Significance
IFN-γ plays an important role in the elimination of liver-stage Plasmodium parasites, but the mechanism involved in this process is unclear. In this study, we demonstrate that IFN-γ treatment induces a noncanonical autophagy pathway in human hepatocytes dubbed an LC3-associated phagocytosis (LAP)-like process, in which the parasitophorous vacuole membrane of the parasites is decorated with LC3, resulting in the colocalization of parasite compartments with lysosomes. Downstream autophagy-related proteins are involved in this pathway, whereas the upstream autophagy-initiating protein is not. Our work shows that a LAP-like process serves as a previously unidentified downstream effector of IFN-γ in elimination of the liver-stage human malarial parasite Plasmodium vivax.
Keywords: autophagy, LC3-associated phagocytosis, IFN-γ, malaria
Abstract
IFN-γ is a major regulator of immune functions and has been shown to induce liver-stage Plasmodium elimination both in vitro and in vivo. The molecular mechanism responsible for the restriction of liver-stage Plasmodium downstream of IFN-γ remains uncertain, however. Autophagy, a newly described immune defense mechanism, was recently identified as a downstream pathway activated in response to IFN-γ in the control of intracellular infections. We thus hypothesized that the killing of liver-stage malarial parasites by IFN-γ involves autophagy induction. Our results show that whereas IFN-γ treatment of human hepatocytes activates autophagy, the IFN-γ–mediated restriction of liver-stage Plasmodium vivax depends only on the downstream autophagy-related proteins Beclin 1, PI3K, and ATG5, but not on the upstream autophagy-initiating protein ULK1. In addition, IFN-γ enhanced the recruitment of LC3 onto the parasitophorous vacuole membrane (PVM) and increased the colocalization of lysosomal vesicles with P. vivax compartments. Taken together, these data indicate that IFN-γ mediates the control of liver-stage P. vivax by inducing a noncanonical autophagy pathway resembling that of LC3-associated phagocytosis, in which direct decoration of the PVM with LC3 promotes the fusion of P. vivax compartments with lysosomes and subsequent killing of the pathogen. Understanding the hepatocyte response to IFN-γ during Plasmodium infection and the roles of autophagy-related proteins may provide an urgently needed alternative strategy for the elimination of this human malaria.
Several hundred million cases of human malaria are reported annually, and nearly 600,000 people die from the disease each year (1). Of the five species that infect humans, Plasmodium vivax is not only the most geographically widespread, but also the most prevalent malarial parasite in areas outside Africa. As such, it has caused massive morbidity in these regions of the world. Although malaria caused by P. vivax was previously regarded as benign compared with that caused by Plasmodium falciparum, the recent alarming increase in both the severity and the drug resistance of P. vivax infection has raised concern (2).
The widespread distribution of P. vivax has been attributed to the parasite’s ability to remain dormant in the liver for years before reactivation (3). The molecular mechanism responsible for P. vivax dormancy is unknown, and knowledge of Plasmodium–hepatocyte interactions remains very limited. Nonetheless, because the number of liver-stage parasites is in the range of 100, whereas in the blood stage as many as 1013 organisms may be found (4), intervention at the liver stage would seem to offer a better strategy for parasite elimination. A prerequisite to this route of malaria control and the development of novel therapies is a better understanding of liver-stage Plasmodium and its interactions with host hepatocytes.
IFN-γ was previously shown to exhibit antimalarial activity against the liver stages of the mouse malarial species Plasmodium berghei and Plasmodium yoelli, in both cultured hepatocytes and murine models (5–7). In chimpanzees, IFN-γ was shown to protect against liver-stage infection by human malarial P. vivax (8). Studies in mice suggested that IFN-γ prevents liver-stage Plasmodium infection by inducing the expression of inducible nitric oxide synthase (iNOS), an enzyme required for the production of nitric oxide (NO), and hence the reactive nitrogen intermediates (RNIs) thought to cause parasite damage inside hepatocytes (9, 10). However, when infected mice were cotreated with the NO inhibitor NGMMLA, only 50% of the parasites were rescued from IFN-γ–mediated elimination (6). In addition, the role of NO in human host defense against infections remains controversial (11). These results infer the involvement of an as-yet unexplored pathway independent of NO and downstream of IFN-γ. Elucidating this pathway may provide the urgently needed innovative measures to fight malaria. Recently, IFN-γ was shown to induce autophagy, an immune mechanism that results in the killing of intracellular pathogens, including Toxoplasma gondii, an apicomplexan parasite closely related to Plasmodium (12).
Autophagy is a cell-autonomous homeostatic process that normally occurs inside eukaryotic cells at a basal level (basal autophagy) and allows cells to degrade cytoplasmic substances for use as nutrients (13). In addition to basal autophagy, autophagy and autophagy-related processes can be induced by such conditions as starvation, drug exposure, and immune mediators (14). In canonical autophagy, unwanted cytosolic substrates are sequestered into double-membrane–bound organelles, called autophagosomes, and delivered to lysosomes for destruction (13). These substrates include aggregated or long-lived proteins and defunct organelles (15). On responding to an upstream signal, autophagy-related (ATG) proteins are organized into complexes that facilitate autophagosome formation. These complexes consist of (i) the upstream initiating complex ULK1 protein kinase, which activates (ii) the downstream complex Beclin 1/PI3K/ATG14, to generate PI3P for the recruitment of downstream effectors; (iii) the ATG12-ATG5-ATG16 conjugation complex to lipidate LC3, which is required for autophagosome elongation and closure; and (iv) Beclin 1/PI3P/UVRAG, a complex that facilitates the fusion of autophagosomes with lysosomes, resulting in the delivery of lysosomal hydrolases to degrade the engulfed contents (13, 14, 16, 17).
Over the past decade, autophagy has also been increasingly appreciated for its role in immunity against bacteria, parasites, and viruses (14, 16). In response to these infections, autophagy directly sequesters the pathogens into double-membrane autophagosomes and then delivers them to lysosomes, which not only results in their destruction, but also enhances antigen presentation by phagocytes (18). In the case of T. gondii, autophagy induction in host cells by IFN-γ treatment also results in parasite restriction (12), but whether IFN-γ–induced autophagy plays a role in the elimination of liver-stage Plasmodium had not been tested.
In addition to the above-described role of canonical autophagy in the digestion of intracellular pathogens, ATG proteins participate in the elimination of invading microorganisms through a noncanonical autophagy process called LC3-associated phagocytosis (LAP) (19). LAP can be triggered by the engagement of pathogens with cell surface receptors, such as Toll-like receptor 2 and 4 (TLR2 and TLR4) and Dectin-1, resulting in the recruitment of ATG proteins, such as ATG5, to single-membrane phagosomes (20). Unlike canonical autophagy, LAP does not require the autophagy-initiating protein ULK1. In addition, LAP induction leads to the decoration of single-membrane phagosomes with LC3 and their subsequent fusion with lysosomes, resulting in the digestion of the phagocytosed materials (20). Burkholderia pseudomallei, Mycobacterium marinum, Aspergillus fumigatus, and Salmonella typhimurium are among the LAP-restricted pathogens identified thus far (21–24).
In this work, we investigated the roles of autophagy-related proteins in the IFN-γ–induced elimination of liver-stage P. vivax. We found that IFN-γ–mediated killing of liver-stage P. vivax depends on the downstream autophagy-related proteins Beclin 1, PI3K, and ATG5, but not on the upstream protein ULK1. In response to IFN-γ, the enhanced decoration of LC3 onto the PVM of P. vivax and increased colocalization of lysosomes with pathogen-containing compartments are observed. Although this process resembles LAP, it differs from LAP in that in response to IFN-γ, LC3 decorates the PVM, not the phagosomal membrane. These data are the first, to our knowledge, to demonstrate the existence of a LAP-like process acting as a downstream effector of IFN-γ in the control of an intracellular pathogen.
Results
IFN-γ–Mediated Control of Liver-Stage P. vivax in HC04 Cells Is Independent of NO.
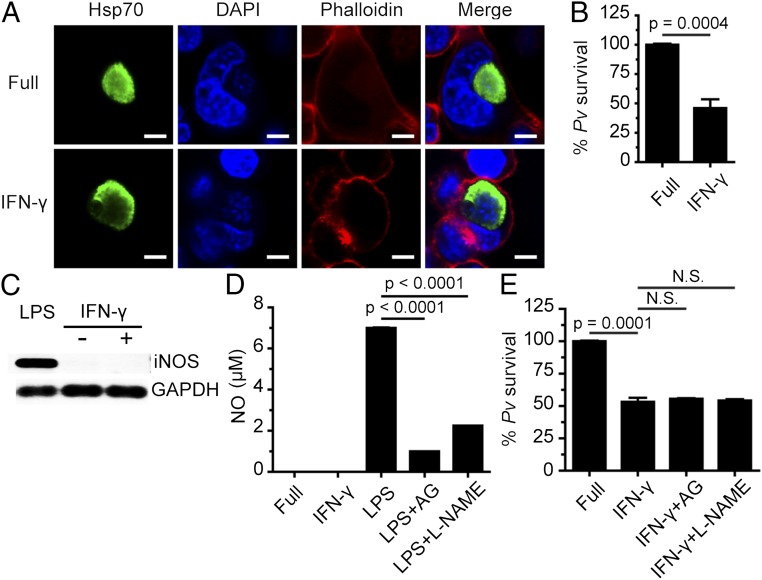
The above-described effect of IFN-γ on the liver stage of P. berghei and P. yoelli in cultured mouse hepatocytes and whole-animal models (5, 7), and the ability of this cytokine to limit the liver stage of human malarial P. vivax replication in chimpanzees (8), suggested a role for IFN-γ in liver-stage P. vivax in humans. Therefore, in this study we asked whether IFN-γ treatment of cultured human hepatocytes infected with P. vivax sporozoites would promote parasite clearance. Accordingly, cells of the hepatocyte cell line HC04 cells were treated with IFN-γ (400 U/mL) for 4 h after P. vivax invasion. The IFN-γ was then washed out and the cells were cultured for 4 more days, at which point the number of surviving intracellular parasites was determined by immunofluorescence analysis (IFA). The results showed a 53.9 ± 7.5% decrease in intracellular P. vivax survival (Fig. 1 A and B). Despite the decreased parasite numbers in the IFN-γ–treated cells, there was no difference in the morphology of surviving parasites harvested on day 4 from treated cultures vs. untreated cultures. In addition, 4 h of IFN-γ treatment (400 U/mL) did not result in a change in HC04 cell survival, as determined by trypan blue exclusion tests of cells harvested at various time points after IFN-γ treatment (Fig. S1). Hence it was highly unlikely that the observed decrease in intracellular P. vivax survival was the result of host cell death.
Fig. 1.
IFN-γ treatment leads to liver-stage P. vivax elimination in cultured human HC04 hepatocytes via an NO-independent process. (A and B) Human HC04 hepatocytes were infected with P. vivax sporozoites (MOI = 1) for 4 h, washed three times with complete medium to remove noninvading sporozoites, and treated with 400 U IFN-γ/mL for 4 h. They were then washed three times and maintained in complete medium until they were harvested on day 4, when they were processed for IFA to quantify the number of surviving parasites. Data are mean ± SEM from at least three independent experiments; the results are expressed relative to the full control, defined as 100%. (Scale bar: 5 µm.) (C) HC04 cells were treated or not with IFN-γ (400 U/mL) for 4 h, after which iNOS mRNA expression was analyzed by RT-PCR. GAPDH served as the internal control. mRNAs from HC04 cells treated with LPS served as the positive control. (D) HC04 cells were treated or not with IFN-γ (400 U/mL) for 4 h, after which NO production in the supernatant was measured using the Griess reaction assay. RAW264.7 cells treated with LPS in the presence or absence of the iNOS inhibitors aminoguanidine (AG) and L-NAME served as the positive control. Data are mean ± SEM from at least three independent experiments. (E) HC04 cells were infected with P. vivax sporozoites as in A and B, washed, treated with IFN-γ with or without the iNOS inhibitors for 4 h, washed again, and maintained in complete medium until day 4, when the surviving parasites were quantified by IFA. Data are mean ± SEM of at least three independent experiments; the results are expressed relative to the full control, defined as 100%. N.S., not significant.
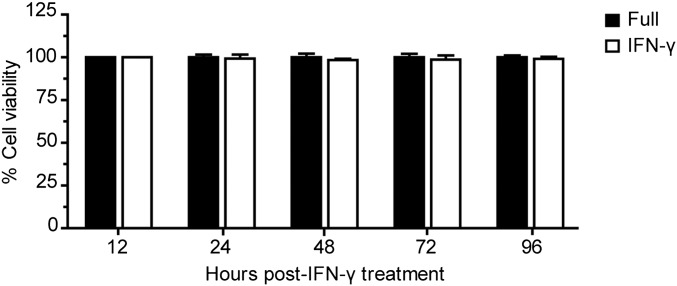
Fig. S1.
IFN-γ treatment does not affect HC04 cell survival. After 4 h of IFN-γ treatment, HC04 cells were harvested at the indicated time points, and their viability was determined using the trypan blue exclusion test. Data are mean ± SEM of three independent experiments; the results are expressed relative to the full control, defined as 100%.
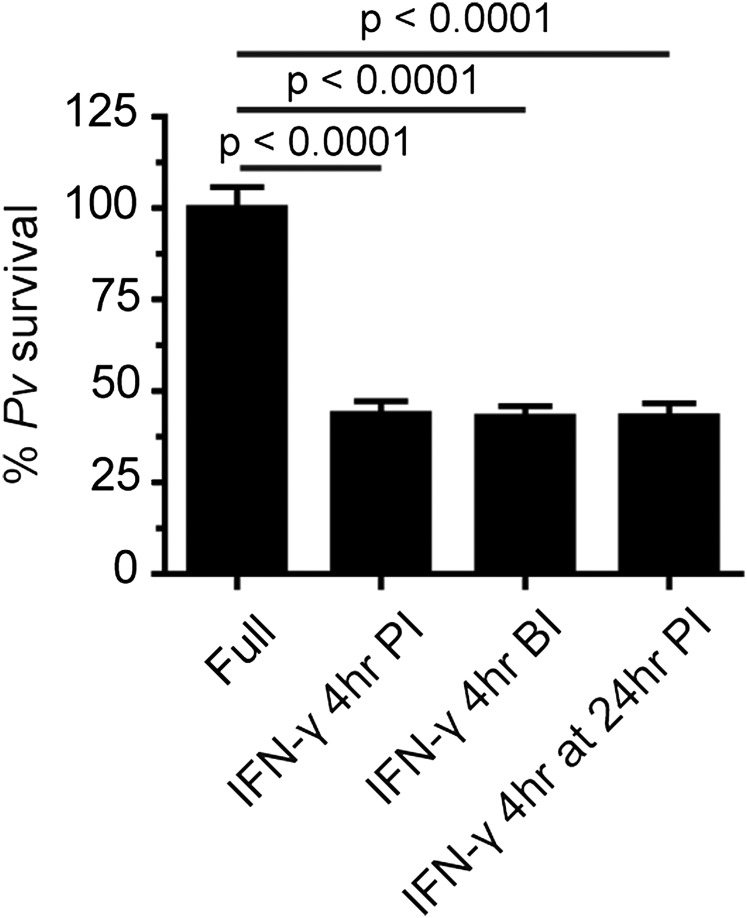
We also examined the effects of different IFN-γ treatment regimens by additionally treating the cells with IFN-γ at 24 h postinfection or at 4 h preinfection (Fig. S2). At least at the IFN-γ dose used in this study (400 U/mL), there was no difference in the level of induced liver-stage P. vivax elimination between the different regimens. Thus, in subsequent experiments, infected HC04 cells were treated with 400 U IFN-γ/mL at 4 h postinfection.
Fig. S2.
Intracellular P. vivax survival in response to different regimens of IFN-γ treatment. Infected HC04 cells were treated with IFN-γ (400 U/mL) for 4 h postinvasion (4 h PI) or for 4 h at 24 h postinvasion (4 h at 24 h PI). Alternatively, the cells were pretreated with IFN-γ (400 U/mL) for 4 h before infection (4 h BI) with P. vivax sporozoites. After IFN-γ washout, the liver-stage parasites were allowed to grow until day 4, at which time the infected cells were harvested for IFA of intracellular P. vivax survival. Data are mean ± SEM of two independent experiments carried out in triplicate; the results are expressed relative to the full control, defined as 100%.
We then investigated the molecular mechanism responsible for the IFN-γ–mediated killing of liver-stage P. vivax in human HC04 cells. Because NO was previously shown to be involved in the IFN-γ–mediated control of liver-stage P. yoelli in mice (6), we examined its effect downstream of IFN-γ in the elimination of liver-stage P. vivax from HC04 cells. However, we found no induction of iNOS, the enzyme responsible for NO synthesis, in HC04 cells treated with IFN-γ for 4 h, as determined by RT-PCR (Fig. 1C). Similarly, a Griess reaction assay showed that, in contrast to the increased NO production in lipopolysaccharide-treated Raw264.7 macrophages, used as positive control cells (Fig. 1D, column 3), and the inhibition of this response on the addition of two iNOS inhibitors, aminoguanidine and L-NAME (Fig. 1D, columns 4 and 5), there was no evidence of IFN-γ–mediated NO production in HC04 cells (Fig. 1D, column 2). Moreover, the IFN-γ–mediated elimination of liver-stage P. vivax was not inhibited by aminoguanidine and L-NAME (Fig. 1E). These results show that NO is not involved downstream of IFN-γ in the reduction of liver-stage P. vivax in cells of the human hepatocyte line HC04.
IFN-γ–Induced Protection Against Liver-Stage P. vivax Depends on Beclin 1 and PI3K.
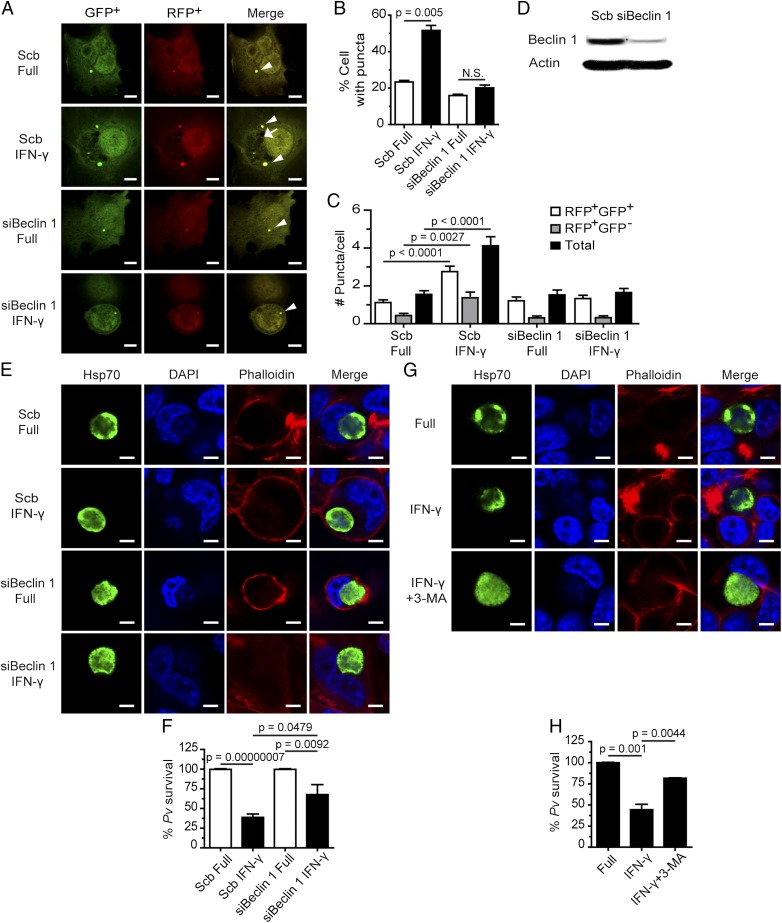
IFN-γ stimulation induces autophagy in various cell types (14). Because liver-stage P. vivax infection of human HC04 cells is attenuated by IFN-γ treatment (Fig. 1 A and B), we asked whether the autophagy pathway and/or ATG proteins are involved in this response. Because Beclin 1 and PI3K were previously shown to be important in both the canonical (autophagosome nucleation and maturation) (25) and noncanonical (LAP) (20) autophagy pathways, we asked whether these proteins are needed in the IFN-γ–mediated restriction of liver-stage P. vivax. To answer this question, we used siRNA technology to knockdown Beclin 1 expression in HC04 cells. The knockdown efficiency was verified both in the LC3 puncta formation assay and by Western blot analysis (Fig. 2 A–D). The former is a functional assay that has been used to demonstrate the requirement for Beclin 1 in IFN-γ–mediated autophagosome formation in other cell types (26).
Fig. 2.
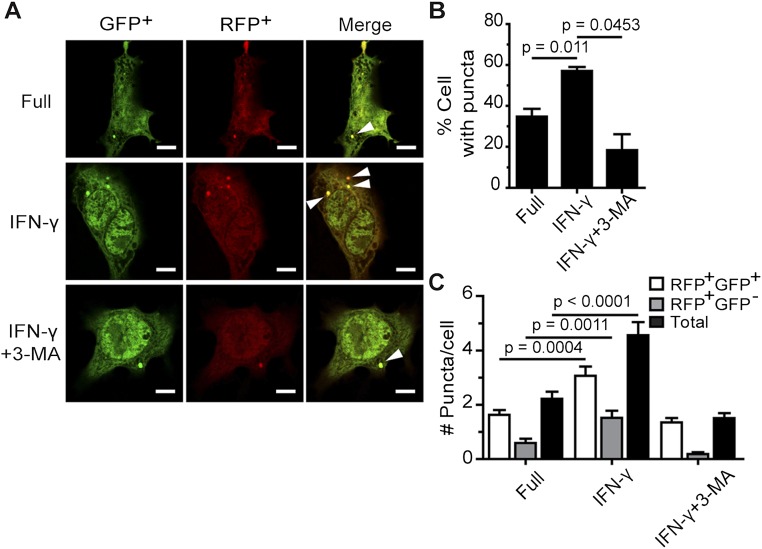
IFN-γ–induced liver-stage P. vivax elimination depends on Beclin 1 and PI3K. (A–C) HC04 cells were transfected with scramble control siRNAs or siRNAs against Beclin 1 and cDNAs encoding RFP-GFP-LC3. At 48 h posttransfection, the cells were treated or not with IFN-γ for 4 h and then processed for fluorescence microscopy. RFP+GFP+-LC3 (autophagosomes) and RFP+GFP−-LC3 (autolysosomes) were quantified, and the percentage of puncta-containing cells was determined by analyzing at least 100 cells per condition from three independent experiments. Only puncta ≥0.25 µm in size were counted. The number of puncta per cell was also quantified in Z-stack images of at least 30 cells per condition per independent experiment. Data are mean ± SEM relative to the full control. N.S., not significant. (Scale bar: 5 µm.) (D) Beclin 1 can be successfully depleted in HC04 cells. HC04 cells were transfected with scramble control siRNAs or siRNAs against Beclin 1 and then harvested at 48 h posttransfection for immunoblot analysis. Actin served as an internal loading control. (E and F) Beclin 1-deficient or -proficient HC04 cells were infected with P. vivax sporozoites at an MOI of 1, followed by IFN-γ treatment for 4 h. The cells were washed and then maintained in complete medium until being processed for IFAs on day 4. Data are mean ± SEM of at least three independent experiments; the results are expressed relative to the full control, defined as 100%. (Scale bar: 5 µm.) (G and H) HC04 cells infected with P. vivax sporozoites were treated with IFN-γ with or without the PI3K inhibitor 3-MA for 4 h, washed, and maintained in complete medium until being processed for IFA on day 4. Data are mean ± SEM of at least three independent experiments; the results are expressed relative to the full control, defined as 100%. (Scale bar: 5 µm.)
After confirming that a 4-h treatment with IFN-γ enhances autophagosome formation in HC04 cells (Fig. S3), we examined this effect in the Beclin 1 knockdown cells and found a decrease in autophagosome formation in response to IFN-γ (Fig. 2 A–C). Immunoblot analysis directly verified the reduced Beclin 1 expression in the knockdown cells (Fig. 2D).
Fig. S3.
IFN-γ treatment increases LC3 puncta formation in HC04 cells. (A–C) HC04 cells were transfected with cDNAs encoding RFP-GFP-LC3. At 48 h posttransfection, they were treated with IFN-γ for 4 h in the presence or absence of 3-MA. RFP+GFP+-LC3 (autophagosomes) and RFP+GFP−-LC3 (autolysosomes) were quantified by determining the percentage of puncta-containing cells among at least 100 cells per condition from three independent experiments. Only puncta ≥0.25 µm in size were counted. The number of puncta per cell was also examined in Z-stack images of at least 30 cells per condition per independent experiment. Data are mean ± SEM. The results are expressed relative to the full control. (Scale bar: 5 µm.)
With the ability to effectively suppress Beclin 1 expression in HC04 cells, we examined whether this protein is involved in the IFN-γ–mediated killing of liver-stage P. vivax. When P. vivax-infected, Beclin 1-proficient HC04 cells were treated with IFN-γ for 4 h, parasite survival was efficiently reduced by 60.7 ± 3.2% (Fig. 2 E and F). In Beclin-1–deficient cells, however, the elimination of liver-stage P. vivax mediated by IFN-γ was partially rescued, to 35.4 ± 10.4%. These findings demonstrate the involvement of Beclin 1 in the IFN-γ–mediated restriction of liver-stage P. vivax.
We also investigated whether PI3K activity is needed for the IFN-γ–mediated control of P. vivax. As in the previous experiment, IFN-γ treatment of HC04 cells infected with P. vivax sporozoites for 4 h resulted in a 55.6 ± 6.4% decrease in parasite survival; however, on cotreatment of the cells with 3-MA, a PI3K inhibitor, a partial rescue of parasite survival, to 18.3 ± 0.4%, was observed (Fig. 2 G and H). Taken together, our results show that the activity of autophagy-related protein Beclin 1 and PI3K is involved in the IFN-γ–mediated elimination of liver-stage P. vivax.
ATG5 Is Involved in the IFN-γ–Mediated Killing of Liver-Stage P. vivax.
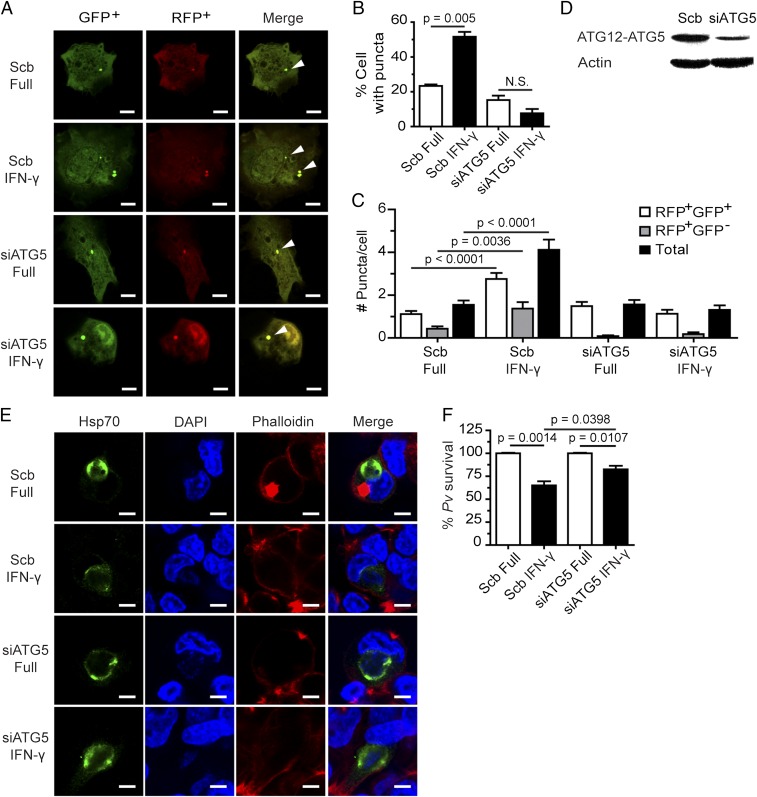
Because ATG5 plays an important role in both autophagosome elongation (canonical autophagy) (17) and LAP (noncanonical autophagy) (20), we asked whether the control of liver-stage P. vivax mediated by IFN-γ depended on ATG5. A reduction in ATG5 expression was achieved using siRNA. Because ATG5 is important for LC3 puncta formation in response to IFN-γ (27), knockdown efficiency was first confirmed in an LC3 puncta formation assay. The number of LC3 puncta in response to IFN-γ was lower in the ATG5 knockdown cells than in cells treated with scramble control siRNAs (Fig. 3 A–C). Immunoblot analysis confirmed the decreased ATG5 expression in the knockdown cells (Fig. 3D). We then investigated the involvement of ATG5 in the killing of liver-stage P. vivax in response to IFN-γ. The percentage of surviving parasite in cells subjected to IFN-γ treatment was higher in ATG5-deficient cells than in control cells (82.6 ± 3.8% vs. 65.1 ± 4.4%) (Fig. 3 E and F). These results highlight a role of ATG5 in the IFN-γ–mediated elimination of liver-stage P. vivax.
Fig. 3.
ATG5 is involved in the IFN-γ–mediated elimination of liver-stage P. vivax. (A–C) HC04 cells were depleted of ATG5 by siRNA-mediated knockdown and cotransfected with cDNAs encoding RFP-GFP-LC3. At 48 h posttransfection, the cells were treated with IFN-γ for 4 h and then processed for fluorescence microscopy. RFP+GFP+-LC3 (autophagosomes) and RFP+GFP−-LC3 (autolysosomes) were quantified by determining the percentage of puncta-containing cells among at least 100 cells per condition from three independent experiments. Only puncta ≥0.25 µm in size were counted. The number of puncta per cell was also analyzed in Z-stack images of at least 30 cells per condition per independent experiment. Data are mean ± SEM; the results are expressed relative to the full control. N.S., not significant. (Scale bar: 5 µm.) (D) Western blot analysis of ATG5 levels after knockdown of the protein. HC04 cells were transfected with scramble control siRNAs or siRNAs against ATG5. At 48 h posttransfection, the cells were harvested for immunoblot analysis. (E and F) ATG5-depleted or control HC04 cells infected with P. vivax sporozoites were treated with IFN-γ for 4 h, washed, and then maintained in complete medium until being harvested on day 4 for IFA. Data are mean ± SEM from at least three independent experiments; the results are expressed relative to the full control, defined as 100%. (Scale bar: 5 µm.)
Elimination of Liver-Stage P. vivax by IFN-γ Does Not Require ULK1.
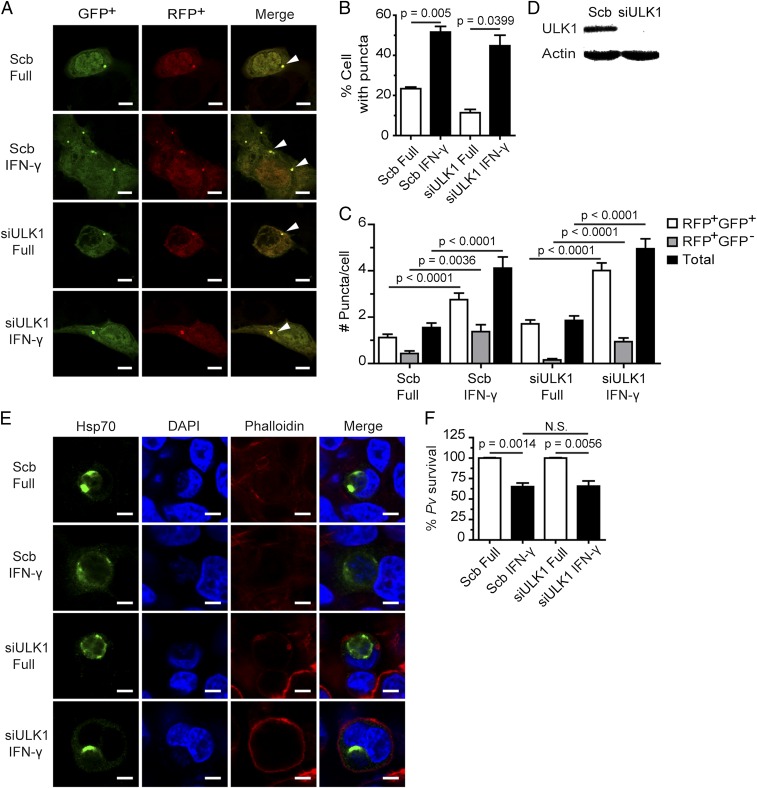
ULK1, a yeast Atg1 ortholog, is required for canonical autophagy induction in amino acid-starved cells (28), but not for LAP (20). To determine whether ULK1 is involved in IFN-γ–mediated P. vivax restriction, we silenced its expression in HC04 cells using siRNA, and verified the efficiency of ULK1 knockdown by Western blot analysis (Fig. 4D). Although ULK1 expression in HC04 cells was effectively reduced, assessment of autophagosome formation in response to IFN-γ–treated, ULK-1–deficient cells found that the number of LC3 puncta was the same as in control cells. This result indicates that IFN-γ–induced autophagosome formation in HC04 cells does not involve ULK1 (Fig. 4 A–C). Examination of the survival of liver-stage P. vivax in IFN-γ–treated HC04 cells revealed no difference in the percentage of surviving parasite in ULK1-deficient cells and that in control cells (Fig. 4 E and F). Thus, unlike Beclin 1, PI3K, and ATG5, ULK1 does not play a role in the IFN-γ–induced elimination of liver-stage P. vivax.
Fig. 4.
ULK1 is dispensable for the killing of liver-stage P. vivax mediated by IFN-γ. (A–C) ULK1 is not required for IFN-γ–induced LC3 puncta formation in HC04 cells. The cells were cotransfected with cDNAs encoding RFP-GFP-LC3 and either siRNAs against ULK1 or scramble control siRNAs. At 48 h posttransfection, they were treated with IFN-γ for 4 h and then processed for fluorescence microscopy analysis. RFP+GFP+-LC3 (autophagosomes) and RFP+GFP−-LC3 (autolysosomes) were quantified by determining the percentage of puncta-containing cells among at least 100 cells per condition from three independent experiments. Only puncta ≥0.25 µm in size were counted. The number of puncta per cell was also examined in Z-stack images of at least 30 cells per condition per independent experiment. Data are mean ± SEM; the results are expressed relative to the full control. (Scale bar: 5 µm.) (D) ULK1 immunoblot analysis after depletion of the protein. HC04 cells were transfected with either scramble control siRNAs or siRNAs against ULK1. At 48 h posttransfection, they were harvested for Western blot analysis. (E and F) ULK1-deficient or control HC04 cells were infected with P. vivax sporozoites at an MOI of 1, treated with IFN-γ for 4 h, washed, and then maintained in complete medium until being harvested on day 4 for IFA. Data are mean ± SEM of at least three independent experiments; the results are expressed relative to the full control, defined as 100%. N.S., not significant. (Scale bar: 5 µm.)
Liver-Stage P. vivax Colocalizes with LC3 and LTR+ Vesicles on IFN-γ Treatment.
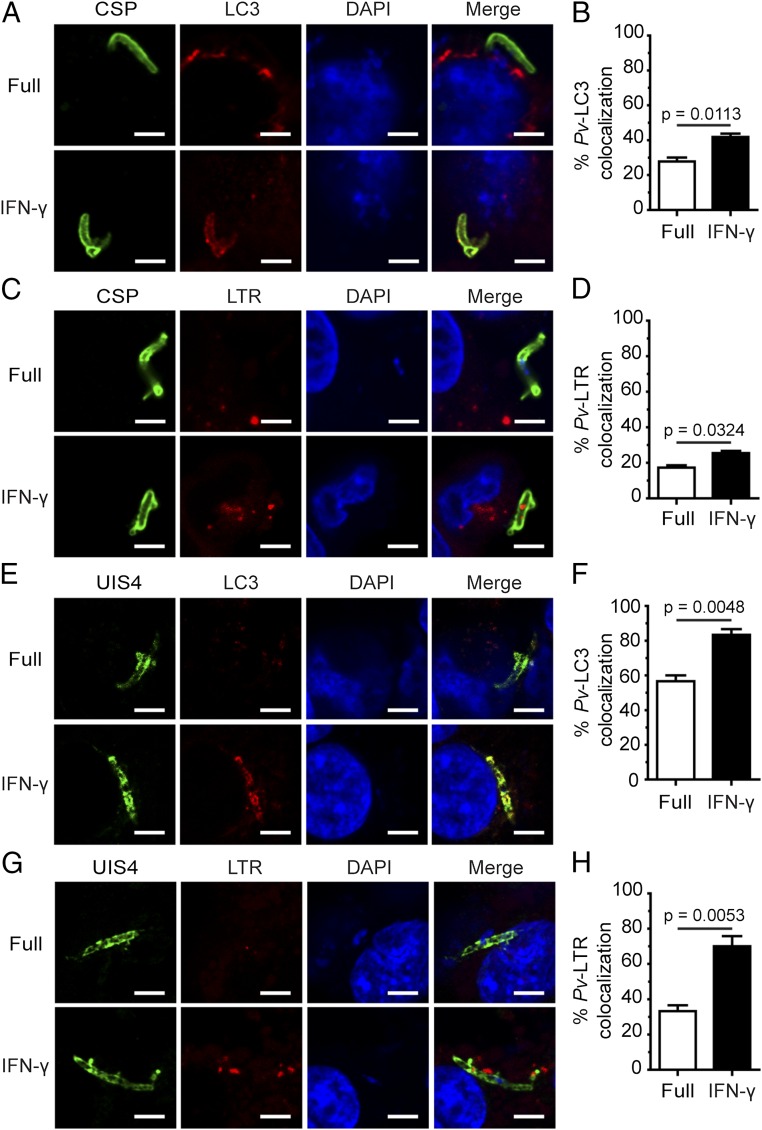
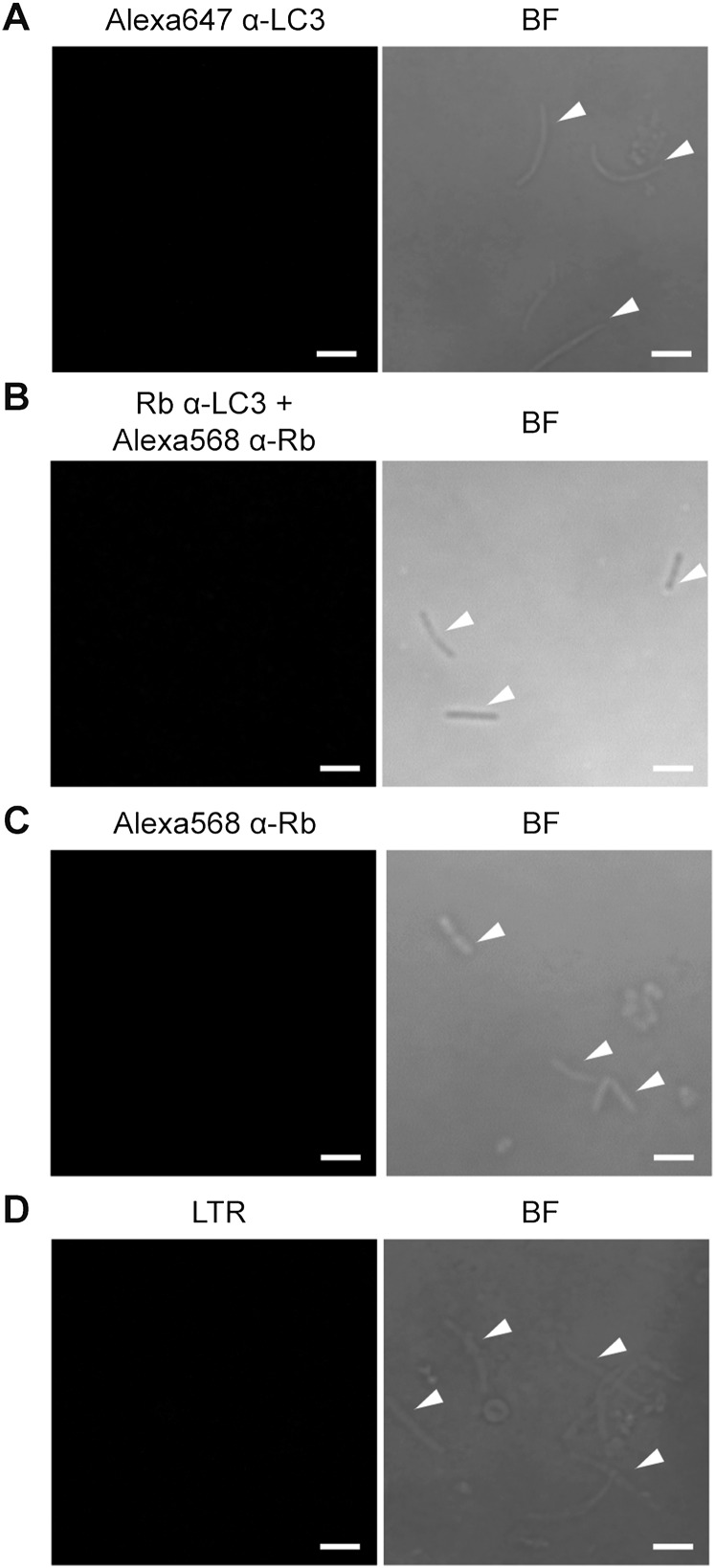
The involvement of Beclin 1, PI3K, and ATG5, but not of ULK1, is characteristic of the noncanonical autophagy process known as LAP, in which single-membrane phagosomes are decorated with LC3 and then fuse with lysosomes, resulting in degradation of the phagocytosed contents (20). Plasmodium liver-stage parasites enter host cells not via phagocytosis, but rather through an active invasion process that leads to the formation of parasite-containing parasitophorous vacuoles (29). Thus, we determined whether LC3 similarly decorates the PVM of P. vivax in response to IFN-γ, by staining the P. vivax surface with CSP and the PVM with UIS4. Both CSP and UIS4 are markers for P. vivax; previous studies in P. berghei showed decreased expression of CSP, but increased expression of UIS4, over the course of hepatocyte infection (30). The results show increased interaction between LC3 and P. vivax during IFN-γ treatment for 4 h postinvasion. Specifically, LC3 colocalized with liver-stage P. vivax (labeled with CSP) and prominently decorated the PVM (labeled with UIS4) in HC04 cells in response to IFN-γ (Fig. 5 A, B, E, and F), indicating that IFN-γ induces LC3 recruitment onto the PVM, similar to LC3 labeling of single-membrane phagosomes in LAP. Note that intracellular liver-stage P. vivax at 4 h postinvasion appears as an elongated form. As a control, isolated P. vivax sporozoites were stained with anti-LC3 antibodies or secondary antibody alone. Under these conditions, the antibodies do not recognize the parasites (Fig. S4 A–C). In addition, because LAP vesicles eventually fuse with lysosomes, we searched for the colocalization of acidic vesicles, which include lysosomes, with intracellular liver-stage P. vivax. The increased recruitment of acidic vesicles to the liver-stage P. vivax during a 4-h treatment with IFN-γ was visualized by Lysotracker Red (LTR) staining (Fig. 5 C, D, G, and H). As a control, isolated P. vivax sporozoites were stained with LTR, and no signal was detected inside the parasites (Fig. S4D).
Fig. 5.
Liver-stage P. vivax colocalization with LC3 and LTR+ vesicles increases in response to IFN-γ. (A and B) HC04 cells were infected with P. vivax sporozoites at an MOI of 1, treated with IFN-γ for 4 h, and then processed for confocal microscopy analysis of the colocalization of liver-stage P. vivax (labeled with CSP) and LC3. Data are mean ± SEM from at least three independent experiments. At least 100 liver-stage P. vivax parasites were quantified per condition. The results are expressed relative to the full control. (C and D) HC04 cells were infected with P. vivax sporozoites as in A and B and then incubated in complete medium containing LTR to stain acidic compartments in the presence or absence of IFN-γ treatment for 4 h. Cells were then processed for confocal microscopy analysis of the colocalization of liver-stage P. vivax (labeled with CSP) and LTR+ vesicles. Data are mean ± SEM of at least three independent experiments. At least 100 liver-stage P. vivax parasites per condition were quantified. The results are expressed relative to the full control. (E and F) HC04 cells were infected with P. vivax sporozoites at an MOI of 1, treated with IFN-γ for 4 h, and processed for confocal microscopy analysis for the colocalization of the PVM of UIS4-labeled P. vivax and LC3. Data are mean ± SEM of at least three independent experiments. At least 50 parasites per condition were quantified. The results are expressed relative to the full control. (G and H) HC04 cells were infected with P. vivax sporozoites and then incubated in complete medium containing LTR to stain acidic compartments in the presence or absence of IFN-γ treatment for 4 h. Cells were then processed for confocal microscopy analysis for the colocalization of the UIS4-labeled PVM and LTR+ vesicles. Data are mean ± SEM of at least three independent experiments. At least 50 parasites per condition were quantified. The results are expressed relative to the full control. (Scale bar: 5 µm.)
Fig. S4.
Anti-LC3 antibodies and LTR do not recognize isolated P. vivax sporozoites. (A–C) Isolated P. vivax sporozoites were spotted onto a 10-well antigen slide, fixed with 4% paraformaldehyde, and permeabilized with 0.1% saponin/3% BSA in PBS. The sporozoites were then stained with Alexa Fluor 647-conjugated rabbit anti-LC3 antibody at room temperature for 2 h (A). Alternatively, the sporozoites were stained with rabbit anti-LC3 antibody at 4 °C overnight, followed by Alexa Fluor 568-conjugated anti-rabbit secondary antibody at room temperature for 2 h (B) or with Alexa Fluor 568-conjugated anti-rabbit secondary antibody alone (C). The samples were then mounted and analyzed by confocal microscopy. (D) Isolated P. vivax sporozoites were incubated with LTR dye for 1 h at room temperature, washed twice with PBS, and spotted onto a 10-well antigen slide. The mounted samples were analyzed by confocal microscopy. Sporozoites are marked with white arrowheads. (Scale bar: 5 µm.)
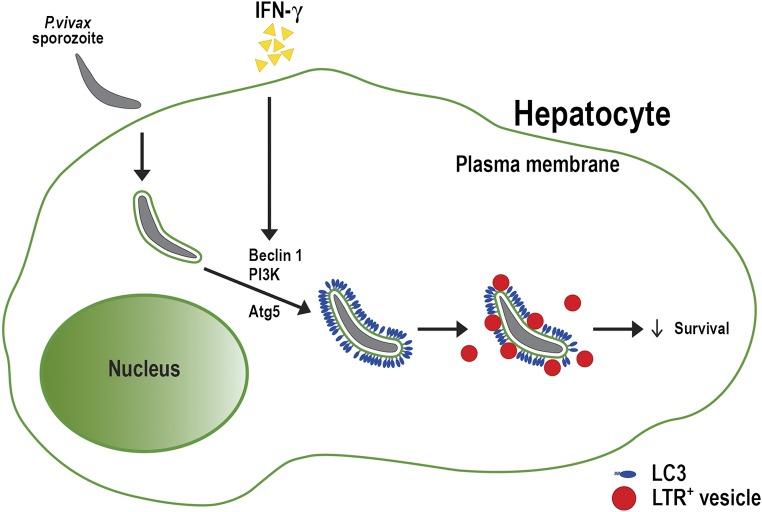
In summary, our results show that IFN-γ treatment restricts liver-stage P. vivax in HC04 cells via a process that involves the autophagy proteins Beclin 1, PI3K, and ATG5, but not ULK1. Moreover, in response to IFN-γ, enhanced decoration of LC3 onto the PVM of intracellular P. vivax and increased colocalization of LTR+ vesicles with the parasite were observed. These results support a model in which IFN-γ induces a LAP-like process in human HC04 hepatocytes that leads to decoration of the PVM with LC3, subsequent fusion of P. vivax compartments with lysosomes, and elimination of the parasite (Fig. S5).
Fig. S5.
Schematic model depicting the IFN-γ–mediated LAP-like process against liver-stage P. vivax. P. vivax sporozoites actively invade human hepatocytes and then reside within the parasitophorous vacuoles. IFN-γ treatment leads to the decoration of the parasitophorous vacuole membrane (PVM) with host LC3, in a process involving Beclin 1, PI3K, and Atg5. The LC3-decorated PVMs then fuse with lysosomes, which kills the parasite.
Discussion
IFN-γ is a critical cytokine used by the human immune system to fight against pathogen infections. In both cultured hepatocytes and animal models, IFN-γ exhibits antimalarial activity against liver-stage Plasmodium (5–8), but the molecular mechanism underlying this activity was unclear. The aim of this study was to identify the factors involved in the IFN-γ–mediated reduction of liver-stage Plasmodium in human cells using P. vivax as a model.
In contrast to a previous report showing the involvement of NO in IFN-γ–mediated Plasmodium elimination in murine models (6), neither iNOS nor NO played a role in the response to IFN-γ treatment of cultured human hepatocyte HC04 cells infected with P. vivax. Moreover, addition of iNOS inhibitors to the cultures had no effect on the IFN-γ response. These results suggest that IFN-γ mediates the elimination of liver-stage P. vivax from HC04 cells through an NO-independent mechanism. Further experiments demonstrated a role for a LAP-like process that depends on the downstream autophagy proteins Beclin 1, PI3K, and ATG5, but not on the upstream autophagy-initiating protein ULK1. Both the induction of LC3 decoration of the P. vivax PVM and the increased colocalization of P. vivax compartments with lysosomes were detected in response to IFN-γ. This is the first report, to our knowledge, to describe a LAP-like process as an IFN-γ downstream effector in the elimination of an intracellular pathogen.
Note that in this study, we investigated an autophagy/autophagy-related process induced by IFN-γ, which is different from the recently described basal autophagy process that was shown to be hijacked by mouse malarial parasites to facilitate their development in murine models (31, 32). This is similar to the effect seen with T. gondii, in which basal autophagy is subverted by the parasite for its own growth (33), whereas the parasite can be eliminated by induced autophagy/autophagy-related process in response to IFN-γ (12, 34–37). Also note that in this study we observed only roughly one-half of the rescue activity with the reduction in Beclin 1 or ATG5 expression and PI3K activity, and thus the contribution of additional factors leading to liver-stage P. vivax elimination remain to be identified.
LAP mediates the clearance of phagocytosed materials such as pathogens, dead cells, and entotic bodies (19). Unlike the canonical autophagy, in which LC3 is recruited onto double-membrane autophagosomes, in LAP single-membrane phagosomes are decorated with the protein (20). The fate of both LC3-associated compartments (autophagosomes and phagosomes) is similar in the two processes, however, in that both are subsequently delivered to lysosomes, where the engulfed substrates are digested. LAP can be distinguished from canonical autophagy by its requirement for downstream autophagy proteins such as Beclin 1, PI3K, and ATG5, but not for the upstream autophagy-initiating protein ULK1 (20). In agreement with the LAP process, our results show that IFN-γ–mediated killing of liver-stage P. vivax depends on Beclin 1, PI3K, and ATG5, but not on ULK1. In addition, we observed the increased decoration of LC3 onto the PVM of P. vivax and the enhanced colocalization of P. vivax compartments with lysosomes. Consequently, we term this phenomenon a LAP-like process, even though LC3 decoration occurs on the parasite PVM and not on the phagosomal membrane.
Note that we observed ∼20% colocalization of the parasites with LC3 and lysosomes after 4 h of IFN-γ treatment postinvasion, but a 50% reduction in parasite survival on day 4. Whether this increase is related to the slow kinetics of parasite elimination by the IFN-γ–induced LAP-like process or to the early digestion/disintegration of a fraction of the parasites, rendering them undetectable, is unclear.
As noted above, the IFN-γ–mediated growth restriction of T. gondii, a closely related apicomplexan parasite, in human cells also requires autophagy-related proteins, including ATG7 and ATG16L, involved in LC3 conjugation (36), as well as the increased recruitment of ubiquitin and the autophagy adaptor proteins p62 and NDP52 (36). Ubiquitin, p62, and NDP52 play important roles in the autophagy-mediated antimicrobial effects used by cells to combat intracellular pathogens (38, 39). We are currently investigating whether these three proteins play a role in the IFN-γ–mediated reduction of liver-stage P. vivax from human cells. Interestingly, in contrast to our findings, IFN-γ treatment of T. gondii-infected human cells leads only to parasite growth inhibition, not parasite clearance, because T. gondii can be seen within multimembrane compartments that do not acquire LAMP1 (36). In our study, the treatment of cultured human hepatocytes with IFN-γ for 4 h resulted in a decrease in P. vivax survival to approximately 50%, which supports a “cidal” role for IFN-γ, and not merely growth inhibition. Note that with the dosage of IFN-γ used to treat infected HC04 cells in this study (400 U/mL), the LAP-like process appears to explain approximately one-half of the parasite elimination, and a contribution by another, as-yet unidentified pathway cannot be ruled out. Alternatively, because only 50% of liver-stage P. vivax can be eliminated with a 4-h treatment of IFN-γ (400 U/mL), whether increasing the treatment time and pulse frequency and/or dosage of IFN-γ can increase the level of parasite elimination remains to be determined.
In addition to autophagy-related proteins, the LAP process triggered by a subset of cell surface receptors, including FcγR and TLR2, requires the recruitment of NADPH oxidases to phagosomes (20). For example, LC3 recruitment onto phagosomes requires ROS produced by NOX2 NADPH oxidase to rescue nonphagocytic cells infected with S. typhimurium (40). Recent studies also have demonstrated that the protein Rubicon, a negative regulator of canonical autophagy, binds to the p22phox subunit of NADPH oxidase and facilitates the recruitment of p22phox and LC3 to phagosomes, where it mediates the subsequent restriction of Listeria monocytogenes and A. fumigatus (24, 41). Whether NADPH oxidase and Rubicon participate in the LAP-like process of P. vivax in response to IFN-γ requires further investigation.
Studies of the expression profiles of genes induced in IFN-γ–treated human hepatocytes infected with P. vivax may allow the identification of additional factors involved in the restriction of liver-stage Plasmodium and thus to the development of new drug targets. Alternatively, inhibiting these key factors to increase the infectivity of human hepatocytes by human malarial Plasmodium sporozoites will facilitate high-throughput in vitro drug testing. In addition, the effect of IFN-γ–mediated LAP-like process on P. vivax hypnozoite survival is of high interest. Because we have codeveloped a humanized mouse model for P. vivax hypnozoite persistence (4), this question can now be further explored and will be a subject of our future study. Moreover, a recent study in P. yoelii by Risco-Castillo and colleagues (42) showed that parasites that fail to egress from their transient traversal vacuoles are colocalized with host lysosomes. Whether the LAP-like noncanonical autophagy process described here with P. vivax is involved in this process is of interest and may be a subject of further study.
Materials and Methods
Cells, Inhibitors, Antibodies, Fluorescent Dye, DNA Construct, and siRNAs.
Human hepatocyte HC04 cells (43) were maintained in HamF12/MEM (Invitrogen), 10% (vol/vol) FBS (Invitrogen), 2 mM l-glutamine (Sigma-Aldrich), and 1% penicillin/streptomycin (Invitrogen) (full medium) at 37 °C and 5% CO2. Raw264.7 macrophages (American Type Culture Collection) were maintained in DMEM, 10% (vol/vol) FBS, and 4 mM l-glutamine (all from HyClone). Recombinant human IFN-γ (Sigma-Aldrich) was used at 400 U/mL, LPS at 10 ng/mL, and the PI3K inhibitor 3-MA (Sigma-Aldrich) at 10 mM. The iNOS inhibitors aminoguanidine (Sigma-Aldrich) and L-NAME (Sigma-Aldrich) were used at 1 mM each. For immunoblotting, the polyclonal antibodies were anti-Beclin 1 (1:500; Santa Cruz Biotechnology), anti-ATG5 (1:1,000; Novus Biologicals), and anti-ULK1 (1:500; Cell Signaling Technology). The mAb against actin (EMD Millipore) was used at a 1:20,000 dilution.
For the IFAs, an mAb against Hsp70 (44) was used at a 1:2 dilution. The polyclonal antibodies used in the IFAs were anti-LC3 (1:500; MBL International) and Alexa Fluor 647-conjugated anti-LC3 (1:250; Novus Biologicals). A monoclonal antibody against CSP (Kirkegaard & Perry Laboratories) was used at 5 µg/mL The polyclonal antiserum against UIS4 (a gift from Dr. Tomoko Ishino, Ehime University, Matsuyama, Japan) was used at a 1:250 dilution. The fluorescent dye phalloidin (Invitrogen) was used at a 1:1,000 dilution, LTR (Invitrogen) at 0.25 µM, and DAPI (Invitrogen) at a 1:1,000 dilution. The plasmid construct used in this study has been described previously (45). All siRNAs used in this study were obtained from Dharmacon.
Hepatocyte Transfection.
For transfection with cDNAs (5 μg) or siRNAs (1.5 μg), 10–15 × 106 HC04 cells were resuspended in 100 µL of electroporation buffer V (Amaxa). Plasmid DNAs or siRNAs were then mixed with the cell suspension, transferred to a cuvette, and nucleoporated using the Amaxa Nucleofector apparatus and the D-032 program. The cells were then transferred to a new flask containing complete medium and incubated at 37 °C. The medium was changed at 6 h posttransfection. The cells were used in the assays at 48 h posttransfection.
Sporozoite Preparation, Hepatocyte Infection, and Immunofluorescence Microscopy.
Blood samples were obtained from P. vivax-infected patients at malaria clinics in Sai Yok District, Kanchanaburi Province, and Tha Song Yang District, Tak Province, Thailand. The study protocol (no. TMEC 11–033) was approved by the Ethical Review Committees of the Faculty of Tropical Medicine, Mahidol University and the Ministry of Public Health, Bangkok. Informed consent was obtained from each patient before sample collection.
Anopheles dirus A (Bangkok colony) mosquitoes were allowed to feed on the collected blood by membrane feeding, as described previously (46). At 15–20 d postfeeding, the mosquitoes were washed with 70% ethanol, 10% penicillin/streptomycin, and 25 µg fungizone (amphotericin B)/mL. The mosquitoes’ salivary glands were then isolated under a stereomicroscope and washed twice with dissecting medium (HamF12/MEM, 10% penicillin/streptomycin, 25 µg fungizone/mL), and once with complete medium. The glands were then ground with a sterile plastic pestle to release the sporozoites. HC04 hepatocytes were infected with P. vivax sporozoites by plating 3 × 105 cells onto coverslips in 24-well plates containing complete medium. Twelve hours later, the cells were infected with P. vivax sporozoites at a multiplicity of infection (MOI) of 1 at 37 °C and 5% CO2 for 4 h. They were then washed three times with complete medium to remove noninvading sporozoites before being treated for 4 h with complete medium supplemented with IFN-γ with or without the indicated inhibitors. After three washes with complete medium, the cells were maintained in complete medium at 37 °C and 5% CO2 until being harvested on day 4.
For parasite survival determination, infected HC04 cells were fixed in 4% (wt/vol) paraformaldehyde/PBS for 10 min and permeabilized with 0.1% Triton X-100/PBS. The cells on the coverslips were then blocked with 3% (wt/vol) BSA/PBS (blocking buffer) and stained with primary antibody at 4 °C overnight. After three washes with PBS, the cells were incubated with the appropriate secondary antibody (Invitrogen), then with phalloidin and DAPI. The coverslips were then washed three times in PBS, mounted using ProLong Gold Antifade Mountant (Invitrogen), and observed using a Zeiss A1 fluorescence microscope. All parasite-harboring cells were counted per coverslip, and the number was compared with that of the untreated control, defined as 100%. All assays were performed in triplicate with at least three independent experiments.
In the LC3 colocalization experiments, the infected cells were fixed as described above but permeabilized with 0.1% saponin in blocking buffer. They were then stained with mouse anti-CSP and rabbit anti-LC3 antibodies at 4 °C overnight, followed by incubation with the appropriate secondary antibodies (Invitrogen) at room temperature for 2 h. Alternatively, the samples were stained with rabbit anti-UIS4 antiserum at 4 °C overnight, followed by Alexa Fluor 568-conjugated anti-rabbit secondary antibody at room temperature for 2 h. The samples were then washed three times and stained with Alexa Fluor 647-conjugated rabbit anti-LC3 antibody (Novus Biologicals) at room temperature for 2 h. For LTR labeling, the cells were stained in complete medium containing 0.25 µM LTR during the 4-h treatment with IFN-γ, then fixed and processed for IFA as described above. All assays were performed in triplicate with at least three independent experiments.
Colocalization was quantified using a Zeiss LSM-700 laser scanning confocal microscope. The percent P. vivax colocalization with LC3 or LTR was the fraction of total liver-stage P. vivax (as determined by CSP or UIS4 staining) examined score as positive when one or more puncta or homogeneously distributed marker were observed overlapping on or in contact with the P. vivax compartments. RFP-GFP-LC3–transfected cells were fixed and mounted as described above. At least 50 cells per experimental condition in three independent experiments were quantified using a Zeiss LSM-700 laser scanning confocal microscope.
Immunoblotting.
The cells were lysed in lysis buffer containing 20 mM Tris, 100 mM NaCl, and 1% Nonidet P-40. The lysates were separated by SDS/PAGE on an 8% or 15% (wt/vol) polyacrylamide gel, after which the proteins were transferred onto a nitrocellulose membrane (Pall). The membranes were blocked with 5% (vol/vol) blocking solution (Roche Diagnostics) for 1 h, followed by the addition of primary antibodies against Beclin 1, ATG5, ULK1, or actin at 4 °C overnight. After four washes with 0.1% PBST, the membranes were incubated with the appropriate HRP-conjugated secondary antibodies (Pierce) for 1 h at room temperature, washed again four times with 0.1% PBST, and then incubated with a chemiluminescence substrate (Roche Diagnostics). The proteins were detected using the enhanced chemiluminescence method.
RT-PCR.
Total RNAs were extracted from the cells according to the manufacturer’s instructions (GE Healthcare). cDNAs were synthesized using the avian myeloblastosis virus reverse-transcription enzyme (Promega). PCR was performed using primer pairs specific for iNOS and GAPDH as the control housekeeping gene. The primer sequences for iNOS were 5′-AAG CCC CAA GAC CCA GTG CC-3′ (sense), 5′-CCA GCA TCT CCT CCT GGT AGA T-3′ (antisense), 5′-AAT GCC AGC GGC TTC CAC CT-3′ (sense), and 5′-GGC ACC CAA ACA CCA AGG TC-3′ (antisense), and those for GAPDH were 5′-ATG GGG AAG GTG AAG GTC G-3′ (sense) and 5′-GGG GTC ATT GAT GGC AAC A-3′ (antisense). The amplified products were electrophoresed on a 1.5% (wt/vol) agarose gel and stained with ethidium bromide before visualization under a UV lamp.
Griess Reaction Assay.
Cell supernatants were harvested and mixed with solution A (sulfanilamide), followed by solution B (N-naphthyl-ethylenediamine). The nitrite level was then determined by measuring the absorbance at 540 nm.
Statistical Analysis.
All experiments were conducted at least three times, and the data were pooled for determination of mean ± SEM. All data were analyzed using Prism software (GraphPad) and a two-tailed unpaired Student’s t test. P values < 0.05 were considered to indicate statistical significance.
SI Materials and Methods
HC04 Cell Viability.
HC04 cells were treated or not with IFN-γ (400 U/mL) for 4 h, washed three times with complete medium, and cultured until they were harvested at the indicated time points. HC04 cell viability was determined by staining cells with trypan blue (Invitrogen). The live cells were counted. Data are mean ± SEM of three independent experiments; the results are expressed relative to the full control, defined as 100%.
Immunofluorescence Microscopy of Isolated P. vivax Sporozoites.
Isolated P. vivax sporozoites were spotted onto a 10-well antigen slide, fixed with 4% (wt/vol) paraformaldehyde, and permeabilized with 0.1% saponin/3% (wt/vol) BSA in PBS. The sporozoites were then stained with Alexa Fluor 647-conjugated rabbit anti-LC3 antibody (Novus Biologicals) at room temperature for 2 h. Alternatively, the sporozoites were stained with rabbit anti-LC3 antibody (MBL Laboratories) at 4 °C overnight, followed by Alexa Fluor 568-conjugated anti-rabbit secondary antibody (Invitrogen) at room temperature for 2 h or with the secondary antibody alone at room temperature for 2 h. The samples were then mounted and analyzed by confocal microscopy.
For LTR staining, isolated P. vivax sporozoites were incubated with complete medium containing 0.25 µM LTR for 1 h at room temperature, washed twice with PBS, and spotted onto a 10-well antigen slide. The mounted specimens were analyzed by confocal microscopy.
Acknowledgments
We thank Chularat Luangjindarat and all members of the Mahidol Vivax Research Unit for their assistance. This work was supported by grants from the Development and Promotion of Science and Technology Talents Project (Research Grant 023/2557) and the Faculty of Science, Mahidol University (to M.P.) and the Bill & Melinda Gates Foundation (to J.S.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 6813.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525606113/-/DCSupplemental.
References
- 1.World Health Organization . World Malaria Report 2014. World Health Organization; Geneva, Switzerland: 2014. [Google Scholar]
- 2.Price RN, et al. Vivax malaria: Neglected and not benign. Am J Trop Med Hyg. 2007;77(6) Suppl:79–87. [PMC free article] [PubMed] [Google Scholar]
- 3.Wells TN, Burrows JN, Baird JK. Targeting the hypnozoite reservoir of Plasmodium vivax: The hidden obstacle to malaria elimination. Trends Parasitol. 2010;26(3):145–151. doi: 10.1016/j.pt.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Mikolajczak SA, et al. Plasmodium vivax liver stage development and hypnozoite persistence in human liver-chimeric mice. Cell Host Microbe. 2015;17(4):526–535. doi: 10.1016/j.chom.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schofield L, Ferreira A, Altszuler R, Nussenzweig V, Nussenzweig RS. Interferon-gamma inhibits the intrahepatocytic development of malaria parasites in vitro. J Immunol. 1987;139(6):2020–2025. [PubMed] [Google Scholar]
- 6.Sedegah M, Finkelman F, Hoffman SL. Interleukin 12 induction of interferon gamma-dependent protection against malaria. Proc Natl Acad Sci USA. 1994;91(22):10700–10702. doi: 10.1073/pnas.91.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vergara U, Ferreira A, Schellekens H, Nussenzweig V. Mechanism of escape of exoerythrocytic forms (EEF) of malaria parasites from the inhibitory effects of interferon-gamma. J Immunol. 1987;138(12):4447–4449. [PubMed] [Google Scholar]
- 8.Ferreira A, et al. Inhibition of development of exoerythrocytic forms of malaria parasites by gamma-interferon. Science. 1986;232(4752):881–884. doi: 10.1126/science.3085218. [DOI] [PubMed] [Google Scholar]
- 9.Klotz FW, et al. Co-localization of inducible-nitric oxide synthase and Plasmodium berghei in hepatocytes from rats immunized with irradiated sporozoites. J Immunol. 1995;154(7):3391–3395. [PubMed] [Google Scholar]
- 10.Nahrevanian H, Dascombe MJ. Nitric oxide and reactive nitrogen intermediates during lethal and nonlethal strains of murine malaria. Parasite Immunol. 2001;23(9):491–501. doi: 10.1046/j.1365-3024.2001.00406.x. [DOI] [PubMed] [Google Scholar]
- 11.Nahrevanian H. Involvement of nitric oxide and its up/down stream molecules in the immunity against parasitic infections. Braz J Infect Dis. 2009;13(6):440–448. doi: 10.1590/s1413-86702009000600010. [DOI] [PubMed] [Google Scholar]
- 12.Ling YM, et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med. 2006;203(9):2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24(1):69–79. doi: 10.1038/cr.2013.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 18.Romao S, Gannage M, Münz C. Checking the garbage bin for problems in the house, or how autophagy assists in antigen presentation to the immune system. Semin Cancer Biol. 2013;23(5):391–396. doi: 10.1016/j.semcancer.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Lai SC, Devenish RJ. LC3-associated phagocytosis (LAP): Connections with host autophagy. Cells. 2012;1(3):396–408. doi: 10.3390/cells1030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehta P, Henault J, Kolbeck R, Sanjuan MA. Noncanonical autophagy: One small step for LC3, one giant leap for immunity. Curr Opin Immunol. 2014;26:69–75. doi: 10.1016/j.coi.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Gong L, et al. The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS One. 2011;6(3):e17852. doi: 10.1371/journal.pone.0017852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kageyama S, et al. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell. 2011;22(13):2290–2300. doi: 10.1091/mbc.E10-11-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerena MC, Colombo MI. Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system. Cell Microbiol. 2011;13(6):814–835. doi: 10.1111/j.1462-5822.2011.01581.x. [DOI] [PubMed] [Google Scholar]
- 24.Martinez J, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2, and autophagy proteins. Nat Cell Biol. 2015;17(7):893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Levine B, Liu R, Dong X, Zhong Q. Beclin orthologs: Integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol. 2015;25(9):533–544. doi: 10.1016/j.tcb.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu SP, et al. IFN-γ inhibits gastric carcinogenesis by inducing epithelial cell autophagy and T-cell apoptosis. Cancer Res. 2011;71(12):4247–4259. doi: 10.1158/0008-5472.CAN-10-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang YP, et al. Autophagy facilitates IFN-gamma–induced Jak2-STAT1 activation and cellular inflammation. J Biol Chem. 2010;285(37):28715–28722. doi: 10.1074/jbc.M110.133355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aly AS, Vaughan AM, Kappe SH. Malaria parasite development in the mosquito and infection of the mammalian host. Annu Rev Microbiol. 2009;63:195–221. doi: 10.1146/annurev.micro.091208.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silvie O, Briquet S, Müller K, Manzoni G, Matuschewski K. Post-transcriptional silencing of UIS4 in Plasmodium berghei sporozoites is important for host switch. Mol Microbiol. 2014;91(6):1200–1213. doi: 10.1111/mmi.12528. [DOI] [PubMed] [Google Scholar]
- 31.Prado M, et al. Long-term live imaging reveals cytosolic immune responses of host hepatocytes against Plasmodium infection and parasite escape mechanisms. Autophagy. 2015;11(9):1561–1579. doi: 10.1080/15548627.2015.1067361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thieleke-Matos C, et al. Host cell autophagy contributes to Plasmodium liver development. Cell Microbiol. 2016;18(3):437–450. doi: 10.1111/cmi.12524. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Weiss LM, Orlofsky A. Host cell autophagy is induced by Toxoplasma gondii and contributes to parasite growth. J Biol Chem. 2009;284(3):1694–1701. doi: 10.1074/jbc.M807890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi J, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014;40(6):924–935. doi: 10.1016/j.immuni.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohshima J, et al. Role of mouse and human autophagy proteins in IFN-γ–induced cell-autonomous responses against Toxoplasma gondii. J Immunol. 2014;192(7):3328–3335. doi: 10.4049/jimmunol.1302822. [DOI] [PubMed] [Google Scholar]
- 36.Selleck EM, et al. A noncanonical autophagy pathway restricts Toxoplasma gondii growth in a strain-specific manner in IFN-γ–activated human cells. MBio. 2015;6(5):e01157–e15. doi: 10.1128/mBio.01157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Wilson D, Matthews S, Yap GS. Rapid elimination of Toxoplasma gondii by gamma interferon-primed mouse macrophages is independent of CD40 signaling. Infect Immun. 2007;75(10):4799–4803. doi: 10.1128/IAI.00738-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyle KB, Randow F. The role of “eat-me” signals and autophagy cargo receptors in innate immunity. Curr Opin Microbiol. 2013;16(3):339–348. doi: 10.1016/j.mib.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang J, et al. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci USA. 2009;106(15):6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang CS, et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe. 2012;11(3):264–276. doi: 10.1016/j.chom.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Risco-Castillo V, et al. Malaria sporozoites traverse host cells within transient vacuoles. Cell Host Microbe. 2015;18(5):593–603. doi: 10.1016/j.chom.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Sattabongkot J, et al. Establishment of a human hepatocyte line that supports in vitro development of the exo-erythrocytic stages of the malaria parasites Plasmodium falciparum and P. vivax. Am J Trop Med Hyg. 2006;74(5):708–715. [PubMed] [Google Scholar]
- 44.Tsuji M, Mattei D, Nussenzweig RS, Eichinger D, Zavala F. Demonstration of heat-shock protein 70 in the sporozoite stage of malaria parasites. Parasitol Res. 1994;80(1):16–21. doi: 10.1007/BF00932618. [DOI] [PubMed] [Google Scholar]
- 45.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 46.Sattabongkot J, et al. Comparison of artificial membrane feeding with direct skin feeding to estimate the infectiousness of Plasmodium vivax gametocyte carriers to mosquitoes. Am J Trop Med Hyg. 2003;69(5):529–535. [PubMed] [Google Scholar]