B., M.G.T., and J. Burger. C.P., J. Burger and S.K. received funding from DFG (BU 1403/6-1). C.P. and J. Burger received funding from the Alexander von Humboldt Foundation. C.S. and M.S. were supported by the European Union (EU) SYNTHESYS/Synthesis of Systematic Resources GA 226506-CP-CSA-INFRA, DFG: (BO 4119/1) and Volkswagenstiftung (FKZ: 87161). L.M.C. is funded by the Irish Research Council (GOIPG/2013/1219). A.S. was supported by the EU CodeX Project 295729. K. Kotsakis, S.T., D.U.-K., P.H., and C.P. were cofinanced by the EU Social Fund and Greek national funds research funding program THALES. C.P., M.U., K. Kotsakis, S.T., and D.U.-K. were cofinanced by the EU Social Fund and the Greek national funds research funding program ARISTEIA II. M.C. was supported by Swiss NSF Grant 31003A_156853. A.K. and D.W. were supported by Swiss NSF Grant 31003A_149920. S.L. is supported by the BBSRC (Grant BB/L009382/1). L.v.D. is supported by CoMPLEX via EPSRC (Grant EP/F500351/1). G.H. is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant 098386/Z/12/Z) and by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. M.G.T. and Y.D. are supported by a Wellcome Trust Senior Research Fellowship Grant 100719/Z/12/Z (to M.G.T.). J. Burger is grateful for support by the University of Mainz and the HPC cluster MOGON (funded by DFG; INST 247/602-1 FUGG). F.G. was supported by Grant 380-62-005 of the Netherlands Organization for Scientific Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Mitochondrial genome sequences have been deposited in the GenBank database (KU171094–KU171100). Genomic data are available at the European Nucleotide Archive under the accession no. PRJEB11848 in BAM format.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523951113/-/DCSupplemental.
References
- 1.Bramanti B, et al. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science. 2009;326(5949):137–140. doi: 10.1126/science.1176869. [DOI] [PubMed] [Google Scholar]
- 2.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513(7518):409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522(7555):207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skoglund P, et al. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 2014;344(6185):747–750. doi: 10.1126/science.1253448. [DOI] [PubMed] [Google Scholar]
- 5.Weninger B, et al. Neolithisation of the Aegean and Southeast Europe during the 6600–6000 calBC period of Rapid Climate Change. Documenta Praehistorica. 2014;41:1–31. [Google Scholar]
- 6.Özdoğan M. A new look at the introduction of the Neolithic way of life in Southeastern Europe. Changing paradigms of the expansion of the Neolithic way of life. Documenta Praehistorica. 2014;41:33–49. [Google Scholar]
- 7.Perlès C, Quiles A, Valladas H. Early seventh-millennium AMS dates from domestic seeds in the Initial Neolithic at Franchthi Cave (Argolid, Greece) Antiquity. 2013;87(338):1001–1015. [Google Scholar]
- 8.Kotsakis K. Domesticating the periphery: New research into the Neolithic of Greece. Pharos. 2014;20(1):41–73. [Google Scholar]
- 9.Maniatis Y. 2014. Radiocarbon dating of the major cultural changes in Prehistoric Macedonia: Recent developments. 1912–2012. A Century of Research in Prehistoric Macedonia. Proceedings of the International Conference, Archaeological Museum of Thessaloniki, 22–24 November 2012, eds Stefani E, Merousis N, Dimoula A (Archaeological Museum of Thessaloniki, Thessaloniki, Greece), pp 205–222.
- 10.Lacan M, et al. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc Natl Acad Sci USA. 2011;108(24):9788–9791. doi: 10.1073/pnas.1100723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller A, et al. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 2012;3:698. doi: 10.1038/ncomms1701. [DOI] [PubMed] [Google Scholar]
- 12.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528(7583):499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamba C, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522(7555):167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 15.Itan Y, Jones BL, Ingram CJ, Swallow DM, Thomas MG. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol Biol. 2010;10(1):36. doi: 10.1186/1471-2148-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgason A, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39(2):218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 17.Raj T, et al. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am J Hum Genet. 2013;92(4):517–529. doi: 10.1016/j.ajhg.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hellenthal G, et al. A genetic atlas of human admixture history. Science. 2014;343(6172):747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Busby GB, et al. The role of recent admixture in forming the contemporary West Eurasian genomic landscape. Curr Biol. 2015;25(19):2518–2526. doi: 10.1016/j.cub.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones ER, et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat Commun. 2015;6:8912. doi: 10.1038/ncomms9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Q, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514(7523):445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seguin-Orlando A, et al. Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346(6213):1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 23.Olalde I, et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature. 2014;507(7491):225–228. doi: 10.1038/nature12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olalde I, et al. A common genetic origin for early farmers from Mediterranean Cardial and Central European LBK cultures. Mol Biol Evol. 2015;32(12):3132–3142. doi: 10.1093/molbev/msv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omrak A, et al. Genomic evidence establishes Anatolia as the source of the European Neolithic gene pool. Curr Biol. 2016;26(2):270–275. doi: 10.1016/j.cub.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Raghavan M, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014;505(7481):87–91. doi: 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8(1):e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslie S, et al. Wellcome Trust Case Control Consortium 2; International Multiple Sclerosis Genetics Consortium The fine-scale genetic structure of the British population. Nature. 2015;519(7543):309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dorp L, et al. Evidence for a common origin of blacksmiths and cultivators in the Ethiopian Ari within the last 4500 years: Lessons for clustering-based inference. PLoS Genet. 2015;11(8):e1005397. doi: 10.1371/journal.pgen.1005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoop UD. 2005. Das anatolische Chalkolithikum. Eine chronologische Untersuchung zur vorbronzezeitlichen Kultursequenz im nördlichen Zentralanatolien und den angrenzenden Gebieten [The Anatolian Chalcolithic Period. A chronological analysis of the Early Bronze Age cultural-sequence in northern Central Anatolia and neighbouring regions]. (Urgeschichtliche Studien I) (Bernhard Albert Greiner, Remshalden, Germany). German.
- 31.Korfmann M, et al. Kumtepe 1993. Bericht über die Rettungsgrabung [Kumtepe 1993. Report on the rescue excavation] Stud Troica. 1995;5:237–289. German. [Google Scholar]
- 32.Whittle AWR. Europe in the Neolithic: The Creation of New Worlds. Cambridge Univ. Press; Cambridge, UK: 1996. pp. 1–460. [Google Scholar]
- 33.Özdoğan M. 2010. Westward expansion of the Neolithic way of life: Sorting the Neolithic package into distinct packages. Proceedings of the 6th International Congress of the Archaeology of the Ancient Near East, eds Matthiae P, Pinnock F, Nigro L, Marchetti N (Harrassowitz, Wiesbaden, Germany), Vol 1, pp 883–897.
- 34.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40(1):e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29(13):1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu Q, et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 2013;23(7):553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vianello D, et al. HAPLOFIND: A new method for high-throughput mtDNA haplogroup assignment. Hum Mutat. 2013;34(9):1189–1194. doi: 10.1002/humu.22356. [DOI] [PubMed] [Google Scholar]
- 41.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ralf A, van Oven M, Zhong K, Kayser M. Simultaneous analysis of hundreds of Y-chromosomal SNPs for high-resolution paternal lineage classification using targeted semiconductor sequencing. Hum Mutat. 2015;36(1):151–159. doi: 10.1002/humu.22713. [DOI] [PubMed] [Google Scholar]
- 43.Wang C, Zhan X, Liang L, Abecasis GR, Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet. 2015;96(6):926–937. doi: 10.1016/j.ajhg.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192(3):1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voight BF, et al. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci USA. 2005;102(51):18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.