Significance
Physical dormancy, often called hardseededness, exists widely in higher plants. This adaptive trait allows plants to maintain a seed bank in soil, and thus survive unfavorable environmental conditions. The molecular mechanism of physical dormancy is largely elusive. We used the model legume plant, Medicago truncatula, to study this trait because this species produces seeds with typical physical dormancy. By screening a large M. truncatula population tagged with a retrotransposon, we isolated mutant seeds that lost physical dormancy. Genetic and molecular analyses revealed that a class II KNOTTED-like homeobox (KNOX) gene, KNOX4, controls physical dormancy by regulating seed-coat cuticle development. The study provides an understanding of the molecular mechanism of physical dormancy and offers a perspective for seed plant evolution.
Keywords: seed physical dormancy, class II KNOX, hardseededness, Medicago truncatula, seed coat
Abstract
Physical dormancy of seed is an adaptive trait that widely exists in higher plants. This kind of dormancy is caused by a water-impermeable layer that blocks water and oxygen from the surrounding environment and keeps embryos in a viable status for a long time. Most of the work on hardseededness has focused on morphological structure and phenolic content of seed coat. The molecular mechanism underlying physical dormancy remains largely elusive. By screening a large number of Tnt1 retrotransposon-tagged Medicago truncatula lines, we identified nondormant seed mutants from this model legume species. Unlike wild-type hard seeds exhibiting physical dormancy, the mature mutant seeds imbibed water quickly and germinated easily, without the need for scarification. Microscopic observations of cross sections showed that the mutant phenotype was caused by a dysfunctional palisade cuticle layer in the seed coat. Chemical analysis found differences in lipid monomer composition between the wild-type and mutant seed coats. Genetic and molecular analyses revealed that a class II KNOTTED-like homeobox (KNOXII) gene, KNOX4, was responsible for the loss of physical dormancy in the seeds of the mutants. Microarray and chromatin immunoprecipitation analyses identified CYP86A, a gene associated with cutin biosynthesis, as one of the downstream target genes of KNOX4. This study elucidated a novel molecular mechanism of physical dormancy and revealed a new role of class II KNOX genes. Furthermore, KNOX4-like genes exist widely in seed plants but are lacking in nonseed species, indicating that KNOX4 may have diverged from the other KNOXII genes during the evolution of seed plants.
Seed dormancy is a complex adaptive trait of higher plants that allows embryos to survive extended periods of unfavorable environmental conditions (1). Over the course of evolution, plant seeds have evolved diverse dormancy forms in response to various climates to maintain viability (2). Physical dormancy, also termed hardseededness, is an exogenous dormancy caused by a water-impermeable hard seed coat. Physical dormancy plays a key role in protecting seeds against microbial attacks and predators and maintaining seed banks in soils (3–5). This type of dormancy has been found in at least 17 plant families and is very common in legume species (2, 6, 7). Seed exhibiting physical dormancy is characterized by specific morphological features, including a water-impermeable palisade cell layer covered by intact cuticles in the hard seed coat. This structure blocks water and oxygen from its surrounding environment and preserves seeds in a viable status for a long time, sometimes for more than hundreds of years (8). Another common type of seed dormancy is physiological dormancy, which is caused by endogenous factors (e.g., abscisic acid) in embryos. Most molecular studies on seed dormancy have focused on physiological dormancy, using model species Arabidopsis thaliana (9, 10). However, A. thaliana seed is water-permeable, and thus not a suitable model to study physical dormancy.
Current knowledge about physical dormancy mainly comes from studies on morphological structure, phenolic content, and cuticle composition in legume species. Typically, wild legumes produce impermeable (hard) seeds, whereas domesticated cultivars tend to produce permeable (soft) seeds, despite a wide range of variation in water imbibition rate (11–14). Careful scraping of a small outer layer of seed coat (deposits and cuticle) or brief disruption of outer cuticle changed the impermeable seeds to permeable ones (15). Morphological observation indicated that seed hardness was associated with the structure of palisade and cuticular layer (16). Numerous minute cracks were observed in permeable cultivated soybean seeds, but not in impermeable cultivated and wild soybean seeds (13, 16). Chemical composition analysis revealed that the impermeable seeds contained a higher amount of hydroxylated fatty acids than permeable seeds (14). At the molecular level, seed hardness has been identified as a quantitative trait in soybean (17, 18). With quantitative trait locus marker analysis, two single nucleotide polymorphism alleles were found to be associated with a seed coat permeability trait in various soybean cultivars (19, 20). The mechanisms that change seed coat properties remain unknown. In contrast to the vast amount of available information regarding hormone-mediated physiological dormancy (9), and partially because of the lack of a good model system, the understanding of the molecular mechanism controlling physical dormancy is very limited.
In recent years, Medicago truncatula has been developed into a model legume and effectively used for studying compound leaf development and nitrogen fixation (21–25). The seeds of M. truncatula belong to a classic hardseededness type. In general, wild-type M. truncatula seeds display strict physical dormancy and do not germinate unless the dormancy is disrupted by scarification. Thus, M. truncatula is an ideal model to study seed physical dormancy. To identify mutants with defects in physical dormancy, we screened a large number of retrotransposon-tagged M. truncatula lines and successfully isolated nondormant seed mutants. Detailed molecular analyses revealed that a class II KNOX (KNOTTED-like homeobox) gene, KNOX4, was responsible for the physical dormancy phenotype. The loss of function of KNOX4 caused dysfunctional palisade cuticles in the mutant seeds, allowing the seeds to absorb water easily. The results demonstrate a novel molecular mechanism underlying physical dormancy and provide implications for understanding seed plant evolution.
Results
Isolation and Characterization of Mutants Without Physical Dormancy.
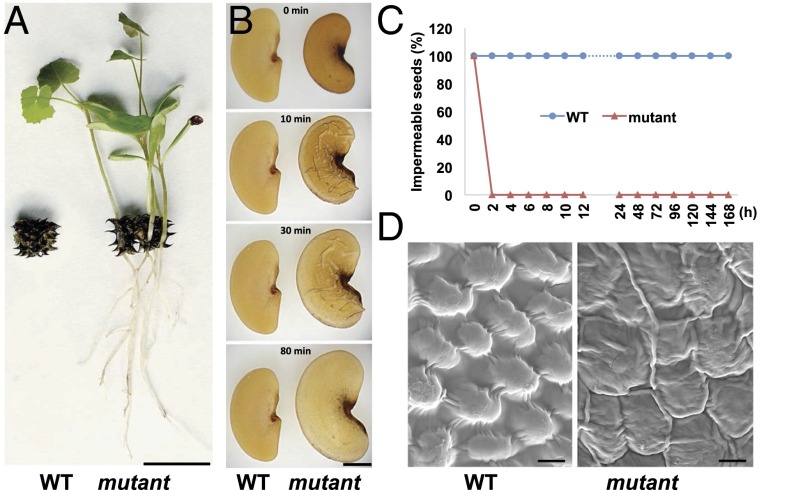
Wild-type M. truncatula seeds do not absorb water or germinate without scarification, even when covered with water. By screening more than 10,000 M. truncatula mutant lines tagged with the transposable element of tobacco (Nicotiana tabacum) cell type 1 (Tnt1 retrotransposon), we identified five mutants that showed no physical dormancy. The mutant seeds were able to germinate directly within intact mature pods and grew into seedlings in the greenhouse (Fig. 1A). To confirm the mutant phenotype, we investigated the seed imbibition process in sterile water and found that the mutant seeds imbibed water very quickly, without any scarification treatment (Movie S1). Seed swelling was observed in the mutants after merely 10 min in water, whereas the wild-type seeds remained static after days in water (Fig. 1 B and C). More than 90% of mutant seeds were able to germinate without scarification, whereas the wild-type seeds showed a similar germination rate only after scarification treatment (SI Appendix, Fig. S1A). These results demonstrated that the mutant seeds lost physical dormancy. Scanning electron microscopy (SEM) was used to detect the differences in seed coat morphology between the mutant and the wild-type. In wild-type seeds, the surface was very smooth, with regularly arranged dome-like epidermal cells. In contrast, the mutant seed surface was wrinkled, with an irregular shape (Fig. 1D).
Fig. 1.
Isolation and characterization of M. truncatula mutant seeds lacking physical dormancy. (A) Wild-type (WT) and mutant seed pods of M. truncatula. Mutant seedlings were able to grow out directly from a mature pod. (Scale bar, 1 cm.) (B) Imbibition of wild-type and mutant seeds in sterile water. (Scale bar, 1 mm.) (C) Percentage of impermeable (hard) seeds in water at multiple points. h, hour. (D) SEMs of mature seeds coat from wild-type and mutant. (Scale bar, 1 µm.)
Physical Dormancy Is Controlled by KNOX4.
To study the genetics of the physical dormancy trait, we crossed the mutants with the wild-type and produced F1 and F2 populations. Seeds from all F1 plants were like the wild-type and showed physical dormancy. In the F2 populations, the segregation ratio of dormant and nondormant seeds was ∼3:1 (SI Appendix, Fig. S1B). The results showed that the loss of physical dormancy in the mutant was caused by a single recessive gene. To identify the gene responsible for physical dormancy, we performed thermal asymmetric interlaced PCR (TAIL-PCR) to recover the flanking sequences from a mutant line, and one sequence was found to be associated with the mutant phenotype. Sequence analysis revealed that the Tnt1 retrotransposon was inserted in the first exon region of the gene Medtr5g011070. On the basis of sequence similarity and structure analyses, Medtr5g011070 codes for a class II KNOX protein named KNOX4. Among the five mutants analyzed, four lines (mtknox4-1, mtknox4-2, mtknox4-3, and mtknox4-4) carried a single Tnt1 retrotransposon insertion (SI Appendix, Fig. S2A). Transcripts were not detectable by RT-PCR in these lines (SI Appendix, Fig. S2B). The fifth line (mtknox4-5) had a single base-pair deletion in the fourth exon and induced a reading-frame shift mutation (SI Appendix, Fig. S2A).
To further confirm the function of KNOX4 in seed physical dormancy, we performed a complementation experiment. The KNOX4 coding sequence driven by its native promoter was transformed into homozygous mtknox4-1 plants. Seeds harvested from the transgenic plants showed physical dormancy similar to that of the wild-type seeds (SI Appendix, Fig. S2C).
Expression Pattern of KNOX4.
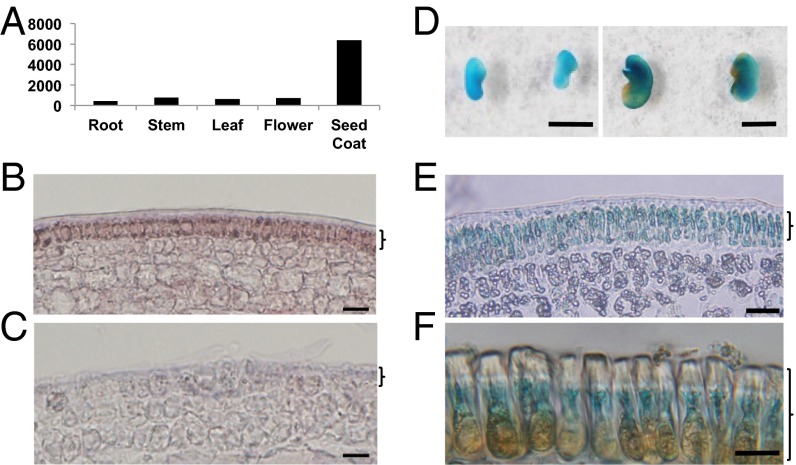
According to the M. truncatula gene expression atlas (mtgea.noble.org/v2), KNOX4 expression is highest in the seed coat compared with other tissues and organs (Fig. 2A). To investigate the detailed expression pattern, we performed in situ hybridization analysis of KNOX4 mRNA in the seed coat tissues. Strong signals were detected in the outermost epidermal cells in the seed coat (Fig. 2 B and C). We also isolated the KNOX4 promoter, fused the promoter with the β-glucuronidase (GUS) gene, and introduced the construct into wild-type M. truncatula. Strong GUS activity was detected in immature seed coat and palisade cells of mature seed coat (Fig. 2 D–F).
Fig. 2.
Expression pattern of KNOX4 in M. truncatula. (A) KNOX4 expression profile in different plant organs. (B and C) In situ hybridization analysis of KNOX4 mRNA in immature seed coat of the wild-type with antisense probe (B) and sense probe (C). (Scale bar, 10 µm.) (D) KNOX4 promoter-GUS expression in immature (Left) and mature (Right) seed coat. (Scale bar, 0.2 cm.) (E and F) KNOX4 promoter-GUS expression in immature seed coat (E) and mature palisade cells of seed coat (F). Brackets B, C, and E indicate epidermis of immature seed coat that will develop into palisade layer (F) in mature seed coat. (Scale bar, 10 µm.)
Loss of Function of KNOX4 Dramatically Altered Seed Coat Architecture.
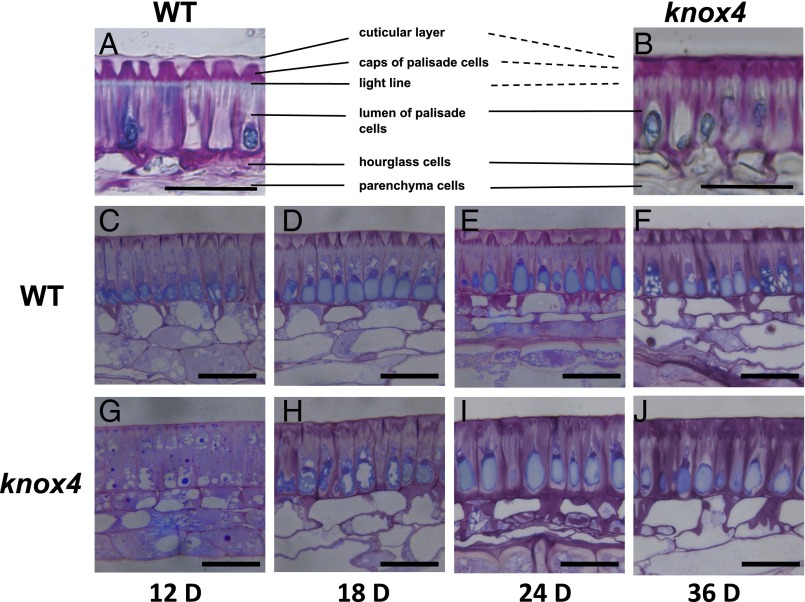
The palisade layer covered by cuticle is the outermost cell layer of seed coat and plays a key role in seed germination (8, 16). We observed that many microcracks formed in the mutant seeds, but not in the wild-type, when the seeds were placed under vacuum for SEM observation (SI Appendix, Fig. S3). This finding suggests that the mutant palisade cuticle was very fragile, and physical change has happened in the mutant seed coat. To further investigate the structural differences between wild-type palisade cuticle layers and those of the mutant, seed coat cryosections stained with toluidine blue were observed. In the wild-type, the palisade cells were well covered with a regularly continuous cuticle layer; however, such a cuticle layer was impaired in the mutant (Fig. 3 A and B). In addition, a clear light line under the cuticle layers was observed in the wild-type (Fig. 3A), whereas the line disappeared in the mutant (Fig. 3B). To track the development process of the palisade cuticle layer, we prepared and examined cross sections of the wild-type and mutant seed coats at different developmental stages. The cuticle layers started to show significant differences at day 12, and the differences became more obvious at later stages (Fig. 3 C–J). Thus, the knox4 mutant developed a structurally anomalous cuticle. These results indicated that KNOX4 was involved in seed coat development.
Fig. 3.
Loss of function of KNOX4 showed developmental defects in seed coat structure in M. truncatula. (A and B) Transverse sections of seed coat at maturation stage of the wild-type (A) and knox4 mutant (B). (Scale bar, 20 µm.) (C–J) Transverse sections of wild-type and knox4 mutant seed coats from different seed developmental stages, stained with 0.05% Toluidine blue O. (Scale bars, 20 µm.) D, days after pollination.
Mutation of KNOX4 Altered Seed Coat Cuticle Composition.
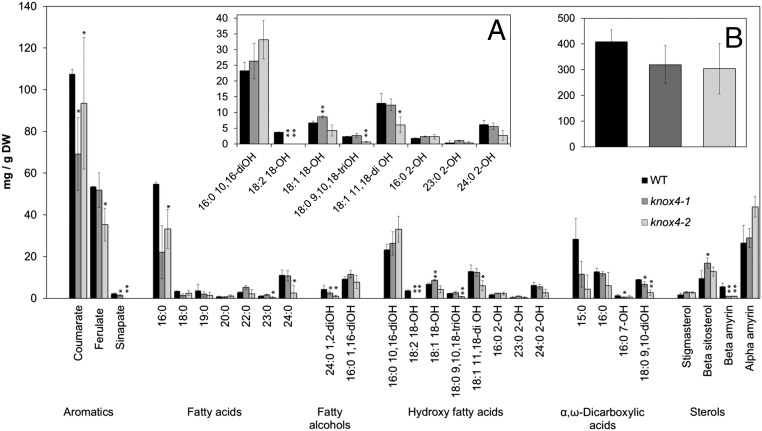
To evaluate the changes that happened in the mutant seed coat cuticle layer, we analyzed cuticle composition. Mature seeds from the wild-type and knox4-1 mutant were first immersed in chloroform to extract cuticular waxes, which were largely composed of 16:0–24:0 fatty acids and lacked components typically associated with plant cuticles; namely, n-alkanes, 1-alkanols, and wax esters. No significant differences were found between the total waxes extracted from the wild-type and knox4-1 seed surfaces (SI Appendix, Fig. S4). However, significant reductions in the loads of very long chain acids and β-amyrin were observed. To determine monomer composition of the polymeric component of the cuticle, delipidated residues from whole seeds were subjected to sodium methoxide-based transmethylation (26). This method bypasses cuticle isolation steps, and therefore determines lipid polyester monomers from seed coat integuments and embryo. Overall, the mutant seeds contained lower amounts of cutin than the wild-type, but such reduction was not significant (t test, two-sided, P < 0.05). At the individual monomer level, the major change was observed in the loads of 18:2 18-OH in both mutants. Differences in the content of aromatics, fatty acids, fatty alcohols, and α, ω-dicarboxylic acids were not so consistent in the two mutants, with some monomers presenting significant differences from the wild-type in the knox4-1 or knox4-2 samples, but not in both (Fig. 4). Deficiency in hydroxylated fatty acids may have altered cuticle layer permeability (14), which is a possible reason for the change in physical dormancy. Changes in some sterols were also observed, but these membrane lipids are not believed to be cutin components.
Fig. 4.
Lipid polyester monomer composition in wild-type and mutant M. truncatula seeds. Mean and SD bars are shown (n = 4). Asterisks denote a significant difference with WT (t test, two-sided, P < 0.05). (A) Expanded view of hydroxy fatty acid monomers. (B) Total loads of identified lipid polyester monomers.
KNOX4 Directly Regulates Cuticle Biosynthetic Genes.
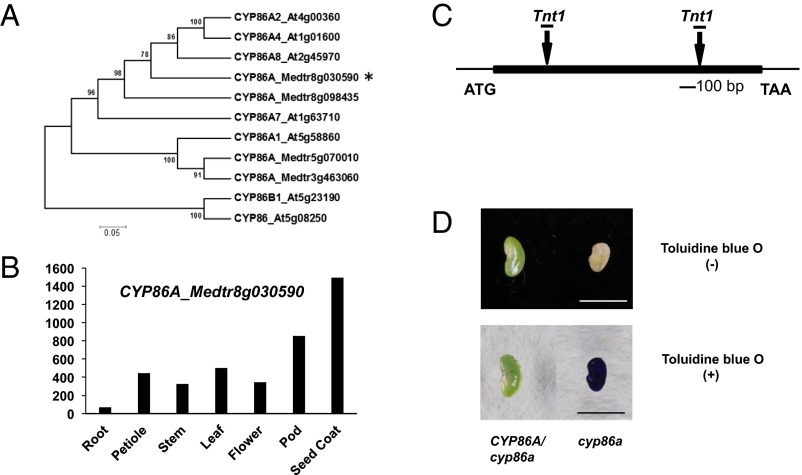
We performed microarray experiments with isolated seed coat tissues from the wild-type and the mutant to investigate potential downstream genes of KNOX4. The results indicated that more than 500 probe sets were down-regulated more than twofold in the knox4 mutants (SI Appendix, Fig. S5). According to Gene Ontology and Mapman analysis, genes involved in lipid metabolism and cell wall pathways were significantly down-regulated (SI Appendix, Fig. S6). Several key genes related to cuticle biosynthesis were identified, such as the cytochrome P450-dependent fatty acid omega-hydroxylase (Medtr8g030590) and fatty acid elongase 3-ketoacyl-CoA synthase (Medtr2g114190) (SI Appendix, Table S1). Phylogenetic analysis indicated that Medtr8g030590 is an ortholog of Arabidopsis CYP86A (Fig. 5A). The CYP86A family plays an important role in the cuticle biosynthetic pathway (27). M. truncatula CYP86A genes showed high expression level in seed coat (Fig. 5B). We isolated two CYP86A loss-of-function mutant alleles from the Tnt1-tagged mutant population (Fig. 5C). The mutant seed coat showed high permeability to toluidine blue staining assay (Fig. 5D), but seeds from cyp86A homozygous plants were aborted. All heterozygous plants still produced seeds with physical dormancy. This observation indicates that CYP86A is critical for seed coat permeability in M. truncatula.
Fig. 5.
Characterization of CYP86A mutants in M. truncatula. (A) Phylogenetic analysis of CYP86A gene family members in M. truncatula and Arabidopsis. CYP86A_Medtr8g030590 is indicated with an asterisk. The scale bar indicates the sequence divergence is 0.05 per unit bar. (B) Transcript level of CYP86A_Medtr8g030590 gene in different plant organs. (C) Schematic diagram of gene structure and the two allele sites of CYP86A (Medtr8g030590). Boxes represent exons and lines represent introns. (D) Permeability of cuticles of CYP86A mutants stained with toluidine blue. Seed coats were immersed in 0.05% toluidine blue O for 2 min and rinsed with water. (Left) Seeds from CYP86A heterozygous plants. (Right) Seeds from CYP86A homozygous plants. (Scale bar, 0.5 cm.)
To investigate in vivo associations between KNOX4 and CYP86A, we performed chromatin immunoprecipitation (ChIP) assay, using a transgenic line that expressed a fusion of green fluorescent protein (GFP) with KNOX4 in the mutant background (SI Appendix, Fig. S7A). The line produced seeds with physical dormancy (SI Appendix, Fig. S7B). With seed coat tissues from the material, our ChIP-PCR data demonstrated that KNOX4 protein directly bound to the promoter regions of MtCYP86A (SI Appendix, Fig. S7C), suggesting KNOX4 regulates the cuticle biosynthesis pathway during M. truncatula seed coat development.
KNOX4-Like Genes Are Involved in Seed Plants Evolution.
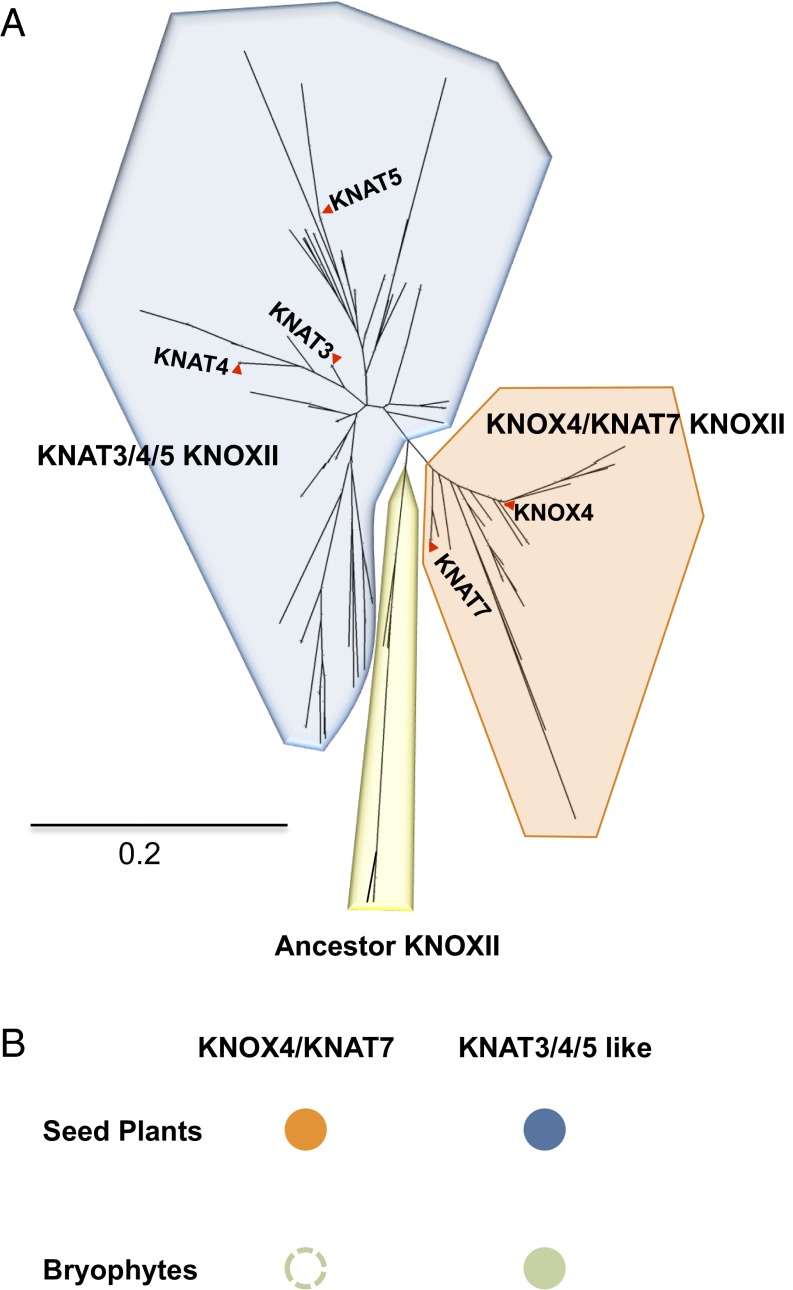
Formation of physical dormancy is a crucial innovation of many seed plants to endure unfavorable conditions of land life. Here, we evaluated the evolutionary history of KNOX4-like genes in seed plants in comparison with nonseed plants bryophytes (SI Appendix, Table S2). In the large phylogeny of class II KNOX transcription factors, KNOX4-like genes form one separate clade. All the remaining KNOXII genes form another clade; many genes in this clade await functional characterization. Interestingly, the analysis showed that KNOX4-like genes only exist in seed plants (Fig. 6A), whereas no genes homologous to KNOX4 were found in nonseed plant species (Fig. 6B). Other KNOXII genes exist in both nonseed and seed plant species. Thus, KNOX4 may have diverged from the other KNOXII genes with the appearance of seed plants and kept a conserved function in the seed coat during the evolution for adaption to land life.
Fig. 6.
Phylogeny of KNOXII homologous proteins from 46 plant species. (A) Phylogenetic analysis of 201 KNOXII protein sequences with neighbor-joining tree in 46 plant species. Representative sequences are highlighted by names. (B) A simplified illustration of KNOXII subclass in bryophytes and seed plants. Presence of a subclass is indicated with a solid closed circle. Absence of a subclass is indicated with an open circle.
Discussion
The presence and absence of physical dormancy in the wild-type and mutant seeds of M. truncatula were distinct and clear, thus setting the basis for detailed genetic analysis and elucidation of the underlying molecular mechanism of this important ecological and agronomic trait. We identified multiple mutant lines and, to our knowledge for the first time, isolated a critical gene controlling physical dormancy in a species that typically produces hard seeds. KNOX4 belongs to the class II KNOX family in plants. Different from class I KNOX genes, which are well known for their roles in maintenance of shoot apical meristem (25, 28–31), class II KNOX genes show diverse expression patterns and few known functions (30, 32, 33). This study revealed a previously unidentified function of this class II KNOX gene.
Anatomical analysis showed that loss of function of KNOX4 altered the integrity of cuticles and cell wall structure. Chemical analysis revealed a difference in cutin monomer composition between the mutants and wild-type seed coats, with 18:2 ω-hydroxy fatty acid being particularly affected. Therefore, loss of function of KNOX4 resulted in abnormal cuticle composition that led to altered seed coat structure and permeability (SI Appendix, Fig. S8).
Microarray and ChIP-PCR analyses identified CYP86A as one of the downstream target genes of KNOX4. CYP86A is a cytochrome P450 monooxygenase and is associated with extracellular lipid polyester biosynthesis. In Arabidopsis, CYP86A2/ATT1, CYP86A4, and CYP86A1 contribute to the modification of aliphatic lipid polyester monomers in leaf, flower, and seed, respectively (34–37), whereas CYP86A8/LCR participates in cutin formation in various organs (34). Loss-of-function mutants of Arabidopsis CYP86A8/LCR and ATT1 presented increased epidermal permeability (34, 36). In tomato (Solanum lycopersicum), a CYP86A mutant showed a thinner cuticle layer, reduced amount of cutin monomers, and faster water loss in fruit than the corresponding wild-type (38). The CYP86A mutant we identified in M. truncatula showed strong seed coat permeability demonstrated by toluidine blue staining. Similarly, seed coat of the knox4 mutant was also stainable by toluidine blue (SI Appendix, Fig. S9). These results indicate that at least one of the mechanisms of KNOX4 in controlling physical dormancy is through the regulation of genes involved in cuticle formation.
It has been shown that seeds of M. truncatula have weak (nondeep) physiological dormancy and that such dormancy is removed during after-ripening (7). The kinetics of physical dormancy release are slower than that of physiological dormancy, making it possible to uncouple the two types of dormancies (7). Graeber et al. (10) summarized all described genes associated with the induction and release of physiological dormancy. None of these genes was affected in our microarray analysis, indicating that KNOX4 is only involved in physical dormancy in M. truncatula.
Physical dormancy is lost or partially lost during the domestication process in legume crops. For example, cultivated soybeans show different degrees of seed coat permeability that are controlled by quantitative trait loci (17, 18). Recently, a quantitative trait locus encoding calcineurin-like protein, GmHs1-1, was isolated by map-based cloning and reported to contribute to hardseededness in soybean with unknown mechanism (20). At the same time, an endo-1,4-β-glucanase gene, qHS1, was cloned by fine mapping and reported to be responsible for the formation of minute cracks by changing cell wall structure (19). Interestingly, these two genes are adjacent to each other in the physical map. The expression levels of the GmHs1-1 and qHS1 homologs in M. truncatula did not show significant changes in our knox4 mutant, suggesting different pathways may control seed physical dormancy.
Compared with lower plants such as bryophytes, seed plants have evolved and developed into abundant species and widely adapted to a range of environments from arid deserts to tropical regions. The seed coat protecting the embryo and other storage tissues provides an evolutionary advantage for seed survival, propagation, and succession. Our phylogenetic analysis revealed that KNOX4-like genes exist widely in seed plants but are lacking in nonseed species. Interestingly, KNOTTED-LIKE FROM ARABIDOPSIS THALIANA7 (KNAT7), a KNOX4-like gene in Arabidopsis, also showed high transcript level in seed coat and modulates mucilage deposition in the seed coat of Arabidopsis (39). Mucilage in plants plays an important role in the storage of water, which is beneficial for seed germination. This indicates that in species without physical dormancy, KNOX4-like genes may still play a role in protecting the embryos. Taken together, the KNOX4 transcription factors may have a conserved function during seed evolution for adaption to land life in seed plants.
In summary, we have shown that M. truncatula is an ideal model for studying physical dormancy of seeds. KNOX4 is a key regulator for seed coat development and controls seed physical dormancy by affecting downstream cuticle genes. This study elucidated a novel molecular mechanism of physical dormancy and revealed a new role of class II KNOX genes. Furthermore, the research shed light on the molecular evolution of seed plants.
Materials and Methods
Plant Materials and Growth Conditions.
M. truncatula ecotype R108 was used as the wild-type for all the experiments in this paper. All knox4 mutant alleles were isolated from a Tnt1 retrotransposon-tagged mutant collection of M. truncatula. TAIL-PCR and RT-PCR were performed to determine the defective gene that caused the nondormant phenotype. Plants were grown under the following conditions: 22 °C/20 °C day/night temperature, 16-h/8-h day/night photoperiod, 70–80% relative humidity, and 150 μmol⋅m−2⋅s−1 light intensity.
Microscopic Observations of Seed Coat.
At least 100 intact seeds were used for imbibition assays. Water uptake process was observed, and pictures were taken under microscope. Seeds were rubbed with sandpaper for scarification. For SEM observation, dry mature seeds were cut with a sharp razor blade at the central region. SEM analysis was performed as described previously (24, 40). Thirty-six-day-old seeds were used for cryosectioning, as seeds at this stage have suitable hardness for cutting with Cryostat Leica CM 1850 without fixation treatment. To investigate seed coat structure during development, seeds from different stages were soaked in sterile water and then fixed with 2.5% (vol/vol) glutaraldehyde and 4% (vol/vol) paraformaldehyde in PBS buffer (pH 7.2) for 12 h at 4 °C. They were then washed with PBS and postfixed in 1% (vol/vol) osmium tetroxide for 2 h at 0 °C in the dark. After washing again with PBS, they were dehydrated in a series of ethanol dilutions and embedded in LR White resin (London Resin) and polymerized at 60 °C for 2 d. Serial 1-μm cross sections (approximately) were cut in the middle of the seed, using Leica EM UC7 Ultramicrotome (Leica Mikrosysteme). Semithin sections were then stained with 1% (wt/vol) toluidine blue O [with 1% (wt/vol) sodium borate] and observed under a Nikon Microphot-2 (Nikon Corporation).
RNA Extraction, RT-PCR, and Microarray Analysis.
Seed coat was harvested from 20-d-old seed for RNA extraction. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen). Two micrograms of RNA were reverse transcribed with SuperScriptIII (Invitrogen), according to the manufacturer’s instructions. For microarray analysis, RNA purification, probe labeling, hybridization, scanning, and microarray data analysis were performed as previously described (40).
In Situ Hybridization Analysis.
The fragments of 503-bp KNOX4 were amplified by PCR and then labeled with digoxigenin-11-UTP (Roche Diagnostics). Seed coat of 12-d-old wild-type seeds was fixed, and RNA in situ hybridization was performed as previously described (40).
GUS Staining.
Immature and mature (before dehydration) seeds were collected for GUS staining. GUS activity was histochemically detected as previously described (41). Cross section was performed to observe the GUS signal in seed coat.
Lipid Polyesters Analysis.
The wild-type and mutant dry seed samples were ground, and dry residues were depolymerized by methanolysis in the presence of sodium methoxide. The depolymerization products extracted by CH2Cl2 were derivatized by treatment with N,O Bis-trimethylsilyl-trifluoroacetamide to form trimethylsilyl ethers, and the monomers were analyzed by gas chromatography (26, 42).
Plasmids and Plant Transformation.
For functional complementation, a 6.5-kb MtKNOX4 genomic sequence including promoter and the coding sequence were amplified from the wild-type, and then inserted into pENTR/d-TOPO cloning vector (Invitrogen). The positive clone was transferred into the pEarleyGate 301 vector, using the Gateway LR recombination reactions (Invitrogen). The resulting construct was introduced into Agrobacterium tumefaciens EHA105 strain. Sterile mutant seedlings were used for stable transformation.
ChIP Assay.
Immunoprecipitation was conducted using the transgenic plants expressing Pro35S::GFP-KNOX in the knox4 mutant background with GFP fluorescence signal and showing normal seed physical dormancy phenotype. The wild-type plants were used as control. Three hundred milligrams fresh seed coat were fixed in 1% formaldehyde and washed three times. ChIP assay was performed according to the method previously described (43, 44). Anti-GFP antibody (Roche) was used for the immunoprecipitation. The precipitated DNA was dissolved in TE buffer (10 mM Tris, 1 mM EDTA at pH 8.0) for quantitative PCR. M. truncatula ubiquitin gene was used as the control.
Phylogenetic Analysis.
The amino acid sequences of KNOXII family members from nonseed plants such as moss and seed plants including different angiosperm species were included for phylogenetic analysis (SI Appendix, Table S2). Alignments were performed with Clustal Mega with default parameters (www.ebi.ac.uk/Tools/msa/clustalo/). The MEG6 program was used to make the phylogenetic tree, using the neighbor joining with 1,000 bootstrap replications (www.megasoftware.net).
Supplementary Material
Acknowledgments
We thank Amy Mason and Courtney Leeper for critical reading of the manuscript; Dr. Kirankumar Mysore for providing the Tnt1 mutants; Xiaofei Cheng, Kuihua Zhang, and Jiangqi Wen for assistance with screening the Tnt1 mutants; and the greenhouse staff for assistance with plant growth. This work was supported by The Samuel Roberts Noble Foundation and the BioEnergy Science Center. The BioEnergy Science Center is a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the Department of Energy Office of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1601256113/-/DCSupplemental.
References
- 1.Koornneef M, Bentsink L, Hilhorst H. Seed dormancy and germination. Curr Opin Plant Biol. 2002;5(1):33–36. doi: 10.1016/s1369-5266(01)00219-9. [DOI] [PubMed] [Google Scholar]
- 2.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171(3):501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 3.Dalling JW, Davis AS, Schutte BJ, Arnold AE. Seed survival in soil: Interacting effects of predation, dormancy and the soil microbial community. J Ecol. 2011;99(1):89–95. [Google Scholar]
- 4.Paulsen TR, et al. Physical dormancy in seeds: A game of hide and seek? New Phytol. 2013;198(2):496–503. doi: 10.1111/nph.12191. [DOI] [PubMed] [Google Scholar]
- 5.Sugden A. A good hiding place. Science. 2013;339(6124):1125. [Google Scholar]
- 6.Smýkal P, Vernoud V, Blair MW, Soukup A, Thompson RD. The role of the testa during development and in establishment of dormancy of the legume seed. Front Plant Sci. 2014;5:351. doi: 10.3389/fpls.2014.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolingue W, Vu BL, Leprince O, Buitink J. Characterization of dormancy behaviour in seeds of the model legume Medicago truncatula. Seed Sci Res. 2010;20(2):97–107. [Google Scholar]
- 8.Rolston MP. Water impermeable seed dormancy. Bot Rev. 1978;44(3):365–396. [Google Scholar]
- 9.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 10.Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012;35(10):1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x. [DOI] [PubMed] [Google Scholar]
- 11.Ranathunge K, et al. Properties of the soybean seed coat cuticle change during development. Planta. 2010;231(5):1171–1188. doi: 10.1007/s00425-010-1118-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhou S, Sekizaki H, Yang Z, Sawa S, Pan J. Phenolics in the seed coat of wild soybean (Glycine soja) and their significance for seed hardness and seed germination. J Agric Food Chem. 2010;58(20):10972–10978. doi: 10.1021/jf102694k. [DOI] [PubMed] [Google Scholar]
- 13.Ma F, Cholewa E, Mohamed T, Peterson CA, Gijzen M. Cracks in the palisade cuticle of soybean seed coats correlate with their permeability to water. Ann Bot (Lond) 2004;94(2):213–228. doi: 10.1093/aob/mch133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao S, Meyer CJ, Ma F, Peterson CA, Bernards MA. The outermost cuticle of soybean seeds: Chemical composition and function during imbibition. J Exp Bot. 2007;58(5):1071–1082. doi: 10.1093/jxb/erl268. [DOI] [PubMed] [Google Scholar]
- 15.Meyer CJ, Steudle E, Peterson CA. Patterns and kinetics of water uptake by soybean seeds. J Exp Bot. 2007;58(3):717–732. doi: 10.1093/jxb/erl244. [DOI] [PubMed] [Google Scholar]
- 16.Vu DT, Velusamy V, Park E. Structure and chemical composition of wild soybean seed coat related to its permeability. Pak J Bot. 2014;46(5):1847–1857. [Google Scholar]
- 17.Hirata K, et al. Identification of quantitative trait loci associated with boiled seed hardness in soybean. Breed Sci. 2014;64(4):362–370. doi: 10.1270/jsbbs.64.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keim P, Diers BW, Shoemaker RC. Genetic analysis of soybean hard seededness with molecular markers. Theor Appl Genet. 1990;79(4):465–469. doi: 10.1007/BF00226154. [DOI] [PubMed] [Google Scholar]
- 19.Jang SJ, et al. A single-nucleotide polymorphism in an endo-1,4-β-glucanase gene controls seed coat permeability in soybean. PLoS One. 2015;10(6):e0128527. doi: 10.1371/journal.pone.0128527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L, et al. GmHs1-1, encoding a calcineurin-like protein, controls hard-seededness in soybean. Nat Genet. 2015;47(8):939–943. doi: 10.1038/ng.3339. [DOI] [PubMed] [Google Scholar]
- 21.Tadege M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54(2):335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 22.Young ND, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480(7378):520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Q, et al. Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proc Natl Acad Sci USA. 2010;107(32):14496–14501. doi: 10.1073/pnas.1009170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou C, et al. Identification and characterization of petiolule-like pulvinus mutants with abolished nyctinastic leaf movement in the model legume Medicago truncatula. New Phytol. 2012;196(1):92–100. doi: 10.1111/j.1469-8137.2012.04268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou C, et al. STM/BP-like KNOXI is uncoupled from ARP in the regulation of compound leaf development in Medicago truncatula. Plant Cell. 2014;26(4):1464–1479. doi: 10.1105/tpc.114.123885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molina I, Bonaventure G, Ohlrogge J, Pollard M. The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry. 2006;67(23):2597–2610. doi: 10.1016/j.phytochem.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Molina I, Ohlrogge JB, Pollard M. Deposition and localization of lipid polyester in developing seeds of Brassica napus and Arabidopsis thaliana. Plant J. 2008;53(3):437–449. doi: 10.1111/j.1365-313X.2007.03348.x. [DOI] [PubMed] [Google Scholar]
- 28.Hake S, Ori N. Plant morphogenesis and KNOX genes. Nat Genet. 2002;31(2):121–122. doi: 10.1038/ng0602-121. [DOI] [PubMed] [Google Scholar]
- 29.Hake S, et al. The role of knox genes in plant development. Annu Rev Cell Dev Biol. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- 30.Hay A, Tsiantis M. KNOX genes: Versatile regulators of plant development and diversity. Development. 2010;137(19):3153–3165. doi: 10.1242/dev.030049. [DOI] [PubMed] [Google Scholar]
- 31.Schneeberger RG, Becraft PW, Hake S, Freeling M. Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev. 1995;9(18):2292–2304. doi: 10.1101/gad.9.18.2292. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, et al. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell. 2014;26(12):4843–4861. doi: 10.1105/tpc.114.128322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Yang X, Zhao W, Lang T, Samuelsson T. Evolution, diversification, and expression of KNOX proteins in plants. Front Plant Sci. 2015;6:882. doi: 10.3389/fpls.2015.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wellesen K, et al. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega -hydroxylation in development. Proc Natl Acad Sci USA. 2001;98(17):9694–9699. doi: 10.1073/pnas.171285998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li-Beisson Y, et al. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA. 2009;106(51):22008–22013. doi: 10.1073/pnas.0909090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao F, et al. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004;23(14):2903–2913. doi: 10.1038/sj.emboj.7600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Höfer R, et al. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. J Exp Bot. 2008;59(9):2347–2360. doi: 10.1093/jxb/ern101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi JX, et al. The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol. 2013;197(2):468–480. doi: 10.1111/nph.12032. [DOI] [PubMed] [Google Scholar]
- 39.Bhargava A, et al. The interacting MYB75 and KNAT7 transcription factors modulate secondary cell wall deposition both in stems and seed coat in Arabidopsis. Planta. 2013;237(5):1199–1211. doi: 10.1007/s00425-012-1821-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhou C, et al. The trans-acting short interfering RNA3 pathway and no apical meristem antagonistically regulate leaf margin development and lateral organ separation, as revealed by analysis of an argonaute7/lobed leaflet1 mutant in Medicago truncatula. Plant Cell. 2013;25(12):4845–4862. doi: 10.1105/tpc.113.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenkin S, Molina I. Isolation and compositional analysis of plant cuticle lipid polyester monomers. J Vis Exp. 2015;(105):e53386. doi: 10.3791/53386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, et al. Control of plant stem cell function by conserved interacting transcriptional regulators. Nature. 2015;517(7534):377–380. doi: 10.1038/nature13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12(16):1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.