Significance
This report is the first publication, to our knowledge, to report the complete mitochondrial genome of an ancient Aboriginal Australian. In addition, it also provides important evidence about the reliability of the only previous publication of this kind. The paper attained international significance, although its conclusions have remained controversial. Using second generation DNA sequencing methods, we provide strong evidence that the DNA sequences reported by Adcock et al. were, indeed, contamination. Our manuscript is also important, because the research was planned and conducted and is published with the support of the Barkindji, Ngiyampaa, and Muthi Muthi indigenous groups.
Keywords: ancient DNA, Aboriginal Australians, mitogenomics, biological sciences, anthropology
Abstract
The publication in 2001 by Adcock et al. [Adcock GJ, et al. (2001) Proc Natl Acad Sci USA 98(2):537–542] in PNAS reported the recovery of short mtDNA sequences from ancient Australians, including the 42,000-y-old Mungo Man [Willandra Lakes Hominid (WLH3)]. This landmark study in human ancient DNA suggested that an early modern human mitochondrial lineage emerged in Asia and that the theory of modern human origins could no longer be considered solely through the lens of the “Out of Africa” model. To evaluate these claims, we used second generation DNA sequencing and capture methods as well as PCR-based and single-primer extension (SPEX) approaches to reexamine the same four Willandra Lakes and Kow Swamp 8 (KS8) remains studied in the work by Adcock et al. Two of the remains sampled contained no identifiable human DNA (WLH15 and WLH55), whereas the Mungo Man (WLH3) sample contained no Aboriginal Australian DNA. KS8 reveals human mitochondrial sequences that differ from the previously inferred sequence. Instead, we recover a total of five modern European contaminants from Mungo Man (WLH3). We show that the remaining sample (WLH4) contains ∼1.4% human DNA, from which we assembled two complete mitochondrial genomes. One of these was a previously unidentified Aboriginal Australian haplotype belonging to haplogroup S2 that we sequenced to a high coverage. The other was a contaminating modern European mitochondrial haplotype. Although none of the sequences that we recovered matched those reported by Adcock et al., except a contaminant, these findings show the feasibility of obtaining important information from ancient Aboriginal Australian remains.
Since the publication in 1863 of Thomas Henry Huxley’s Man’s Place in Nature, there has been considerable interest and debate regarding the origins of the first Australians. It was evolutionary biologist Ernst Haeckel who initially argued that humans originated in South Asia, a theory that enjoyed considerable support for the first half of the 20th century. The Asian origin theory was provided with a “missing link” after the discoveries by Eugene Dubois of the Java Man fossils in Trinil, Indonesia in the 1890s. Although there is once again increasing acceptance that Asia played a significant role in hominin evolution, particularly after the discoveries of new hominin species at Dmanisi and Flores (1), few researchers have suggested that Asia has played a key role in modern human origins. Some paleoanthropologists still argue that Australian origins are closely linked with archaic hominins from Indonesia, such as the Ngandong series of Homo erectus (2). The results of the work by Adcock et al. (3) have also been argued to support a significant Asian contribution to modern human origins. Several recent studies that focus on genomic data, however, suggest that archaic hominins, such as the Denisovans and Neanderthals, only made a small genetic contribution to Aboriginal Australians (4, 5).
Dating methods have established the presence of Aboriginal Australians on the Australian continent at least at 47,000 y B.P. (6). The existence of morphologically gracile and robust forms of Aboriginal Australian people from the Pleistocene subfossil record suggested to some (7) a complex history of the settlement of the Australian continent. For many years, the robust Pleistocene Australian morphology was used to argue for a close phylogenetic relationship to archaic humans from Java, Indonesia, providing evidence for the regional evolution of modern humans in Southeast Asia. Although the basic nature and usefulness of the “robusticity” debate has been questioned (8), it has been, in the past, a significant component of many theories of human evolution in the region.
In recent decades, the debate regarding the origins of the first Australians has largely focused on the mtDNA diversity of contemporary Aboriginal Australians. Generally, this debate has centered on whether this diversity was the result of a single migration or multiple migrations into Australia. In the context of these discussions, the publication by Adcock et al. (3) of mtDNA sequences from ancient robust and gracile forms originating from two sites, Kow Swamp and the Willandra Lakes (Fig. 1), stood as an important test of Thorne’s Dihybrid model for Aboriginal Australian origins. Adcock et al. (3) made four landmark claims. First, they had recovered authentic mitochondrial sequences from 10 sets of ancient Aboriginal Australian remains. Second, most of these recovered ancient sequences fell within the diversity of contemporary human sequences, whereas Mungo Man [Willandra Lakes Hominid (WLH3)] and Kow Swamp 8 (KS8) yielded unique mtDNA sequences. Importantly, these two sequences fell outside the range of contemporary human variation and clustered with a nuclear DNA insert. Moreover, the divergence between the mitochondrial and nuclear inserts predated the divergence of all contemporary modern humans. The implications of these claims were profound, because they suggested that multiple waves of migration to Australia were, in fact, possible, involving population replacement or selective sweeps to explain modern genetic and morphological diversity. Third, to explain the disconnect between the morphological and genetic data, Adcock et al. (3) invoked selective sweeps. Fourth, Adcock et al. (3) suggested that a reinterpretation of the “Out of Africa” theory was required and questioned the use of contemporary mitochondrial data in support of a single origin of modern humans from Africa. Given the obvious importance of these claims, the work by Adcock et al. (3) deserves serious reevaluation using more recent advances in DNA methods and powerful analytical approaches.
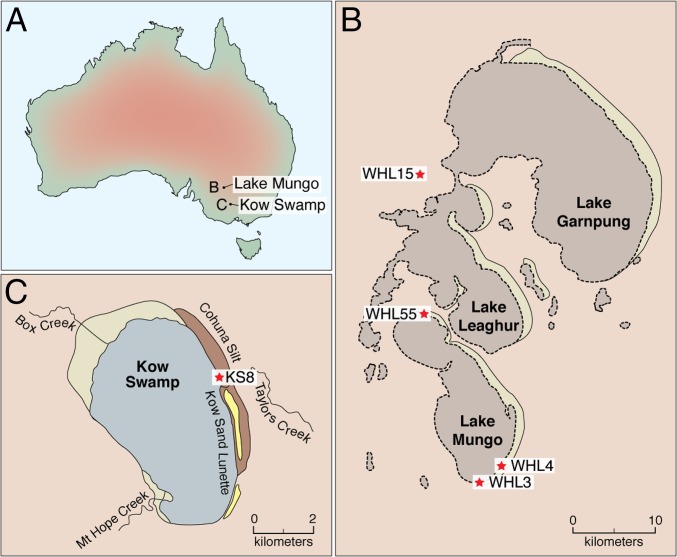
Fig. 1.
The locations of the Willandra Lakes and Kow Swamp where the remains for this study originated. (A) Map of Australia showing the locations of Lake Mungo (B on map) and Kow Swamp (C on map). (B) Map of the Willandra Lakes detailing where each of the remains were excavated. (C) Map of Kow Swamp detailing where the remains were excavated.
The majority of ancient mitochondrial sequences reported by Adcock et al. (3), including those from individuals with both “robust” and “gracile” morphologies, were shown to fall into a single clade that included sequences of living Aboriginal Australians. These mtDNA sequences failed to phylogenetically differentiate individuals with robust or gracile morphologies (3). Two of the sampled individuals, Mungo Man (WLH3) [Lake Mungo 3 (LM3) in the study by Adcock et al. (3)] and KS8, were identified by Adcock et al. (3) as lying outside the range of modern Aboriginal Australian mitochondrial variation. The latter observation was argued to support the highly debated multiregional hypothesis (9, 10).
These controversial findings were further challenged by researchers who questioned the authenticity of the sequences reported (11–13), highlighted problems with the phylogenetic approach (11–15), and expressed concern about the validity (11, 12, 14, 15) of the conclusions by Adcock et al. (3) . It is worth noting that Adcock et al. (16, 17) rebutted some of these criticisms, defending their conclusions about modern human origins, particularly with regard to the phylogenetic placement of Mungo Man (WLH3).
To assess the results obtained by Adcock et al. (3), we were given consent from the Willandra Lakes World Heritage Area Aboriginal Elders Committee (comprising the Barkindji, Ngiyampaa, and Muthi Muthi elders) to resample material from this important fossil series. In addition, we obtained access to several samples and extracts from the original research by Adcock et al. (3), including those from KS8.
These samples are either Late Pleistocene or Holocene in age (Table 1). WLH4 was excavated by Wilfred Shawcross in 1974 but does not have an absolute date. WLH4 is likely to be Holocene in age, because the skeletal remains are not heavily mineralized. The teeth from WLH4 exhibit a pattern of occlusal wear typical of Aboriginal hunter-gatherer populations and includes no evidence of dental caries. Interestingly, they also exhibit a pattern of interproximal tooth wear bilaterally at the second mandibular molars, indicating that some task activity that incorporated repeatedly dragging fibrous material between these teeth had occurred (18). This wear identifies WLH4 as an individual from a traditional hunter-gatherer population and combined with the lack of mineralization in the bone and its position in the stratigraphic sequence at Lake Mungo, indicates that the remains are estimated to be late Holocene in age (∼3,000–500 y B.P.).
Table 1.
Details of archaeological material examined in this study
| Details of remains | WLH3 | WLH4 | WLH15 | WLH55 |
| Age | 41,000 ± 4,000 y B.P. (24) | Late Holocene pre-European (18) | 117 ± 5 y B.P. (25) | 4,000 ± 1,000 y B.P; 6,200 ± 200 y B.P.; 28,400 ± 9,200 y B.P. (25) |
| Average level of human DNA (excluding enriched libraries) | 1.29% | 1.35% | 0.08% | 0.01% |
| Damage (proportion) | 1% | 7% | 1% | 0% |
| Proportion of identified haplogroups | Sample a: H15a1* (69%), U5a* (15%), H1* (8%); sample b: H40b* (57%), H1* (18%), H3* (20%) | S2† (81%); V3c* (18%) | Insufficient coverage | Insufficient coverage |
Haplogroups that are European origin.
Haplogroups of Aboriginal Australian origin.
The investigated remains are shown in Fig. 2. To avoid and monitor previously hypothesized contamination issues and PCR artifacts, we used a range of methods that were independently performed at different locations. We focused our next generation sequencing efforts on all four Willandra Lakes individuals and our PCR and single-primer extension (SPEX) cloning/sequencing efforts on the KS8 and Mungo Man (WLH3) studied by Adcock et al. (3).
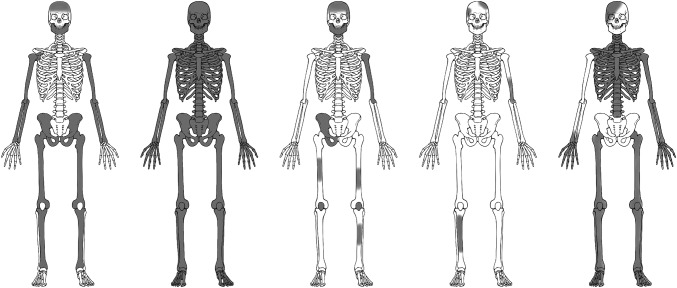
Fig. 2.
Details of five sets of skeletal remains of individuals from the Willandra Lakes that were investigated in this study. These are, from left to right: WLH3, WLH4, WLH15, WLH55 and KS8. Gray indicates skeletal elements that have survived for each individual.
Results and Discussion
We extracted DNA and constructed second generation Illumina DNA sequencing libraries from samples of remains of the Willandra Lakes individuals previously studied by Adcock et al. (3), namely Mungo Man (WLH3), WLH4, WLH15, and WLH55.
Using the resulting data, we addressed three questions in relation to the original study by Adcock et al. (3). First, is there any evidence of human DNA in these samples? Second, do we find any evidence of ancient human DNA sequences in these remains? Third, what is the mitochondrial sequence of any ancient or modern human DNA?
By mapping the sequences that we recovered against the human reference genome, we determined that there were 1.29%, 1.35%, 0.08%, and 0.01% human DNA on average (enriched libraries excluded) in sample libraries from the following remains: Mungo Man (WLH3), WLH4, WLH15, and WLH55, respectively (Table 1 and SI Appendix, Table S1). The very low levels of DNA reported for the latter two samples suggest that there is no human DNA present, and these levels of DNA are typically caused by the mapping of low-complexity regions, repetitive DNA, and highly conserved regions from nonhuman contaminants. Whole-genome capture (19) was successfully performed on two subsamples of Mungo Man (WLH3a and WLH3b), enriching the number of unique reads that mapped to the human reference genome by 4.5× and 27.3×, respectively (SI Appendix, Table S2).
Indicators of the successful recovery of degraded human DNA sequences are short read lengths because of fragmentation resulting from molecular damage and characteristic ancient DNA damage patterns (20). This ancient DNA damage pattern is typically observed as an elevated level of cytosine to thymine misincorporations in the 5′ end of sequences and the complement thereof, an elevated level of guanine to adenine misincorporations in the 3′ end of the DNA sequences (21). This pattern, caused by hydrolytic cytosine deamination, is typical for ancient or damaged sequences and generally not observed in modern DNA, although the levels of damage may vary (22). Because the Australian continent is considered inhospitable for DNA preservation, we can expect characteristic DNA damage to have affected DNA from archaeological remains (23).
To investigate the authenticity of the sequences obtained, we determined read length distributions and calculated appropriate molecular damage statistics (SI Appendix, Fig. S1). WLH4, WLH15, and WLH55 showed average read lengths of 67.8, 55.7, and 60.5 bp, respectively (SI Appendix, Table S3), indicating the presence of highly fragmented, potentially ancient DNA. Mungo Man (WLH3a and WLH3b) showed longer (79.1 and 78.5 bp) average read lengths compared with the other three samples, possibly indicating a modern origin for these sequences. However, only WLH4 showed relatively high proportions of cytosine deamination at the terminal bases of reads (7% vs. 0–1% for the other samples) (24, 25) (Table 1 and SI Appendix, Table S3). Collectively, these results suggested that WLH4 was the only sample that potentially contained endogenous DNA.
Next, the recovered sequences for each sample were mapped to the Cambridge reference mitochondrial genome (rCRS) (26) alone. We used this approach, because it recovers more mitochondrial hits compared with mapping against the entire genome. Because only one and six recovered sequences from WLH15 and WLH55, respectively, mapped to the rCRS, we did not use them for additional analyses. WLH3a, WLH3b, and WLH4, however, presented sufficient reads that mapped to the reference human mitochondrial genome, totaling 47×, 22×, and 126× coverage, respectively. The level of enrichment was greater for the mitochondrial genome sequences than for the nuclear human genome sequences: 9.7× for WLH3a and 109.7× for WLH3b (compared with nuclear enrichment values) (SI Appendix, Table S2). Length and damage statistics reveal that WLH3a and WLH3b have extremely low proportions of terminal base cytosine deamination: 1% for both (SI Appendix, Fig. S2). This pattern is indicative of modern DNA and therefore, contaminating sequences. In contrast, WLH4 shows a higher proportion of terminal base cytosine deamination for the mitochondrial data (13%), being indicative of the presence of ancient sequences that are likely to be endogenous.
For each of the subsamples of Mungo Man (WLH3) as well as for WLH4, we observed a number of variable nucleotide sites compared with the consensus mitochondrial genome. This variability suggested that multiple mitochondrial genomes were recovered for each (sub)sample. By aligning the reads to the Reconstructed Sapiens Reference Sequence—the inferred ancestral sequence for all human mitochondrial haplogroups (27)—we were able to identify the haplotypes and their proportions for each (sub)sample (Table 1 and SI Appendix, Table S4).
The haplogroups observed for WLH3a were H15a1, U5a, and H1. WLH3b showed haplogroups H40b, H1, and H3. All five haplogroups were European in origin. The haplogroups observed for WLH4 were S2 and V3c. The first is Australian in origin, whereas the latter is European. The reads from WLH4 that contained haplotype-specific variants (SI Appendix, Table S4) were separated according to the haplotype of origin. The S2 haplotype dataset showed 14% terminal base cytosine deamination and an average read length of 78.1 bp. The V3c haplotype dataset showed 4% terminal base cytosine deamination. A smaller sample size (476 vs. 2,113 sequence reads for the S2 haplotype) together with an absence of the typical damage pattern suggest that this relatively high level of damage is the result of errors occurring at the end of the sequences rather than resulting from genuine ancient damage (SI Appendix, Fig. S2). An average read length of 88.3 bp was observed. Other than the geographic origin, these read length and damage statistics confirm that the Australian haplotype is endogenous and that the European haplotype is a contaminant. The entire S2 mitochondrial genome of WLH4 was constructed by calling a consensus sequence from all reads that mapped to the Reconstructed Sapiens Reference Sequence.
Compared with all publicly available Aboriginal Australian mitochondrial genomes, the authentic endogenous sequence from WLH4 groups with individuals carrying the S2 haplogroup in the maximum likelihood tree (Fig. 3). The two haplotypes recovered from WLH4 (S2 and V3c) are distinct from the one observed in the work by Adcock et al. (3) (SI Appendix, Table S7). Several haplotypes from this study, H1, H15a1, and U5a from WLH3a and H1, H3, and H40b from WLH3b, have exactly the same sequence after trimmed to the amplicon reported in the work by Adcock et al. (3) (SI Appendix, Table S7).
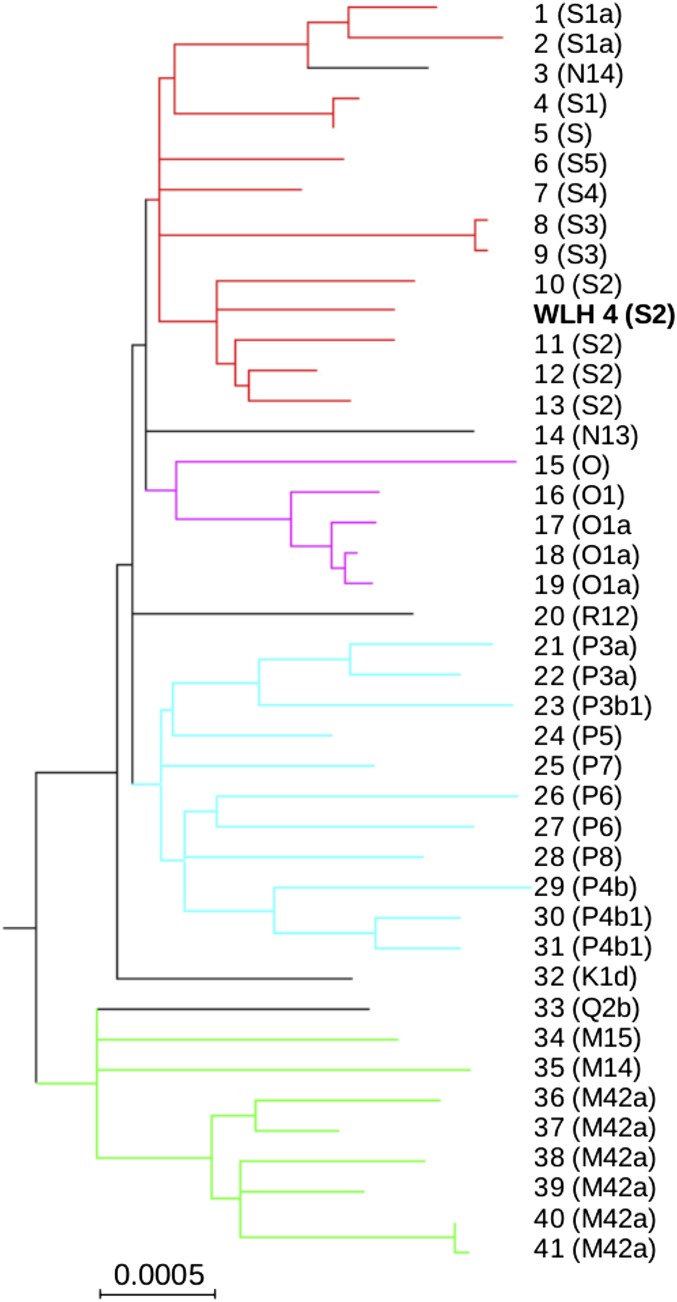
Fig. 3.
Maximum likelihood tree for all publicly available (near) complete Aboriginal Australian mitochondrial genomes and that of WLH4. Major Australian haplogroups are marked as follows: S (red), O (magenta), P (turquoise), and M (green). The sequence from WLH4 clusters with haplogroup S and in particular, haplogroup S2. Previously published sequences are marked with numbers (SI Appendix, Table S6 has more details), and haplogroups are shown in parentheses.
PCR Amplification for Mitochondrial Sequences Using the Original Material Used by Adcock et al. (3).
We present here results of research using the remaining original DNA extracts (WLH3 and KS8) from the work by Adcock et al. (3). We commenced by amplifying sections of the first and second hypervariable sections of the human mitochondrial control region by PCR in an attempt to replicate the original results. For WLH3, all 38 clones had transitions relative to the rCRS at nucleotide positions (nps) 16,224 and 16,311. Ten clones carried transitions at nps 146 and 200 (SI Appendix, Table S7). For KS8, all 23 clones had transitions relative to the rCRS at nps 16,223 and 16,278, 5 of these clones carried an additional transversion at np 16,259, and an additional 8 clones amplified carried transitions at nps 146 and 152.
These results show the presence of a DNA template that is most likely of European origin (haplogroup K) and that matches the sequence from Gregory J. Adcock (Australian National University, Canberra, Australia) in the products amplified from WLH3 (SI Appendix, Table S7). The motifs T16224C and T16311C are diagnostic for haplogroup K (www.Phylotree.org; build 16). These results are not consistent with those returned by Adcock et al. (3) for WLH3 and KS8 from the same extracts. We deliberately chose to use different primers and PCR conditions that those used by Adcock et al. (3) to avoid the likelihood of PCR artifacts previously identified. Rather, they are clearly indicative of copurified contaminants. Because all PCR blanks did not show any amplified product and because the results were repeated in separate PCR assays, we suggest that the DNA was present in the original extracts.
Because the target amplicon length for the HVS1 (178 bp) was much longer than the average length of templates usually observed in highly degraded DNA extracts, we used the SPEX method (28). SPEX is a forerunner of shotgun sequencing, being an early application of ligation-based methods to degraded DNA templates. The use of a single forward primer to target a specific region of the mtDNA genome allows precise strand-specific genotyping of SNPs of interest. This property determined that cytosine deamination was the only genuine type of postmortem damage in ancient DNA templates (29). Each molecule is copied in a single round of primer extension before the addition of a universal reverse primer binding site for subsequent amplification. The PCR, therefore, takes place on synthetic DNA molecules of various lengths and strongly favors the amplification of shorter molecules, resulting in the reduction of nonauthentic recombinant hybrids and modern contaminants to undetectable levels. Unique molecules can be distinguished by means of their length and damage patterns, providing highly accurate and quantitative genotyping from ancient human samples (29). Using SPEX, we applied the primer set targeting np 16,224 to the WLH3 and KS8 DNA extracts. This primer set has the advantage of being able to characterize both the nps 16,224 and 16,223 within a 2-bp read frame.
The results indicate that all 24 reads for WLH3 are without the transition at np 16,224, and none indicate any other differences from the rCRS as far as the polyC tract at np 16,189. For KS8, in 16 molecules recovered using SPEX, all match the rCRS at np 16,224, but 4 have the transition at np 16,223 found in the PCR result. These differences observed between the PCR and SPEX results are consistent with the fact that the former requires longer template molecules to allow for the binding of two primers (28). We hypothesize that the prelaboratory sequence contamination from handling is more fragmented (preferentially targeted by SPEX) than the later introduced contaminating sequences from Gregory J. Adcock (preferentially targeted by PCR). Alternatively, the original extracts for WLH3 contained some endogenous DNA templates of insufficient length to be phylogenetically informative.
Phylogenetic Analyses.
To prove that the hypervariable region 1 (HVR1) sequences were, indeed, from ancient Australians, Adcock et al. (3) performed phylogenetic and molecular evolutionary analyses, and the results were shown in figures 1 and 2 in ref. 3. Adcock et al. (3) reported that the ancient LM3 (WLH3 in this study) sequence belongs to an early diverging mitochondrial lineage. This view was based on a maximum likelihood tree, which showed that WLH3 fell outside of all modern and ancient Australian HVR1 sequences (figure 1 in ref. 3). Furthermore, their tree showed that WLH3 clustered with another divergent sequence, a nuclear insert of HVR1 (figure 1 in ref. 3). To further support their claim, they performed a pairwise distance estimation, which showed distinct distributions for those distances estimated within modern Aboriginal Australian sequences and those estimated between WLH3 and modern Aboriginal Australians (figure 2 in ref. 3). The mode (Mo) of the divergence between WLH3 and modern Aboriginal Australians (Mo = 12) was higher than that estimated within modern Aboriginal Australians (Mo = 6). The higher divergence observed for the comparison between WLH3 and modern Aboriginal Australians is consistent with the maximum likelihood tree showing WLH3 as an outgroup.
We reanalyzed the data presented by Adcock et al. (3) by including more Aboriginal Australian sequences (SI Appendix, SI Methods). We constructed a maximum likelihood tree using HVR1 data from WLH3, WLH4, modern Aboriginal Australians, and the nuclear insert sequence (SI Appendix, Fig. S3). This tree shows that the divergent WLH3 and nuclear insert sequences formed a clade that clearly fell outside of the other ancient and modern Aboriginal Australian sequences (SI Appendix, Fig. S3A), similar to those shown in figure 1 in ref. 3. However, when we excluded the insert sequence, WLH3 became a sister taxon to the remaining Aboriginal Australian sequences (SI Appendix, Fig. S3B). Similarly, the insert sequence had become a sister taxon to other Aboriginal Australian HVR sequences when WLH3 was excluded (SI Appendix, Fig. S3C). These patterns suggest that the divergent WLH3 and the insert sequence attract each other, a well-known problem of long-branch attraction (30). The exclusion of one of the divergent sequences resulted in the displacement of the other to another part of the tree, consistent with long-branch attraction (31).
We performed pairwise distance estimations similar to those in the work by Adcock et al. (3) to examine the validity of their claims. We obtained distinct distributions for WLH3 and modern Aboriginal Australians (SI Appendix, Fig. S4), similar to those found in the work by Adcock et al. (3). The mode of the pairwise divergence estimated for the comparison between WLH3 and modern Aboriginal Australians (Mo = 12) was much higher than that observed within modern Aboriginal Australians (Mo = 7). We also estimated pairwise divergences between three fast-evolving modern Aboriginal Australians and the other remaining contemporary Aboriginal Australian sequences. The distributions of pairwise divergences between these sequences were almost indistinguishable to that of WLH3 (SI Appendix, Fig. S4). The mode of the divergence between the fast evolving and other remaining modern Aboriginal Australians (Mo = 11–13) overlaps with the mode obtained for the comparison between WLH3 and modern Aboriginal Australians (Mo = 12). The higher modes of these pairwise divergences and distinct distribution for WLH3 need not mean that it is an outgroup for modern Aboriginal Australians. Such results could be obtained, because WLH3 has a long branch, while remaining a sister group to modern Aboriginal Australians, which is shown in SI Appendix, Fig. S3B.
To investigate the ability of the locus used by Adcock et al. (3) to correctly infer phylogenetic relationships, we used a worldwide dataset of complete mitochondrial genomes (32) with the addition of the mitochondrial genomes of the gorilla, chimpanzee, bonobo, Neanderthal, and Denisovan. The maximum likelihood tree for these complete mitochondrial genomes was as expected; primates and archaic humans were basal to modern humans, and the oldest modern human haplogroups were African (SI Appendix, Fig. S5A). In contrast, the equivalent dataset trimmed to the amplicon by Adcock et al. (3) failed to infer these placements (SI Appendix, Fig. S5B); for example, apes and archaic humans fell within the diversity of modern humans, and the normally basal human haplogroups (L0-3) appeared elsewhere in the tree. Another concern is illustrated by the fact that some of the Aboriginal Australian sequences appeared more closely related to those of European or African origin.
We estimated the age of the most recent common ancestor (MRCA) of WLH4 and modern mitogenomes belonging to S2 haplogroup using a Bayesian Markov chain Monte Carlo (MCMC)-based analysis. We used the age of the human–Neanderthal split and the ages (tip dates) of WLH4 and Neanderthal genomes to calibrate the Bayesian tree (SI Appendix, SI Methods). This analysis revealed that the age of the MRCA of WLH4 modern S2 haplogroup to be 28,500 y (SI Appendix, Fig. S6). Furthermore, our results produced an age of the MRCA of all available S2 haplogroup genomes (including WLH4) of 35,500 y, which is only slightly younger than that reported by a previous study (38,700 y) (27).
It is clear that a short (353 bp) hypervariable locus of mtDNA is not suitable for resolving the human worldwide phylogeny. We suggest that this lack of power is a result of many key mutations defining the major human mtDNA haplogroups being located outside of this 353-bp region combined with the effects of mutational saturation on many of the informative sites that occur within the mitochondrial HVR.
Conclusion
The Aboriginal Australian individuals referred to as WLH3 and KS8 have played a critical role in the interpretation of modern human evolution outside of Africa. The fact that we could not replicate the original results by Adcock et al. (3) is perhaps not surprising given that they were obtained by PCR-based methods applied to samples recovered from Holocene and Pleistocene deposits in semiarid and temperate Australia. The methods used here present advantages over PCR, in that they are able to target very degraded templates and improve the ability to distinguish authentic sequences from contaminants (19, 28, 29, 33, 34). These approaches have allowed us to clarify the results for the four Willandra Lakes individuals analyzed by Adcock et al. (3); only WLH3 (Mungo Man) and WLH4 contained verifiable human DNA, whereas nothing could be retrieved from extracts of WLH15 and WLH55. However, in principle, we cannot exclude the possibility that these specimens contain endogenous human DNA, which may be differentially preserved within the same individual. It has been shown that some regions of the skeleton, such as the petrous bone, are better suited than other regions for preserving ancient DNA (35). The recovery of an authentic Aboriginal Australian haplotype (S2) from WLH4 together with a European contaminating sequence (V3c) show the advantages of the non–PCR-based approach used here. We did not observe any amplified product for the extraction and library build blanks, suggesting that no contamination occurred during the laboratory procedures. For WLH3, however, we found a total of five European mtDNA contaminants, presumably derived from peopling handling the remains. Our study also suggests that the original 2001 extracts contained modern contaminants, although the haplogroup K sequence found in WLH3 was likely introduced during the extraction process. The SPEX assay, which has the ability to amplify shorter molecules, supports the next generation sequencing results using the new extractions. A similar conclusion can be drawn for KS8, because we were unable to amplify any diagnostic human mtDNA from either the SPEX assay or the next generation sequencing of enriched secondary libraries. The abundance of contamination that can be identified from these remains highlights the necessity for careful precautions and specially constructed laboratory environments now typical of ancient DNA facilities. With ancient DNA studies focusing on increasingly older and more marginal samples, it is important to reduce direct contact with the remains to an absolute minimum. As a further control against contamination within the laboratory, it may also prove beneficial to sequence the genomes of all of those who handled the remains. This procedure would have the advantage of providing data on other loci that may be similar to the individual being studied (e.g., nuclear inserts of the mtDNA in this case).
In relation to the four landmark claims made by Adcock et al. (3), we were unable to verify that the original study recovered any authentic Aboriginal Australian mitochondrial sequences from the four Willandra Lakes samples that they studied (claim 1). The contamination observed for WLH3 and the absence of human DNA for KS8 suggest that the validity of the originally reported sequences should be reappraised in light of the technological advances available to this study. These observations together with the uncertainties of the phylogenetic resolution obtained using HVR1 sequences provide no support for the historical existence of mitochondrial lineages in Australia that fall outside contemporary human variation (claim 2). As a consequence, there is no need to explain the presence of robust and gracile morphologies by invoking population replacement or selective sweeps (claim 3). We suggest that all of the sequences reported by Adcock et al. (3) were either modern contamination or PCR artifacts. As a result, it seems that contemporary mitochondrial data are consistent with the Out of Africa theory (claim 4).
Of the four Willandra Lakes samples, we show that WLH4 does contain authentic Aboriginal Australian DNA sequences, and we report the complete mitochondrial sequence from this individual. This report is the first example, to our knowledge, of DNA being recovered from ancient remains in an Australian archaeological context. The recovery of a complete mitochondrial genome to a high level of coverage of one Willandra Lake sample suggests that future research into First Australian material has the potential to lead to the recovery of complete nuclear genomes from these iconic remains.
Materials and Methods
Sample Collection.
In partnership with the Barkindji, Ngiyampaa, and Muthi Muthi tribal elders, we sampled material from four sets of remains of First Australians. Originally, these remains derived from the Willandra Lakes region (a copy of the approval is supplied in SI Appendix) and were held in storage at the Australian National University. Subsamples were taken from Mungo Man (WLH3), WLH4, WLH15, and WLH55. The following procedure was conducted in a fume hood that had previously not been used for any human-based research. We treated the surface of bones with concentrated sodium hypochlorite to remove any contaminating surface DNA, and ∼200 mg bone shavings were then removed from the treated surface using a sterile Dremel on low speed. This method is minimally invasive and has been generally successful at removing contaminating human DNA, thereby allowing the recovery of authentic endogenous ancient DNA.
All pre-PCR procedures were carried out in the dedicated ancient DNA laboratory at Griffith University or a similar laboratory located at Oxford University (Ancient Biomolecules Centre). DNA was extracted from ∼50 mg bone powder and eluted in 50 μL water using a protocol that retains short DNA fragments and removes inhibitory substances (32). Extraction blanks were included throughout all procedures.
Illumina DNA Libraries were built according to the methods described by Meyer and Kircher (36) and Rasmussen et al. (4). Using the NEBNext DNA Library Prep Master Mix Set for 454 (NEB), 21.25 μL DNA extract was subjected to one-quarter volume end repair reaction. After a MinElute (Qiagen) purification with 10× binding buffer PN (Qiagen) or PB (Qiagen), the resulting solution was subjected to quick ligation in a one-half volume reaction with blunt end adapters. After an additional MinElute purification, the DNA was subjected to a one-half volume fill-in reaction. The libraries were amplified to levels required for sequencing with one of three protocols (SI Appendix, Table S5), and if necessary, a secondary PCR was used to increase quantities. For all three protocols, the primary 100-μL library PCR was done according to the manufacturer’s instructions using the InPE1.0 and indexing primers. The primary PCR was subsequently cleaned using the MinElute PCR Purification Kit (Qiagen) according to the manufacturer’s instructions. The secondary PCR used 5 μL purified primary PCR and primers IS5 and IS6. The volume of the secondary PCR was 33 μL for the AccuPrime Pfx SuperMix (Life Technologies) protocol. Cycling was as follows for both PCR stages: 95 °C for 10 min, cycling (95 °C for 15 s, 60 °C for 30 s, and 68 °C for 30 s), and 10 °C indefinitely. The volume of the secondary PCR was 50 μL for the KAPA HiFi HotStart Uracil+ ReadyMix (Kapa Biosystems) protocol, and cycling was as follows for both PCR stages: 98 °C for 2 min, cycling (98 °C for 30 s, 60 °C for 15 s, and 72 °C for 15 s), 72 °C for 1 min, and 10 °C indefinitely. The volume of the secondary PCR was 50 μL for the Platinum Taq DNA Polymerase High Fidelity (Life Technologies) protocol, and cycling was as follows for both PCR stages: 94 °C for 1 min, cycling (94 °C for 15 s, 60 °C for 30 s, and 68 °C for 30 s], and 10 °C indefinitely.
DNA Enrichment Using Whole-Genome Capture and Bioinformatics.
Details of DNA capture methods, second generation sequencing, and bioinformatic processing of sequence reads together with mitochondrial sequence analyses and phylogenetics can be found in SI Appendix, SI Methods. In addition, details of the SPEX method and related PCR methods can also be found in SI Appendix.
Supplementary Material
Acknowledgments
This research was conducted in partnership with the Barkindji, Ngiyampaa, and Muthi Muthi elders. We thank these people for their permission to study the Lake Mungo remains and their advice and guidance. We thank Manaasa Raghavan and Morten Rasmussen for help with Illumina library construction methods, Hannes Schroeder and Morten Allentoft for help with the DNA capture methods, Greg Baillie for advice regarding DNA extraction methods, and Gregory Adcock for providing the original DNA extracts for WLH3 and KS8 as well as undigested homogenized bone powder from WLH3. We also thank Christina Strobl (Innsbruck Medical University) for second generation DNA sequencing and Paul Brotherton (Department of Zoology, Oxford University) for assistance with SPEX assays. We thank the anonymous reviewers for helpful suggestions and comments. We also thank The Danish National High-Throughput DNA Sequencing Centre for sequencing the samples. We thank the Australian Research Council for support through Discovery and Linkage Grant Projects DP140101405, DP110102635, and LP120200144. J.L.W. thanks Griffith University for a PhD scholarship and the Environmental Futures Research Institute for support. P.E. was funded by the Wenner–Gren Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence (WLH4, haplogroup S2) reported in this paper has been deposited in the GenBank database (accession no. KU659023).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521066113/-/DCSupplemental.
References
- 1.Morwood MJ, et al. Archaeology and age of a new hominin from Flores in eastern Indonesia. Nature. 2004;431(7012):1087–1091. doi: 10.1038/nature02956. [DOI] [PubMed] [Google Scholar]
- 2.Wolpoff MH, Lee SH. WLH 50: How Australia informs the worldwide pattern of Pleistocene human evolution. PaleoAnthropology. 2014;2014:505–564. [Google Scholar]
- 3.Adcock GJ, et al. Mitochondrial DNA sequences in ancient Australians: Implications for modern human origins. Proc Natl Acad Sci USA. 2001;98(2):537–542. doi: 10.1073/pnas.98.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334(6052):94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reich D, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89(4):516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connell JF, Allen J. The process, biotic impact, and global implications of the human colonization of Sahul about 47,000 years ago. J Archaeol Sci. 2015;56(April 2015):73–84. [Google Scholar]
- 7.Thorne AG, Wolpoff MH. Regional continuity in Australasian Pleistocene hominid evolution. Am J Phys Anthropol. 1981;55(3):337–349. doi: 10.1002/ajpa.1330550308. [DOI] [PubMed] [Google Scholar]
- 8.Pardoe C. Global human variation: Polarised positions and alternative perspectives. Before Farming. 2010;2010(3):1–21. [Google Scholar]
- 9.Holden C. Paleoanthropology. Oldest human DNA reveals Aussie oddity. Science. 2001;291(5502):230–231. doi: 10.1126/science.291.5502.230. [DOI] [PubMed] [Google Scholar]
- 10.Relethford JH. Ancient DNA and the origin of modern humans. Proc Natl Acad Sci USA. 2001;98(2):390–391. doi: 10.1073/pnas.98.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colgan DJ. Commentary on GJ Adcock, et al, 2001” Mitochondrial DNA sequences in ancient Australians: Implications for modern human origins. Archaeol Ocean. 2001;36(3):168–169. doi: 10.1073/pnas.98.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper A, et al. Human origins and ancient human DNA. Science. 2001;292(5522):1655–1656. doi: 10.1126/science.292.5522.1655. [DOI] [PubMed] [Google Scholar]
- 13.Trueman JWH. Does the Lake Mungo 3 mtDNA evidence stand up to analysis? Archaeol Ocean. 2001;36(3):163–165. [Google Scholar]
- 14.Groves C. Lake Mungo 3 and his DNA. Archaeol Ocean. 2001;36(3):166–167. [Google Scholar]
- 15.Cameron DW, Groves CP. Bones, Stones, and Molecules: “Out of Africa” and Human Origins. Academic; London: 2004. [Google Scholar]
- 16.Adcock GJ, et al. Human origins and ancient human DNA—response. Science. 2001;292(5522):1656. doi: 10.1126/science.292.5522.1655. [DOI] [PubMed] [Google Scholar]
- 17.Adcock GJ, et al. Lake Mungo 3: A response to recent critiques. Archaeol Ocean. 2001;36(3):170–174. [Google Scholar]
- 18.Durband AC, Westaway MC, Rayner DR. Interproximal grooving of lower second molars in WLH 4. Aust Archaeol. 2012;75(December 2012):118–120. [Google Scholar]
- 19.Carpenter ML, et al. Pulling out the 1%: Whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am J Hum Genet. 2013;93(5):852–864. doi: 10.1016/j.ajhg.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millar CD, Huynen L, Subramanian S, Mohandesan E, Lambert DM. New developments in ancient genomics. Trends Ecol Evol. 2008;23(7):386–393. doi: 10.1016/j.tree.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Dabney J, Meyer M, Pääbo S. Ancient DNA damage. Cold Spring Harb Perspect Biol. 2013;5(7):a012567. doi: 10.1101/cshperspect.a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawyer S, Krause J, Guschanski K, Savolainen V, Pääbo S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS One. 2012;7(3):e34131. doi: 10.1371/journal.pone.0034131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofreiter M, et al. The future of ancient DNA: Technical advances and conceptual shifts. BioEssays. 2015;37(3):284–293. doi: 10.1002/bies.201400160. [DOI] [PubMed] [Google Scholar]
- 24.Olley JM, Roberts RG, Yoshida H, Bowler JM. Single-grain optical dating of grave-infill associated with human burials at Lake Mungo, Australia. Quat Sci Rev. 2006;25(1920):2469–2474. [Google Scholar]
- 25.Gillespie R. Dating the first Australians. Radiocarbon. 2002;44(2):455–472. [Google Scholar]
- 26.Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 27.Behar DM, et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet. 2012;90(4):675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brotherton P, Sanchez JJ, Cooper A, Endicott P. Preferential access to genetic information from endogenous hominin ancient DNA and accurate quantitative SNP-typing via SPEX. Nucleic Acids Res. 2010;38(2):e7. doi: 10.1093/nar/gkp897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brotherton P, et al. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 2007;35(17):5717–5728. doi: 10.1093/nar/gkm588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felsenstein J. Inferring Phylogenies. Macmillan; New York: 2004. [Google Scholar]
- 31.Bergsten J. A review of long branch attraction. Cladistics. 2005;21(2):163–169. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 32.Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408(6813):708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 33.Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29(13):1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heupink TH, van Grouw H, Lambert DM. The mysterious Spotted Green Pigeon and its relation to the Dodo and its kindred. BMC Evol Biol. 2014;14(1):136. doi: 10.1186/1471-2148-14-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamba C, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010(6):t5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.