Significance
The Deleted in Azoospermia (DAZ) family of RNA-binding proteins, consisting of Boule, Daz-like (Dazl), and DAZ, plays important roles in gametogenesis. Here we demonstrate that boule2 in the freshwater planarian Schmidtea mediterranea is necessary for the maintenance of early male germ cells, similar to the function of its vertebrate ortholog, Dazl. Our results are significant in that a premeiotic role for an invertebrate boule homolog has not been described to date. Furthermore, we functionally characterize planarian homologs of human DAZL/DAZ-associated proteins and mRNA targets. Our study alters the current understanding of DAZ family evolution and establishes S. mediterranea as a tractable model organism for the study of premeiotic functions of the DAZ family, and its binding partners and targets.
Keywords: germ cells, spermatogenesis, Deleted in Azoospermia, DAZ, DAZL
Abstract
Mutations in Deleted in Azoospermia (DAZ), a Y chromosome gene, are an important cause of human male infertility. DAZ is found exclusively in primates, limiting functional studies of this gene to its homologs: boule, required for meiotic progression of germ cells in invertebrate model systems, and Daz-like (Dazl), required for early germ cell maintenance in vertebrates. Dazl is believed to have acquired its premeiotic role in a vertebrate ancestor following the duplication and functional divergence of the single-copy gene boule. However, multiple homologs of boule have been identified in some invertebrates, raising the possibility that some of these genes may play other roles, including a premeiotic function. Here we identify two boule paralogs in the freshwater planarian Schmidtea mediterranea. Smed-boule1 is necessary for meiotic progression of male germ cells, similar to the known function of boule in invertebrates. By contrast, Smed-boule2 is required for the maintenance of early male germ cells, similar to vertebrate Dazl. To examine if Boule2 may be functionally similar to vertebrate Dazl, we identify and functionally characterize planarian homologs of human DAZL/DAZ-interacting partners and DAZ family mRNA targets. Finally, our phylogenetic analyses indicate that premeiotic functions of planarian boule2 and vertebrate Dazl evolved independently. Our study uncovers a premeiotic role for an invertebrate boule homolog and offers a tractable invertebrate model system for studying the premeiotic functions of the DAZ protein family.
Human male infertility is often associated with Y chromosome microdeletion (1). In 1976, Tiepolo and Zuffardi proposed the existence of an azoospermia factor (AZF) located on the distal arm of the Y chromosome, which could result in infertility when deleted (2). A strong candidate for AZF is Deleted in Azoospermia (DAZ), a Y chromosome gene (3, 4). Soon after the discovery of DAZ, the mouse and human DAZ homolog, DAZ-like (Dazl/DAZL) (5–7), and the Drosophila DAZ homolog, boule (8), were identified. Phylogenetic analyses showed that boule is the ancestral member of the family (9) and is predicted to be present in most metazoans. Dazl resulted from duplication of boule in an early vertebrate ancestor about 450 million years ago (9). DAZ, the newest member of the family, arose from duplication of its autosomal homolog Dazl about 30 million years ago (9). The DAZ locus is on the Y chromosome and is restricted to humans and Old World monkeys. Thus, in invertebrates, the DAZ family is currently represented only by boule; nonprimate vertebrates contain both boule and Dazl; and humans and Old World monkeys possess boule, DAZL, and DAZ (9).
Structurally, DAZ family members are characterized by a highly conserved RNA recognition motif (RRM) for binding of target mRNA and a DAZ motif for binding of partner proteins. Boule and Dazl have a single DAZ motif, whereas DAZ has multiple DAZ repeats in tandem (3). Functionally, members of the DAZ family are known to play important roles in both male and female germ cell development, although boule, Dazl, and DAZ function at different stages of gametogenesis.
boule appears to function in meiotic or postmeiotic germ cells. Disruption of boule in Drosophila melanogaster results in male germ cell meiotic arrest at the G2/M transition, whereas female flies are unaffected (8). In Caenorhabditis elegans, loss of function of the boule ortholog daz1 causes sterility by blocking oocytes at the pachytene stage of meiosis I (10). Male boule knockout mice are not capable of spermatid maturation, and there is no effect on female gametogenesis (11).
In contrast to boule, Dazl appears to have an earlier role in germ cell maintenance. Xenopus Xdazl is present in the germ plasm (12) and in the absence of functional maternal Xdazl, primordial germ cells (PGCs) in tadpoles are specified, but fail to differentiate (13). In zebrafish, zDazl is expressed in germ plasm of oocytes, activates tudor domain containing protein 7 (tdrd7), and antagonizes miR-430, a microRNA that represses tdrd7 and dazl mRNAs in PGCs (14). In Dazl-deficient mice of mixed genetic background, Aaligned spermatogonia are unable to differentiate (15). In C57BL/6 mice, Dazl is first expressed at embryonic day 11.5 (E11.5) (16) and is essential for the survival of both male and female germ cells (17, 18). In male Dazl null mice, PGCs are specified and reach the gonad, but by E15.5, show reduced expression of typical germ cell markers and undergo apoptosis (17). Thus, in vertebrates Dazl plays a role before meiosis. Finally, Y chromosome deletions spanning the DAZ gene are the best-known molecular cause of human male infertility (3, 19), resulting in a range of male germ-line phenotypes from complete absence of germ cells to sperm maturation defects (3).
Many years of work have led to a consensus with regard to when the functional divergence between meiotic boule and premeiotic Dazl/DAZ occurred (9, 20–22). Based on the roles members of this family play across different phyla, it has long been assumed that vertebrate DAZ homologs acquired a premeiotic function following duplication of boule in a vertebrate ancestor. This hypothesis was proposed based on phylogenetic analysis of both gene families as well as the finding that more exon–intron splicing sites are shared between human BOULE and DAZL than between human BOULE and Drosophila boule. In addition, human BOULE and DAZL have an identical number of exons, suggesting a close relationship between vertebrate DAZ homologs (9). Based on studies performed in C. elegans and D. melanogaster, it was also thought that invertebrates only had a single representative of the DAZ family; however, it was recently shown that the flatworm Macrostomum lignano has three paralogs of boule (macbol1, macbol2, and macbol3) (21). RNA interference (RNAi) against macbol2 yielded no detectable phenotypes, macbol1 RNAi resulted in accumulation of primary spermatocytes and degeneration of more differentiated germ cells of testes, and macbol3 was required for oocyte maturation and female fertility. This study raised several questions: Do other invertebrates have multiple DAZ family members? If so, do any of these invertebrate paralogs play a premeiotic role in germ cell development? Is the premeiotic function of this protein family indeed derived, as currently hypothesized? We addressed these questions using the planarian Schmidtea mediterranea, a freshwater flatworm that has emerged as an important model for studying regeneration and germ cell biology (23–29).
Results and Discussion
S. mediterranea Has Two Homologs of boule That Perform Different Functions in Spermatogenesis.
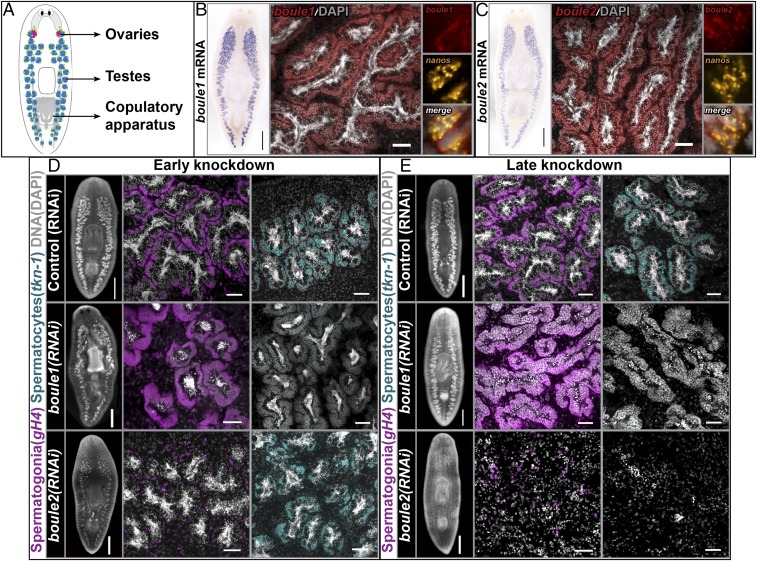
We identified two planarian boule homologs, boule1 and boule2, from the S. mediterranea genome database (30) based on the presence of highly conserved RRMs and DAZ motifs characteristic of DAZ family members. To determine the spatial expression of these genes, we performed colorimetric in situ hybridization (ISH) on sexual adults (illustration in Fig. 1A). Both boule1 and boule2 were expressed in male and female gonads (Fig. 1 B and C and SI Appendix, Fig. S1). To determine which specific cells in the testes expressed these transcripts, we performed fluorescence in situ hybridization (FISH) followed by confocal imaging. boule1 and boule2 mRNAs were detected in spermatogonial stem cells (SSCs) (Fig. 1 B and C and SI Appendix, Fig. S2), spermatogonia (SI Appendix, Fig. S2), and spermatocytes, to a lesser extent in spermatids, and were absent from mature sperm.
Fig. 1.
Planarian boule1 and boule2 perform different functions in spermatogenesis. (A) Illustration of sexual planarian depicting the positions of reproductive structures. Ovaries are in red, testes are in blue, and germ-line stem cells are in green. (B and C) Colorimetric ISH showing boule1 and boule2 mRNA expression in the testes. (Scale bars, 1 mm.) FISH detects boule1 and boule2 expression in spermatogonial stem cells (SSCs), spermatogonia, and spermatocytes. (Scale bars, 50 μm.) Coexpression of boule transcripts with nanos+ SSCs is shown. (D) Animals fixed following two feedings of dsRNA spaced 4–5 d apart. Control (RNAi), boule1(RNAi), and boule2(RNAi) animals labeled with germinal histone H4 (gH4) in magenta to detect mitotic spermatogonia and tektin-1 (tkn-1) in cyan to mark meiotic spermatocytes. boule1(RNAi) animals show absence of meiotic labeling, but expansion of spermatogonia. The spermatogonial layer is reduced in boule2(RNAi) animals, whereas the spermatocyte population is comparable to controls. (E) Animals fixed following four feedings of dsRNA spaced 4–5 d apart. boule1(RNAi) testes contain clusters of SSCs and spermatogonia; meiotic and postmeiotic male germ cells are absent. boule2(RNAi) animals show a loss of all male germ cells. The remaining gH4 label coincides with neoblasts (somatic stem cells). Left in D and E show whole-mount images. (Scale bars, 1 mm.) Middle and Right in D and E show high magnification view of testis lobes. (Scale bars, 50 μm.)
SSCs of S. mediterranea give rise to spermatogonia, which undergo three rounds of mitosis with incomplete cytokinesis to generate cysts containing eight primary spermatocytes. These meiotic spermatocytes generate 32 spermatids that mature into sperm (SI Appendix, Fig. S3A) (25). We will refer to SSCs and spermatogonia as early male germ cells to distinguish them from the more differentiated meiotic and postmeiotic germ cells. We have previously identified markers for various stages of planarian spermatogenesis (SI Appendix, Fig. S3A) (23, 24, 26, 28). RNA ISH using these markers enables us to assess which male germ cell population is affected following gene knockdown experiments.
To determine the roles of boule1 and boule2 in testes, we knocked them down by RNAi and observed effects during homeostasis (in uninjured animals). In early stages of boule1(RNAi) (two feedings, 4–5 d apart), tektin-1+ (tkn-1+) primary spermatocytes (28) were absent (n = 6/6, Fig. 1D). This spermatocyte loss was accompanied by a concomitant increase in the germinal histone H4+ (gH4+) mitotic spermatogonial layer (23, 24) (n = 6/6, Fig. 1D). At this RNAi timepoint, boule1(RNAi) animals showed no discernible changes in the nanos+ SSC population (SI Appendix, Fig. S3B). The protein kinase A+ (pka+) spermatid population is slightly reduced in boule1(RNAi) animals, possibly as a secondary effect of spermatocyte loss (SI Appendix, Fig. S3B). In late stages of boule1(RNAi) (four feedings, 4–5 d apart), the testes contained expanded clusters of spermatogonia, with numbers of SSCs comparable to control animals; more mature, meiotic, and postmeiotic male germ cells were absent (Fig. 1E and SI Appendix, Fig. S3C).
By contrast, in early boule2 knockdown animals, there was a reduction in gH4+ spermatogonia (n = 5/5, Fig. 1D), but tkn-1+ meiotic spermatocytes remained comparable to control animals (n = 5/6, Fig. 1D). Half of boule2(RNAi) animals (n = 3/6) had no nanos+ SSCs (SI Appendix, Fig. S3B); pka+ spermatids appeared unaffected in boule2(RNAi) animals at these early stages (SI Appendix, Fig. S3B). We validated the specificity of the gene knockdowns to ensure that RNAi of either boule1 or boule2 did not directly affect the other paralog (SI Appendix, Fig. S4). To examine whether early germ cells were being lost at least in part due to apoptosis, we performed TUNEL staining on early boule2(RNAi) animals and found that these animals showed an increase in apoptosis compared with control or boule1(RNAi) animals (SI Appendix, Fig. S5). In late stages of boule2(RNAi), there was a complete loss of all male germ cells (Fig. 1E and SI Appendix, Fig. S3C).
From our RNAi experiments, we conclude that boule1 is required for the maintenance and/or formation of meiotic male germ cells. The meiotic role of planarian boule1 is in agreement with known functions of boule orthologs in other systems. However, boule2 is required for the maintenance of premeiotic male germ cells, SSCs and spermatogonia, remarkably similar to the function of mouse Dazl (7, 15, 17). When boule2 expression is inhibited, the early germ cells appear to undergo increased apoptosis (SI Appendix, Fig. S5).
boule2 Is Required for the Maintenance, but Not Specification, of Early Male Germ Cells.
In addition to their remarkable ability to regenerate all body parts and organ systems, planarians are capable of respecifying germ cells from amputated tissue fragments devoid of reproductive structures (24, 25, 29). Thus, like mammals, planarians can specify their germ line via inductive signals. Within 2 wk of regeneration, germ cells are respecified, as determined by the expression of nanos, the earliest known marker expressed in planarian germ cells (schematic in Fig. 2A) (24, 29). We examined whether boule1 or boule2 is required for respecifying germ cells by knocking down the corresponding genes before amputation.
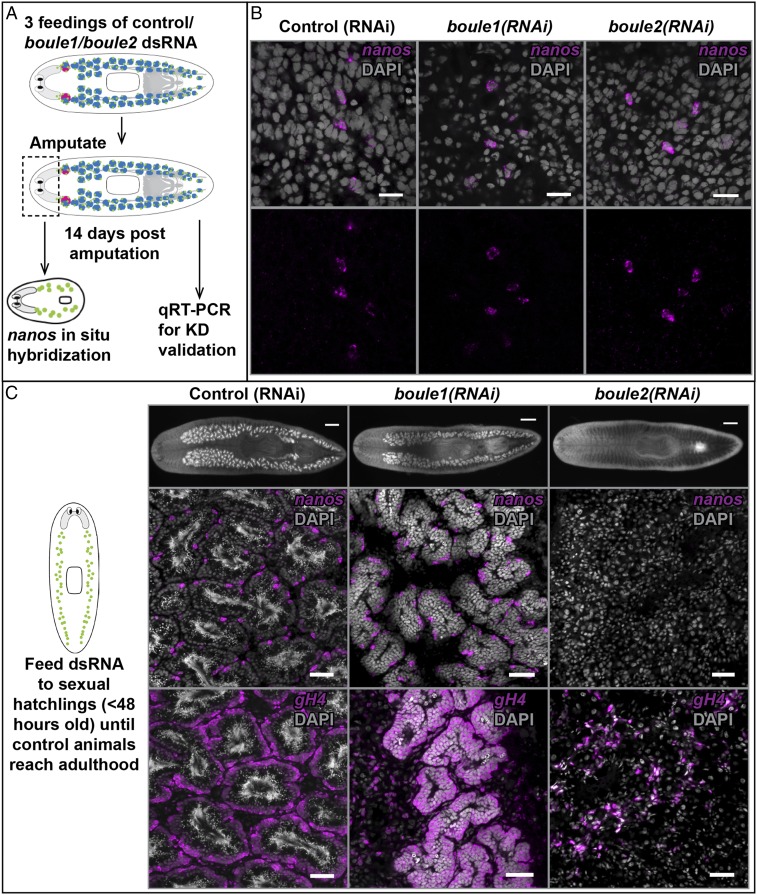
Fig. 2.
boule2 is required for maintenance of early male germ cells but not required for respecification of SSCs. (A) Experimental scheme for testing the requirement of a gene for de novo respecification of SSCs. Animals are fed control/boule1/boule2 dsRNA three times and amputated anterior to the ovaries. Head fragments, lacking reproductive structures, are allowed to regenerate. Tail fragments are also maintained for knockdown validation. At 14 d following amputation, head fragments are fixed for nanos FISH and RNA is extracted from the tail fragment to ensure that test mRNA levels are reduced. nanos labels planarian SSCs. (B) Control (RNAi), boule1(RNAi), and boule2(RNAi) animals all show respecification of nanos+ SSCs. (Scale bars, 100 μm.) (C) Sexual hatchlings (<48 h old) are fed liver containing dsRNA until control animals are sexually mature (∼10–12 feedings over ∼2 mo). SSCs in control (RNAi) animals differentiate and form mature testes. boule1(RNAi) animals have testis lobes with only SSCs (nanos+) and spermatogonia (gH4+). boule2(RNAi) animals lack male germ cells; remnant gH4 signal is due to neoblasts. (Scale bars, 50 μm.)
We found that both boule1 and boule2 were dispensable for the regeneration of nanos+ SSCs (n = 10/10 for both, Fig. 2B). As an additional control, we performed a parallel experiment with dmd1 (SI Appendix, Fig. S6A), a gene previously shown to be required for SSC respecification (29). We confirmed gene knockdowns at 14 d postamputation by quantitative real time-PCR (qRT-PCR) (SI Appendix, Fig. S6B).
To test whether boule1 or boule2 is required for the maintenance and differentiation of early germ cells postspecification, we performed gene knockdowns on sexual hatchlings (<48 h posthatching). At this stage of development, the male gonad of sexual planarians consists of small clusters of nanos+ SSCs and dmd1+ somatic gonadal cells, enabling us to examine the consequences of boule1 or boule2 loss on early male germ cells in the absence of more differentiated cells. When control animals reached adulthood after ∼12 feedings of dsRNA, they exhibited robust spermatogenesis in all samples (n = 14/14, Fig. 2C). SSCs in boule1(RNAi) animals are able to progress through mitosis and form clusters of spermatogonia, but are unable to produce meiotic and postmeiotic cells (n = 13/13, Fig. 2C). boule2(RNAi) animals completely lack male germ cells (n = 14/14, Fig. 2C). We also imaged the hatchlings after two and four feedings of dsRNA to further confirm that the two genes are required for early germ cell maintenance. We found that the knockdown phenotypes are similar to the phenotype seen in sexually mature adults (SI Appendix, Fig. S6 C and D). Experiments on animals regenerating their reproductive system (29) also showed comparable results (SI Appendix, Fig. S7).
Together, these experiments show that neither boule1 nor boule2 is necessary for the specification of male germ cells; however, the two genes perform distinct roles in male germ cells after they are specified. boule1 is required for meiotic progression, and boule2 is required for the maintenance of the earliest male germ cells, nanos+ SSCs. Our observation that boule2 is not necessary for the specification of SSCs, but is required for the maintenance and differentiation of early male germ cells, is similar to the Dazl null phenotype seen in vertebrates (12, 13, 17), further lending support to the hypothesis that planarian boule2 and vertebrate Dazl perform similar functions.
boule1 and boule2 Are Necessary for Oogenesis.
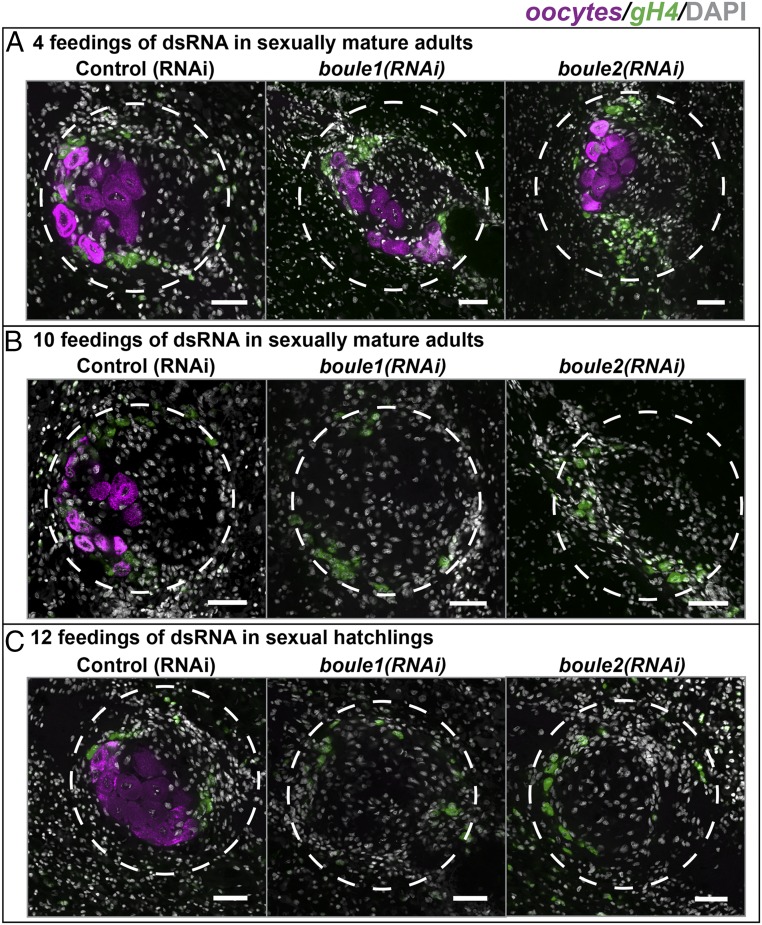
We examined the role of boule1 and boule2 in the ovaries by carrying out gene knockdowns for different lengths of time and during different developmental stages. Following 4 dsRNA feedings (spaced 4–5 d apart), ovaries of boule1(RNAi) and boule2(RNAi) animals appeared comparable to controls (n = 6/6 for all samples, Fig. 3A). However, following prolonged gene knockdown (10 feedings over a period of ∼2 mo), both boule1(RNAi) (n = 4/4) and boule2(RNAi) (n = 6/6) animals lacked oocytes, whereas early gH4+ female germ cells were still present (Fig. 3B). Similarly, when sexual hatchlings were fed boule1 and boule2 dsRNA over a period of 2 mo, the animals lacked mature oocytes, but gH4+ female germ cells were present (n = 4/4 for all samples, Fig. 3C). The dual role of planarian boule genes in both testes and ovaries is especially interesting because, with the exception of Dazl, other members of the DAZ family (boule orthologs in various systems and DAZ) appear restricted in function exclusively to the male or the female germ line.
Fig. 3.
Planarian boule genes play a role in the ovaries. (A) Following 4 feedings of dsRNA, boule1(RNAi) and boule2(RNAi) female gonads appear similar to controls. Oocytes are marked using Contig2621 (magenta) (26), and gH4 (green) labels early female germ cells. (B) Following 10 feedings of dsRNA, boule1(RNAi) and boule2(RNAi) ovaries have early female germ cells, but lack oocytes. (C) A similar absence of oocytes was seen when sexual hatchlings were fed dsRNA over a period of 2 mo. Dashed circles outline the ovaries. (Scale bars, 50 μm.)
Homologs of Vertebrate DAZ-Associated Proteins Are Expressed and Function in the Testes of S. mediterranea.
Yeast two-hybrid screens and other in vitro studies (31–34) have identified several potential DAZL/DAZ-interacting partners using human DAZ as bait. Homologs of these genes have not been described in C. elegans and D. melanogaster, which only possess meiotic boule (Methods). To further investigate the functions of these DAZL/DAZ-interacting partners, we sought to identify planarian homologs of DAZ-binding partners.
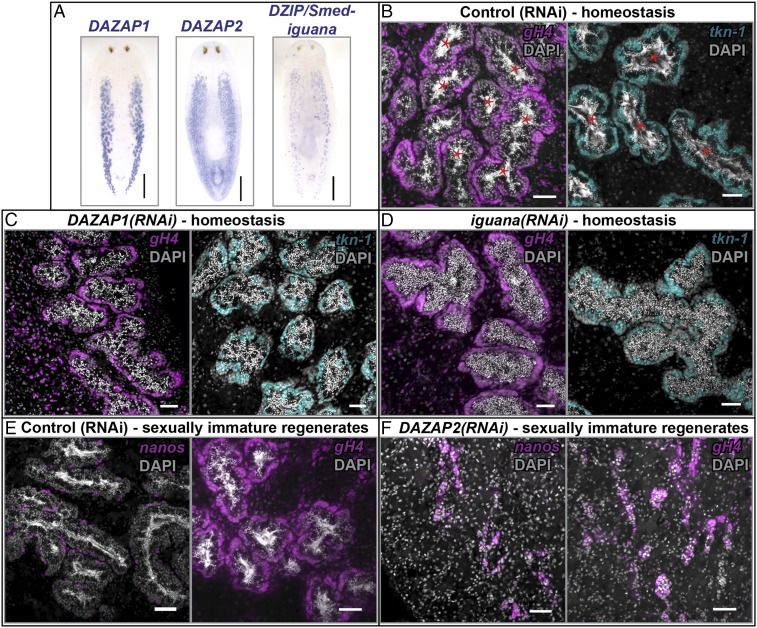
Using BLAST similarity search, we identified planarian homologs of putative DAZL/DAZ-interacting partners—DAZAP1, DAZAP2, and DZIP (Smed-iguana)—and found that these genes were expressed in the testes (Fig. 4A). To determine the role of these genes in spermatogenesis, we performed RNAi during homeostasis (in sexually mature adults), during development (in hatchlings), as well as in sexually immature regenerates (animals fed dsRNA three times, amputated prepharyngeally to induce regression of testes (29), and refed dsRNA during regeneration). DAZAP1(RNAi) animals lacked elongated spermatids and mature sperm, whereas other male germ cells appeared intact in all three experimental conditions (n = 6/6 for all; Fig. 4C and SI Appendix, Fig. S8 C, E, and F and Table S1). Therefore, similar to DAZAP1 knockout mice, which lack mature male gametes (35), DAZAP1 is required for spermiogenesis.
Fig. 4.
Homologs of vertebrate DAZ-associated proteins are expressed and function in the testes of S. mediterranea. (A) DAZAP1, DAZAP2, and DZIP (Smed-iguana) transcripts are detected in the testes by ISH. (Scale bars, 1 mm.) (B) Adults fed control dsRNA in homeostasis show robust spermiogenesis. Thin, threadlike nuclei of mature sperm in the lumen of testis lobes are labeled with DAPI (marked by red asterisk). Both (C) DAZAP1(RNAi) and (D) iguana(RNAi) animals lack mature sperm, but have spermatogonia and spermatocytes similar to control (RNAi) animals. (E) Sexually immature regenerates fed control dsRNA regenerate their testes, whereas (F) DAZAP2 dsRNA-fed regenerates have small testes containing only SSCs and spermatogonia. (Scale bars, 50 μm.)
DAZAP2 did not have a germ cell RNAi phenotype in sexually mature adults (SI Appendix, Fig. S8 A and B) or hatchlings (SI Appendix, Table S1). However, regeneration experiments revealed that DAZAP2(RNAi) regenerates either lacked testes (n = 2/6) or had regressed testis lobes containing only SSCs and spermatogonia (n = 4/6) (Fig. 4 E and F). Understanding this regeneration-specific role of DAZAP2 in male germ cells requires further investigation. DAZAP1 and DAZAP2 are not required for respecification of nanos+ SSCs (SI Appendix, Table S1).
The planarian DZIP gene, known as Smed-iguana, has previously been shown to be required for ciliogenesis in asexual planarians (36). Regenerating iguana(RNAi) asexuals are able to produce normal blastemas, but do not form ciliated epidermis (leading to defects in cilia-driven locomotion) or ciliated protonephridia (resulting in bloating and blistering defects due to disrupted osmoregulatory function) (36). iguana(RNAi) in the sexual strain led to bloating defects similar to the asexual strain (SI Appendix, Table S1). Furthermore, we observed spermiogenesis defects in iguana(RNAi) animals (Fig. 4D and SI Appendix, Fig. S8D). In addition to defects in the testes, sexually immature iguana(RNAi) regenerates underwent lysis during regeneration (SI Appendix, Table S1). This lysis phenotype was not reported in asexual planarians; differences in our observations may be explained by differences in dsRNA-treatment regimes.
Planarian DAZAP1, DAZAP2, and iguana play roles in spermatogenesis, but the knockdown of these genes does not phenocopy boule1(RNAi) or boule2(RNAi), in that these genes appear to be required for later stages of germ cell maturation. Several possibilities may explain this finding. Because there are multiple DAZ binding partners, knockdown of one factor alone may not be sufficient to recapitulate the boule1/2(RNAi) phenotype. iguana could have pleiotropic effects as it is also required for regeneration and osmoregulation. Alternatively, boule1 and boule2 may play a role in postmeiotic spermatid elongation and maturation (similar to DAZAP1 and iguana), but the rapid loss of meiotic and premeiotic germ cells may not allow us to observe these possible secondary, less obvious effects. It is also possible that the gonadal function of these putative binding partners is independent of boule1 or boule2. Together, our data support roles for planarian DAZAP1, DAZAP2, and iguana in male germ cell differentiation (SI Appendix, Table S1).
Transcripts of other broadly conserved DAZ family interacting partners, such as Pumilio and Poly(A) Binding Protein (PABP) (34, 37, 38), are also enriched in planarian testes. pumilio(RNAi) is lethal, consistent with a similar observation in the planarian Dugesia japonica (39), and specific germ cell defects were not detected before death (SI Appendix, Table S1). Knockdown of planarian PABPC has been described previously (26) and is remarkably similar to the boule1(RNAi) phenotype in that meiotic and postmeiotic male germ cells are lost with a concomitant accumulation of spermatogonia. The identification of these homologs of vertebrate DAZ-associated proteins in S. mediterranea is promising as it allows functional studies of these genes and other putative DAZ-associated proteins in a tractable invertebrate model system.
Knockdown of Putative Planarian DAZ Family Targets Phenocopies boule2(RNAi).
Several in vitro studies have identified presumptive mRNA targets for the DAZ protein family, but to what extent these targets overlap between different orthologs (Boule, Dazl, and DAZ) is uncertain (40–44). We identified and cloned a number of planarian homologs of putative DAZ family targets (Fig. 5A and SI Appendix, Fig. S9A and Table S2), but we will focus on the putative targets with germ cell RNAi phenotypes.
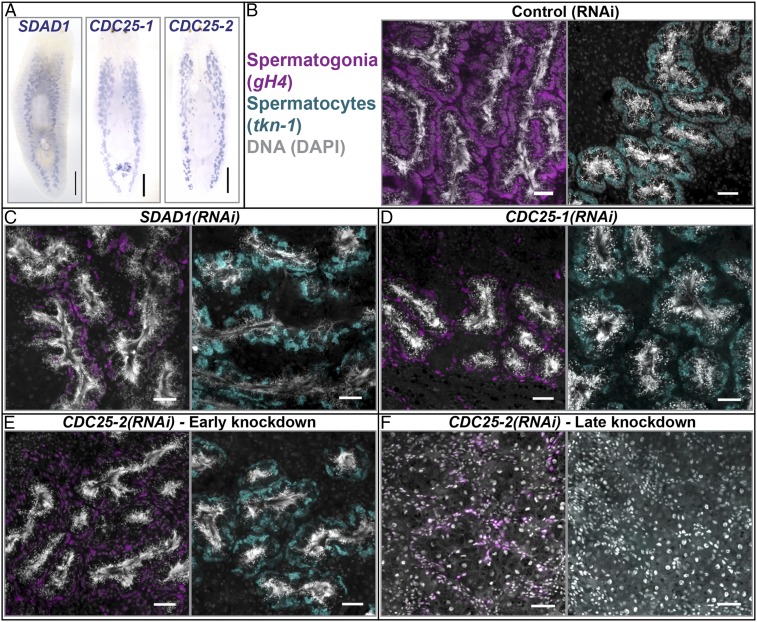
Fig. 5.
Knockdown of putative DAZ family targets phenocopies boule2(RNAi). (A) SDAD1, CDC25-1, and CDC25-2 are enriched in planarian testes. (Scale bars, 1 mm.) (B–E) Animals fed control, SDAD1, CDC25-1, and CDC25-2 dsRNA (three feedings spaced 4–5 d apart) labeled with gH4 and tkn-1. Similar to boule2(RNAi), RNAi knockdown of these putative targets results in animals having fewer spermatogonia, and the spermatocyte layer remains intact. Continued SDAD1(RNAi) and CDC25-1(RNAi) results in lysis, whereas (F) CDC25-2(RNAi) animals lose all male germ cells over time. (Scale bars, 50 μm.)
SDAD1, a homolog of the yeast gene severe depolymerization of actin, is a putative target of human DAZL and PUMILIO 2 (44). A function for SDAD1 in spermatogenesis has not been reported previously. By ISH, we find that Smed-SDAD1 was detected in the testes as well as soma (Fig. 5A). RNAi experiments showed that SDAD1 is required for maintenance of SSCs (n = 3/6) and spermatogonia (n = 6/6), similar to boule2(RNAi) (Fig. 5C and SI Appendix, Fig. S9C). SDAD1(RNAi) animals undergo lysis upon continued knockdown or when amputated (SI Appendix, Table S2), indicating a possible somatic function and precluding the possibility of testing if SDAD1 is necessary for specification of early germ cells.
The CDC25 homolog twine is a known target of Boule in D. melanogaster (40). Two of the planarian homologs of CDC25 (a somatic planarian CDC25 homolog has been described previously (45) and will not be discussed here), designated CDC25-1 and CDC25-2, were expressed in testes (Fig. 5A). Interestingly, following three feedings of CDC25-1 or CDC25-2 dsRNA in adults, animals showed defects similar to boule2(RNAi): the spermatogonial layer was reduced, whereas the spermatocyte layer appeared intact (n = 6/6 for both knockdowns, Fig. 5 D and E). The numbers of SSCs and spermatids were largely unaffected at the initial stages of knockdown (SI Appendix, Fig. S9 D and E); at later stages, all male germ cells were absent (Fig. 5F and SI Appendix, Table S2). We next tested the requirement of CDC25-1 and CDC25-2 for specification and maintenance of early germ cells in sexual regenerates. CDC25-1(RNAi) animals do not regenerate, and undergo lysis, but there are no male germ cells present in regenerates before lysis (SI Appendix, Table S2). CDC25-2(RNAi) sexual regenerates phenocopy boule2(RNAi) regenerates—these animals respecify their SSCs (n = 9/9; SI Appendix, Table S2), but cannot maintain early germ cell clusters (n = 6/6; SI Appendix, Fig. S9 F and G). The in vitro prediction that these transcripts are DAZ family targets in other systems, combined with the similarity of RNAi phenotypes between these genes and boule2, makes these transcripts strong candidates for putative targets regulated by planarian Boule2.
Premeiotic Functions of the DAZ Family Evolved Independently in Planarians and Vertebrates.
Vertebrate Dazl, which plays a premeiotic role in germ cells, arose either during vertebrate evolution, or was present in a last common bilaterian ancestor and was subsequently lost in some invertebrates. Based on the presence of a single DAZ family representative, boule, in both C. elegans and D. melanogaster, phylogenetic analyses, and comparison of gene structure and intron/exon counts, it has been proposed that Dazl arose through duplication of boule in the vertebrate stem lineage (9). Our identification of multiple paralogs of boule in an invertebrate model system, combined with the premeiotic germ cell function for one of these paralogs, provides us valuable tools for testing this hypothesis in a phylogenetic context.
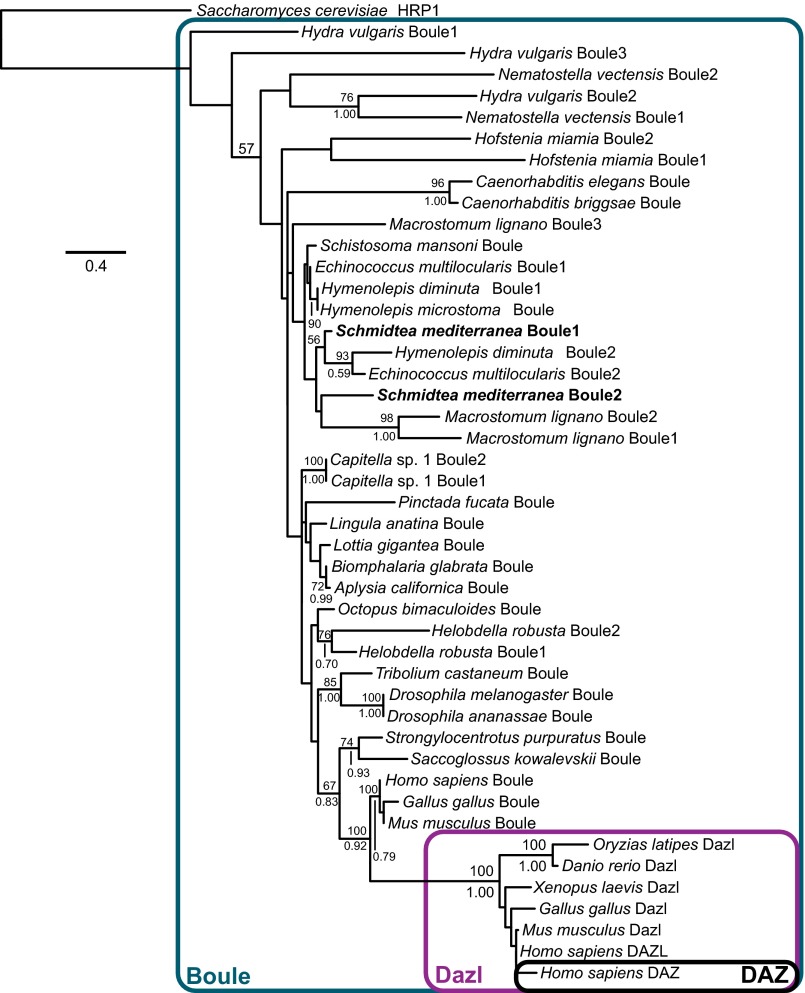
We obtained multiple Boule sequences from diverse animal phyla (accession nos. in SI Appendix, Table S3; alignments in SI Appendix, Fig. S10A), placing special focus on invertebrates with multiple annotated Boule homologs. We performed both maximum likelihood (ML) (46) and Bayesian inference (BI) (47) analyses and found that S. mediterranea Boule paralogs were recovered in a clade formed by other platyhelminth Boule orthologs (Fig. 6). The short patristic distance between S. mediterranea paralogs suggests lineage-specific differentiation of premeiotic and meiotic functions of DAZ family members in flatworms and vertebrates. When we enforced a topological constraint to render a single origin of premeiotic Boule function, forcing monophyly of planarian Boule2 and vertebrate Dazl/DAZ clade, the constrained tree was significantly less likely than the optimal ML tree (Methods). The phylogenetic distance between S. mediterranea Boule paralogs and their vertebrate orthologs supports a scenario of independent origins of premeiotic DAZ family members in planarians and vertebrates.
Fig. 6.
Phylogenetic analysis reveals independent origins of planarian Boule2 and vertebrate Dazl. Phylogenetic tree topology of DAZ gene family from ML and BI analysis. Numbers above nodes indicate ML bootstrap resampling frequencies (500 replicates). Numbers below nodes indicate Bayesian posterior probability values.
To infer whether the premeiotic planarian Boule had diverged from its ancestral sequence (an independent test of neofunctionalization) (48, 49), we examined the ratio of branch lengths (sequence divergences) of premeiotic and meiotic DAZ family members in two planarians and three vertebrates, with branch lengths drawn from the Bayesian postburnin tree set (SI Appendix, Fig. S10B). For both S. mediterranea and the vertebrates, the distributions of ratios of premeiotic paralog branch lengths to meiotic paralog branch lengths were highly comparable, in contrast to the ratio distribution for the Boule proteins of Macrostomum lignano. This result is consistent with neofunctionalization of planarian and vertebrate premeiotic Boule derivatives.
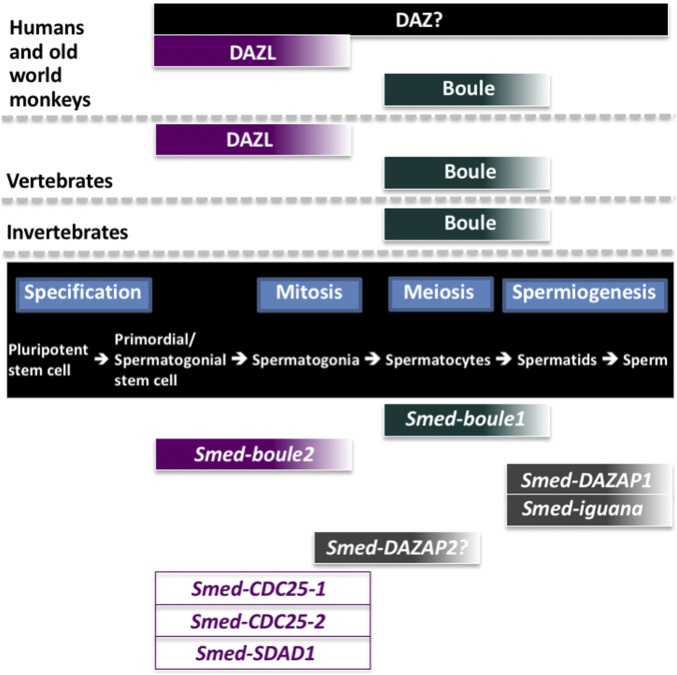
boule, Dazl, and DAZ play crucial and conserved roles in gametogenesis across the animal kingdom (Fig. 7). However, there is considerable phenotypic diversity caused by defects in the DAZ family of proteins, and our present study adds another dimension to the understanding of these genes. Our study also raises many interesting questions. For instance, have vertebrate DAZ-associated proteins evolved independently in planarians, or have they been lost in Ecdysozoans such as C. elegans and D. melanogaster, especially in light of the finding that a DAZAP-like protein has been described in the flatworm D. japonica (50)? Another interesting question is why some invertebrates have multiple boule homologs and others do not. Functional and phylogenetic studies of boule genes in other species with multiple boule paralogs will open the field to further address these questions and will help illuminate the entire range of functions of the DAZ protein family.
Fig. 7.
Summary of DAZ family functions in planarians and other systems. (Center) Different stages of spermatogenesis. (Upper) Known functions of the DAZ family in male germ cell development in other systems. (Lower) Summary of the functions of planarian boule genes, their putative-associated proteins, and targets.
Methods
Planarian Culture.
Sexual planarians were maintained in 0.75× Montjuïc salts at 18 °C (24). Animals were fed organic calf liver and starved for 1 wk before use.
Identification and Cloning of boule Homologs, Putative Binding Partners, and Targets.
Planarian boule homologs were identified by the presence of RRM and DAZ motifs and cloned into pJC53.2 (27). The full-length sequence for boule2 was obtained from PlanMine v1.0 (51). Planarian homologs of putative binding partners and targets were identified from the Smed genome database (30), based on sequence similarity to human counterparts. More specifically, the amino acid sequence of human/vertebrate DAZ-associated proteins and targets was obtained from National Center for Biotechnology Information (NCBI) and tblastn analysis was performed in PlanMine v1.0. The top genes obtained from this search were subjected to a reciprocal blastp against NCBI protein databases to ensure that the planarian gene was indeed a homolog of the human gene. BLAST analysis comparing human DAZAP1 to FlyBase and WormBase revealed a heterogeneous nuclear ribonucleoprotein, the reciprocal protein blast of which to NCBI protein databases did not yield DAZAP1 as the highest hit. No sequences corresponding to DAZAP2 and DZIP/iguana were found. Cloning primers are in SI Appendix, Table S4.
dsRNA Synthesis and RNAi.
cDNAs corresponding to boule1 and boule2 cloned in pJC53.2 (27) were used as template to generate dsRNA by in vitro transcription (IVT). The 20-μL IVT reaction contains 2 μL 10× high yield transcription buffer (0.4 M Tris pH 8.0, 0.1 M MgCl2, 20 mM spermidine, 0.1 M DTT), 5 μL 25 mM rNTPs (Promega), 1 μL T7 polymerase, 1 μL thermostable inorganic pyrophosphatase (TIPP; 2,000 units/mL; New England Biolabs), 0.5 μL recombinant ribonuclease inhibitor (RNasin; 2,500 units/mL; Promega), and 0.5–2.5 μg of PCR product. Reactions were incubated at 37 °C overnight, then treated with 1 μL of RQ1 RNase-free DNase (Fisher Scientific) for 20 min at room temperature. Each reaction was brought up to 100 μL, followed by denaturating and annealing at the following temperatures: 95 °C (3 min), 75 °C (3 min), 50 °C (3 min), and room temperature (5 min). dsRNA was precipitated using ammonium acetate (2.5 M final concentration) plus two volumes of 100% ethanol. dsRNA (0.4–1 μg) was mixed with 10 μL of 3:1 liver:Montjuïc salts mix. Control animals were fed dsRNA synthesized from a nonplanarian gene inserted in pJC53.2.
Riboprobe Synthesis.
boule1 and boule2 cDNA cloned in pJC53.2 (27) were used as templates to generate riboprobes. Each 20-μL reaction contained 2 μL 10× high yield transcription buffer (0.4 M Tris pH 8.0, 0.1 M MgCl2, 20 mM spermidine, 0.1 M DTT), 1 μL 10/6 mM rNTPs (CTP, ATP, and GTP 10 mM final, UTP 6 mM final) (Promega), 0.4 μL of Digoxigenin-12-UTP (Roche), 0.6 μL recombinant ribonuclease inhibitor (RNasin, 2,500 units/mL) (Promega), 2 μL of SP6/T3 RNA polymerase, and 0.5–2.5 μg of PCR product. Riboprobes were synthesized for 4–5 h at 37 °C, treated with 1 μL of RQ1 RNase-free DNase (Fisher Scientific) for 20 min at room temperature, and precipitated with ammonium acetate (2.5 M final concentration) plus two volumes of 100% EtOH.
ISH.
ISH was performed as described previously (52). Detailed methods are provided in SI Appendix, SI Methods.
TUNEL on Sections.
The planarian whole-mount TUNEL protocol was modified for cryosections (53, 54). Detailed methods are provided in SI Appendix, SI Methods.
Multiple Sequence Alignment and Phylogenetic Analysis.
Peptide sequences of 46 Boule, Dazl, and DAZ RRMs (accession nos. in SI Appendix, Table S3) were aligned using MUSCLE v. 3.8 (55) with default alignment parameters. HRP1 of Saccharomyces cerevisiae was used as an outgroup. The sequence alignment is provided as SI Appendix, Fig. S10A. Tree topologies were inferred using ML and BI. ML analysis was done using RAxML, 100 independent searches, 500 bootstraps, using LG+Gamma model of evolution (46). BI analysis was done using MrBayes v. 3.2 (47). Four runs, each with four chains and a default distribution of chain temperatures, were run for 2 × 106 generations, with sampling every 2,000th iteration. A mixed+I+G model (56) was implemented, following model selection with ProtTest v.3 (57). Convergence was independently assessed using average split frequency and with Tracer v. 1.6 (58). As a conservative treatment, 5 × 105 generations (25%) were discarded as burnin.
Likelihood Ratio Tests.
The strength of phylogenetic evidence for independent origins of premeiotic DAZ family representatives in vertebrates and the planarian was assessed using Shimodaira–Hasegawa (59) and approximately unbiased (60) tests in RAxML v. 7.7.5 (46). Topological constraint to render a single origin of premeiotic function was enforced and the resulting tree topology was compared to our unconstrained ML tree. Per-site log likelihood values were computed using the -f g command in RAxML v. 7.7.5. The resulting likelihoods were analyzed using CONSEL v. 0.1i (61). using 10,000 bootstrap replicates to conduct the tests of monophyly.
Supplementary Material
Acknowledgments
We thank Tracy Chong for the illustrations; Mansi Srivastava for Hofstenia miamia boule sequences; Umair Khan for the oocyte marker clone; and Tracy Chong, Jayhun Lee, Rachel Roberts-Galbraith, and Rachel Smith-Bolton for their valuable comments on the manuscript. This work was supported by Damon Runyon Cancer Research Foundation Postdoctoral Fellowship Award 2135-12 (to M.I.), NSF Award DBI-1202751 (to P.P.S.), NSF Grant IOS-1257217 (to C.G.E.), and NIH Grant R01 HD043403 (to P.A.N.). P.A.N. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KU519616 (boule1), KU519617 (boule2), KU852687 (CDC25-1), KU852688 (CDC25-2), KU852689 (CDC25-3), KU852669 (DAZAP1), KU852670 (DAZAP2), KU852671 (DZIP), KU852676 (GRSF1-1), KU852677 (GRSF1-2), KU852680 (PAM), KU852681 (Pumilio), KU852682 (Ringo/SPY), KU852686 (SDAD1), KU852672 (TPX1), KU852673 (TRF2-1), KU852674 (TRF2-2), KU852675 (TRF2-3), KU852683 (TSSK), KU852684 (Vasa1), and KU852685 (Vasa2)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521341113/-/DCSupplemental.
References
- 1.Reynolds N, Cooke HJ. Role of the DAZ genes in male fertility. Reprod Biomed Online. 2005;10(1):72–80. doi: 10.1016/s1472-6483(10)60806-1. [DOI] [PubMed] [Google Scholar]
- 2.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34(2):119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 3.Reijo R, et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet. 1995;10(4):383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 4.Reijo R, Alagappan RK, Patrizio P, Page DC. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet. 1996;347(9011):1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- 5.Reijo R, et al. Mouse autosomal homolog of DAZ, a candidate male sterility gene in humans, is expressed in male germ cells before and after puberty. Genomics. 1996;35(2):346–352. doi: 10.1006/geno.1996.0366. [DOI] [PubMed] [Google Scholar]
- 6.Shan Z, et al. A SPGY copy homologous to the mouse gene Dazla and the Drosophila gene boule is autosomal and expressed only in the human male gonad. Hum Mol Genet. 1996;5(12):2005–2011. doi: 10.1093/hmg/5.12.2005. [DOI] [PubMed] [Google Scholar]
- 7.Ruggiu M, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389(6646):73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 8.Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381(6585):783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 9.Xu EY, Moore FL, Pera RA. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci USA. 2001;98(13):7414–7419. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karashima T, Sugimoto A, Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127(5):1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- 11.VanGompel MJW, Xu EY. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum Mol Genet. 2010;19(12):2360–2369. doi: 10.1093/hmg/ddq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125(2):171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- 13.Houston DW, King ML. A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development. 2000;127(3):447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 14.Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4(10):e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrans-Stassen BH, Saunders PT, Cooke HJ, de Rooij DG. Nature of the spermatogenic arrest in Dazl-/- mice. Biol Reprod. 2001;65(3):771–776. doi: 10.1095/biolreprod65.3.771. [DOI] [PubMed] [Google Scholar]
- 16.Seligman J, Page DC. The Dazh gene is expressed in male and female embryonic gonads before germ cell sex differentiation. Biochem Biophys Res Commun. 1998;245(3):878–882. doi: 10.1006/bbrc.1998.8530. [DOI] [PubMed] [Google Scholar]
- 17.Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev Biol. 2005;288(2):309–316. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 18.Gill ME, Hu YC, Lin Y, Page DC. Licensing of gametogenesis, dependent on RNA binding protein DAZL, as a gateway to sexual differentiation of fetal germ cells. Proc Natl Acad Sci USA. 2011;108(18):7443–7448. doi: 10.1073/pnas.1104501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooke HJ, Elliott DJ. RNA-binding proteins and human male infertility. Trends Genet. 1997;13(3):87–89. doi: 10.1016/s0168-9525(97)01066-4. [DOI] [PubMed] [Google Scholar]
- 20.Haag ES. Rolling back to BOULE. Proc Natl Acad Sci USA. 2001;98(13):6983–6985. doi: 10.1073/pnas.141237898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuales G, et al. Boule-like genes regulate male and female gametogenesis in the flatworm Macrostomum lignano. Dev Biol. 2011;357(1):117–132. doi: 10.1016/j.ydbio.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, et al. Differential conservation and divergence of fertility genes boule and dazl in the rainbow trout. PLoS One. 2011;6(1):e15910. doi: 10.1371/journal.pone.0015910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zayas RM, et al. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: Analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci USA. 2005;102(51):18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Zayas RM, Guo T, Newmark PA. nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci USA. 2007;104(14):5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newmark PA, Wang Y, Chong T. Germ cell specification and regeneration in planarians. Cold Spring Harb Symp Quant Biol. 2008;73(0):573–581. doi: 10.1101/sqb.2008.73.022. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Stary JM, Wilhelm JE, Newmark PA. A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes Dev. 2010;24(18):2081–2092. doi: 10.1101/gad.1951010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins JJ, et al. Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 2010;8(10):e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong T, Stary JM, Wang Y, Newmark PA. Molecular markers to characterize the hermaphroditic reproductive system of the planarian Schmidtea mediterranea. BMC Dev Biol. 2011;11:69. doi: 10.1186/1471-213X-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chong T, Collins JJ, 3rd, Brubacher JL, Zarkower D, Newmark PA. A sex-specific transcription factor controls male identity in a simultaneous hermaphrodite. Nat Commun. 2013;4:1814. doi: 10.1038/ncomms2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robb SMC, Gotting K, Ross E, Sánchez Alvarado A. SmedGD 2.0: The Schmidtea mediterranea genome database. Genesis. 2015;53(8):535–546. doi: 10.1002/dvg.22872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsui S, et al. Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics. 2000;65(3):266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 32.Moore FL, et al. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc Natl Acad Sci USA. 2003;100(2):538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore FL, Jaruzelska J, Dorfman DM, Reijo-Pera RA. Identification of a novel gene, DZIP (DAZ-interacting protein), that encodes a protein that interacts with DAZ (deleted in azoospermia) and is expressed in embryonic stem cells and germ cells. Genomics. 2004;83(5):834–843. doi: 10.1016/j.ygeno.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Urano J, Fox MS, Reijo Pera RA. Interaction of the conserved meiotic regulators, BOULE (BOL) and PUMILIO-2 (PUM2) Mol Reprod Dev. 2005;71(3):290–298. doi: 10.1002/mrd.20270. [DOI] [PubMed] [Google Scholar]
- 35.Hsu LC-L, et al. DAZAP1, an hnRNP protein, is required for normal growth and spermatogenesis in mice. RNA. 2008;14(9):1814–1822. doi: 10.1261/rna.1152808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glazer AM, et al. The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev Biol. 2010;337(1):148–156. doi: 10.1016/j.ydbio.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24(14):2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brook M, Smith JWS, Gray NK. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction. 2009;137(4):595–617. doi: 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- 39.Salvetti A, et al. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development. 2005;132(8):1863–1874. doi: 10.1242/dev.01785. [DOI] [PubMed] [Google Scholar]
- 40.Maines JZ, Wasserman SA. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat Cell Biol. 1999;1(3):171–174. doi: 10.1038/11091. [DOI] [PubMed] [Google Scholar]
- 41.Venables JP, Ruggiu M, Cooke HJ. The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res. 2001;29(12):2479–2483. doi: 10.1093/nar/29.12.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao X, Trifillis P, Kiledjian M. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol Reprod. 2002;66(2):475–485. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- 43.Kee K, Angeles VT, Flores M, Nguyen HN, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462(7270):222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fox M, Urano J, Reijo Pera RA. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics. 2005;85(1):92–105. doi: 10.1016/j.ygeno.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Wenemoser D, Lapan SW, Wilkinson AW, Bell GW, Reddien PW. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 2012;26(9):988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 47.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Töpel M, Ling Q, Jarvis P. Neofunctionalization within the Omp85 protein superfamily during chloroplast evolution. Plant Signal Behav. 2012;7(2):161–164. doi: 10.4161/psb.18677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Assis R, Bachtrog D. Neofunctionalization of young duplicate genes in Drosophila. Proc Natl Acad Sci USA. 2013;110(43):17409–17414. doi: 10.1073/pnas.1313759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higuchi S, et al. Expression and functional analysis of musashi-like genes in planarian CNS regeneration. Mech Dev. 2008;125(7):631–645. doi: 10.1016/j.mod.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Brandl H, et al. PlanMine: A mineable resource of planarian biology and biodiversity. Nucleic Acids Res. 2016;44(D1):D764–D773. doi: 10.1093/nar/gkv1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.King RS, Newmark PA. In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev Biol. 2013;13(1):8. doi: 10.1186/1471-213X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pellettieri J, et al. Cell death and tissue remodeling in planarian regeneration. Dev Biol. 2010;338(1):76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forsthoefel DJ, Park AE, Newmark PA. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Dev Biol. 2011;356(2):445–459. doi: 10.1016/j.ydbio.2011.05.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol. 1996;11(9):367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 57.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27(8):1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014 Tracer v1.6. Available at beast.bio.ed.ac.uk/Tracer. Accessed March 1, 2016.
- 59.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16(8):1114–1116. [Google Scholar]
- 60.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51(3):492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 61.Shimodaira H, Hasegawa M. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17(12):1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.