Significance
The evolutionarily conserved RNA-binding protein Musashi-2 (MSI2) regulates mRNA translation and influences multiple biological processes, including maintenance of stem cell identity. This work for the first time, to our knowledge, identifies that more aggressive patient tumors have higher MSI2 levels. We define a critical role for MSI2 in supporting non-small cell lung cancer (NSCLC) invasiveness and further define claudins 3, 5, and 7 (CLDN3, CLDN5, and CLDN7), TGF-β receptor 1 (TGFβR1), and the small mothers against decapentaplegic homolog 3 (SMAD3) as targets through which MSI2 regulates this process. The observation that MSI2 expression is progressively elevated from an early stage in human NSCLC tumors suggests that this protein may play an essential role in the reprogramming of TGF-β signaling from growth-inhibiting to invasion-promoting during oncogenesis.
Keywords: MSI2, NSCLC, metastasis, lung cancer, claudins
Abstract
Non-small cell lung cancer (NSCLC) has a 5-y survival rate of ∼16%, with most deaths associated with uncontrolled metastasis. We screened for stem cell identity-related genes preferentially expressed in a panel of cell lines with high versus low metastatic potential, derived from NSCLC tumors of KrasLA1/+;P53R172HΔG/+ (KP) mice. The Musashi-2 (MSI2) protein, a regulator of mRNA translation, was consistently elevated in metastasis-competent cell lines. MSI2 was overexpressed in 123 human NSCLC tumor specimens versus normal lung, whereas higher expression was associated with disease progression in an independent set of matched normal/primary tumor/lymph node specimens. Depletion of MSI2 in multiple independent metastatic murine and human NSCLC cell lines reduced invasion and metastatic potential, independent of an effect on proliferation. MSI2 depletion significantly induced expression of proteins associated with epithelial identity, including tight junction proteins [claudin 3 (CLDN3), claudin 5 (CLDN5), and claudin 7 (CLDN7)] and down-regulated direct translational targets associated with epithelial–mesenchymal transition, including the TGF-β receptor 1 (TGFβR1), the small mothers against decapentaplegic homolog 3 (SMAD3), and the zinc finger proteins SNAI1 (SNAIL) and SNAI2 (SLUG). Overexpression of TGFβRI reversed the loss of invasion associated with MSI2 depletion, whereas overexpression of CLDN7 inhibited MSI2-dependent invasion. Unexpectedly, MSI2 depletion reduced E-cadherin expression, reflecting a mixed epithelial–mesenchymal phenotype. Based on this work, we propose that MSI2 provides essential support for TGFβR1/SMAD3 signaling and contributes to invasive adenocarcinoma of the lung and may serve as a predictive biomarker of NSCLC aggressiveness.
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related deaths in the world (1). Approximately 7% of individuals born in the United States in 2013 will ultimately be diagnosed with lung cancer, and 160,000 die from this disease each year (1). The 5-y survival rate for lung cancer is around 16% of diagnosed cases (2). Much of the lethality of lung cancer is due to frequent diagnosis of the malignancy at the metastatic stage, when fundamental changes in tumor biology cause the disease to be refractory to many treatments. A better understanding of the biological processes that promote NSCLC metastasis promises to further improve clinical care of the patients.
KrasLA1/+;P53R172HΔG/+ (KP) mice provide a useful and well-validated model for the study of NSCLC metastasis. These mice combine a mutant p53 allele (p53R175HΔG) with an activating KrasG12D allele (KrasLA1) (3), leading to development of adenocarcinomas resembling human NSCLC, which are often characterized by mutation of KRAS (∼30%) (4) and loss of TP53 (∼60%) (5). Many of the KP tumors metastasize to sites commonly seen in NSCLC patients (3). These features make the KP murine model a useful tool with which to evaluate factors that underlie NSCLC metastasis. Among the pathways activated in metastasis, a significant number are associated with tumor-initiating progenitor cell populations (6–12). In this study, using cell lines with high or low metastatic potential derived from multiple independent tumors arising in KP mice (13), we evaluated a set of genes associated with progenitor cell identity as candidate regulators of the invasive and metastatic properties of NSCLC tumors. As described later, this work for the first time, to our knowledge, identifies elevated expression of the Musashi-2 (MSI2) protein as a common driver of metastasis in NSCLC and defines its mechanism of action in this disease.
Results
Up-Regulation of MSI2 Accompanies Metastasis in Mouse NSCLC Cells and Human NSCLC Tumors.
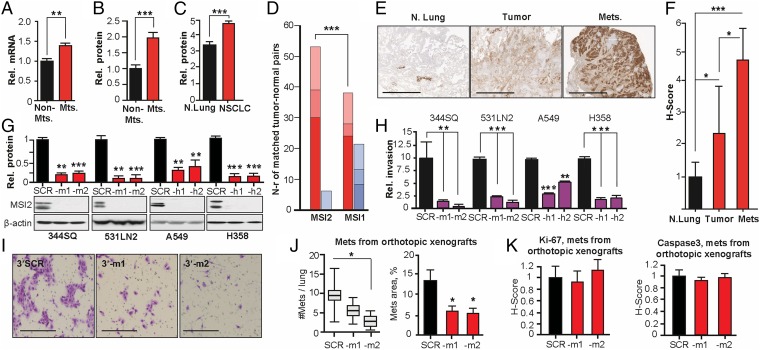
Using quantitative RT-PCR (qRT-PCR) (see SI Appendix, SI Methods and Table S1), we compared the mRNA expression of a candidate set of stem cell marker genes in a highly metastatic (344SQ) versus a nonmetastatic (393P) NSCLC cell line (SI Appendix, Fig. S1A), both derived from primary tumors that developed spontaneously in KP mice (13). Genes with a significant difference in expression between these two cell lines were further assessed in a panel of seven metastatic and seven nonmetastatic cell lines derived from additional independent KP tumors. Msi2 expression was consistently elevated in metastatic cell lines at both the mRNA and the protein level (Fig. 1 A and B and SI Appendix, Fig. S1B). Msi1, a paralogue of Msi2, also displayed a trend toward elevated expression in metastatic murine cell lines at the mRNA level, but this was not observed at the protein level (SI Appendix, Fig. S1 C and D).
Fig. 1.
Elevated MSI2 expression supports NSCLC invasion. (A and B) Averaged values from qRT-PCR (A) and Western blot (B) analysis of MSI2 mRNA and protein expression in seven metastatic versus seven nonmetastatic murine NSCLC cell lines. (C) Quantification of MSI2 expression in TMA of 22 normal and 123 NSCLC specimens; significance was determined by Mann–Whitney test. (D) Analysis of TCGA data for MSI2 and MSI1 mRNA in 59 NSCLC specimens versus paired normal lung samples. Dark red, percent of samples with tumor–up-regulated mRNA with a z score >3; red, up-regulation with a z score of 2–3; light red, up-regulation with a z score >1. Dark blue, percent of tumor samples with an mRNA down-regulation z score >3; blue, down-regulation with a z score of 2–3; light blue, down-regulation with a z score >1. (E and F) Immunohistochemical (IHC) assessment of MSI2 levels in normal lung, primary tumor, or lymph nodes, with representative data (E) and averaged H scores (F). (G) Western validation of MSI2 depletion with two independent shRNAs (–m1, –m2, –h1, and –h2) in indicated cell lines, referenced to control shRNA (SCR)-depleted cells. (H and I) Quantified (H) and representative (I; for 344SQ cells) data for Matrigel invasion for models shown in J. (J) Pathologist-quantified number of independent metastases per lung (Left) and relative area of lung metastases (Right) 28 d after injection of orthotopic 344SQ xenografts in 129Sv immunocompetent mice. (K) Quantification of Ki-67 (Left) and cleaved caspase (Right) in orthotopic xenografts; caspase levels were low in all specimens. For all graphs, *P < 0.05, **P < 0.01, and ***P < 0.001 relative to controls.
To assess whether MSI2 overexpression is physiologically relevant in the context of human NSCLC, we first used Automated Quantitative Analysis to analyze the protein expression of MSI2 in tissue microarrays (TMAs) containing 123 primary human NSCLC tumors and normal lung tissue (SI Appendix, Table S2). This analysis indicated highly significant elevation of MSI2 in tumors compared with normal tissue (Fig. 1C and SI Appendix, Fig. S1E). In contrast, parallel assessment of MSI1 expression did not reveal differences in expression between normal lung tissue and primary NSCLC tumor specimens (SI Appendix, Fig. S1F). Independent analysis of 59 primary NSCLC tumors versus matching normal lung tissue data from The Cancer Genome Atlas (TCGA) Research Network (cancergenome.nih.gov/) indicated frequently elevated expression of MSI2 but not MSI1 mRNA levels in tumor specimens (Fig. 1D), whereas Kaplan–Meier plots analysis suggested higher levels of MSI2 were associated with reduced overall survival (SI Appendix, Fig. S1G). In addition, analysis of an independent cohort of matched NSCLC specimens containing normal lung, primary tumor, and tumor-positive lymph nodes from 14 individuals (SI Appendix, Table S3) demonstrated significant 2.4-fold elevation of MSI2 levels in the primary tumor and highly 4.5-fold elevation in the lymph nodes versus normal lung tissue (Fig. 1 E and F). The progressive increase in MSI2 expression as human lung tumors metastasized from the primary site, combined with data from the KP model, suggested a potential functional and clinical relevance for MSI2 in regulating tumor progression.
MSI2 Depletion in Metastatic NSCLC Cells Inhibits Invasion in Vitro and Tumor Dissemination in Vivo.
Based on expression profiles of NSCLC cell lines available through the Cancer Cell Line Encyclopedia (14), we identified the human NSCLC cell lines A549 (KRASmut) and H358 (KRASmut;TP53−/−) as metastasis-competent adenocarcinoma cell lines with high expression of MSI2 (SI Appendix, Fig. S1H). We used shRNA depletion of these and two metastatic murine NSCLC cell lines (344SQ and 531LN2) to further study the role of MSI2 in metastasis. Effective MSI2 mRNA and protein depletion were confirmed for four MSI2-depleted cell lines, in reference to matching scrambled shRNA control cell lines (Fig. 1G and SI Appendix, Fig. S1I). MSI2 depletion consistently and significantly reduced invasion through Matrigel for all lines (Fig. 1 H and I and SI Appendix, Fig. S1J). Similar results were obtained using transient siRNA transfections to deplete MSI2 (SI Appendix, Fig. S1 K and L). For three of the four cell lines (344SQ, A549, and H358), depletion of MSI2 had no effect on cell proliferation in vitro (SI Appendix, Fig. S1 M and N), and Msi2 depletion did not affect in vitro growth of 344SQ spheroids in the extracellular matrix (ECM) (SI Appendix, Fig. S2).
We next performed orthotopic lung injections of metastasis-competent 344SQ cells with shRNA vector control or shRNA targeting Msi2 into syngeneic, immunocompetent 129Sv mice. Msi2 depletion significantly reduced the burden of lung metastases following injection, predominantly affecting the total number of metastases and to a lesser extent the size of individual metastases (Fig. 1J), whereas no statistically significant difference in Ki-67 or cleaved caspase staining was observed (Fig. 1K). In a complementary experiment, an s.c. xenograft of 344SQ also indicated Msi2 depletion induced no statistically significant difference in the growth of primary xenografts or associated Ki-67 or cleaved caspase staining (SI Appendix, Fig. S3 A and B). However, there was a significantly higher metastatic burden in the lungs of mice bearing control versus Msi2-depleted tumors (SI Appendix, Fig. S3D). In this case, the difference in metastatic area predominantly reflected differences in numbers of metastatic foci, with Ki-67 and caspase staining comparable between control and MSI2-depletion groups (SI Appendix, Fig. S3 E and F). In a reciprocal approach, we overexpressed MSI2 in 393P nonmetastatic cells, which have low endogenous levels of Msi2. MSI2 overexpression did not affect 393P proliferation (SI Appendix, Fig. S4A) but greatly increased invasion through Matrigel (SI Appendix, Fig. S4 B and C). In vivo analysis of orthotopic xenografts indicated a nonstatistically significant increase in metastasis in MSI2-overexpressing cells (SI Appendix, Fig. S4D). Finally, analysis of migration independent of invasion (SI Appendix, Fig. S4 E and F) showed limited effects of MSI2 depletion. We therefore focused subsequent analyses on mechanistic analysis of invasion signaling.
Unbiased and Candidate-Based Investigation of MSI2-Regulated Signaling.
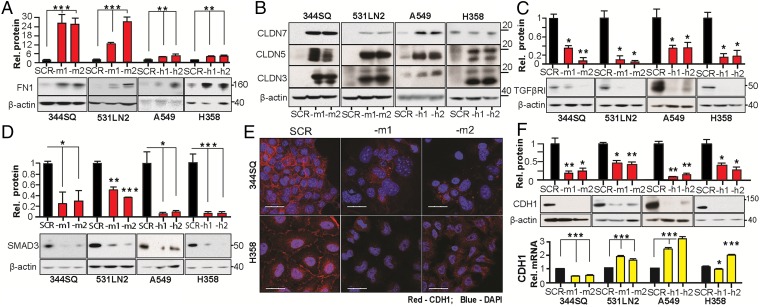
We used Reverse Protein Phase Array (RPPA) to query 171 total proteins and phosphoproteins for expression changes associated with Msi2 knockdown using control shRNA and Msi2-targeted shRNA derivatives of 344SQ cells (SI Appendix, Fig. S5). This work suggested a number of previously unidentified candidates associated with Msi2 expression and relevant to control of epithelial–mesenchymal transition (EMT) and invasion. Proteins with the greatest magnitude of response to Msi2 depletion that were subsequently validated by low throughput Western analysis included the tight junction (TJ)-associated protein claudin 7 (CLDN7) (15–17), elevated 19.4-fold, and the ECM protein fibronectin (FN1) (18–21), elevated 2.5-fold. Subsequent independent evaluation confirmed these RPPA results, as MSI2 depletion significantly elevated FN1 mRNA (4.0–9.4-fold) and protein (2.4–23 fold) in all four cell lines and elevated CLDN7 protein (2.5–28-fold) in three of the four cell lines (Fig. 2 A and B and SI Appendix, Fig. S6 A and B). Results were independently confirmed using transient siRNAs to deplete MSI2 (SI Appendix, Fig. S6C). Although RPPA data also suggested the neurogenic locus notch homolog protein 1 (NOTCH1) expression may be affected by MSI2 (SI Appendix, Fig. S5) and although some studies suggested expression of the NOTCH regulator NUMB is influenced by MSI1/2 (22, 23), we found no consistent and significant differences in NUMB in MSI2-depleted cells (SI Appendix, Fig. S6D), supporting the idea that regulation of NUMB by MSI1/2 may depend on cellular context (24, 25).
Fig. 2.
MSI2 depletion controls the expression of indirect and direct targets relevant to invasion and EMT. (A and B) Western analysis of FN1 (A) and CLDN7, CLDN5, and CLDN3. (B) In murine and human NSCLC cell lines in the context of MSI2 depletion with independent targeting shRNAs (–m1/–m2, –h1/–h2). SCR, control scrambled shRNA. Graphs represent data from four independent runs. (C and D) Western analysis of expression of TGFβR1 (C) and SMAD3 (D) in murine and human NSCLC cell lines with stably depleted MSI2. Graphs represent data from three independent runs. For all graphs,*P < 0.05, **P < 0.01, and ***P < 0.001 relative to SCR (scrambled shRNA) controls. (E) Immunofluorescence analysis of E-Cadherin staining in 344SQ murine (Top) and H358 human (Bottom) NSCLC cell lines, with or without depleted MSI2. Blue, DAPI; red, E-Cadherin. (Scale bars, 30 µm.) (F) Western (Top) and qRT-PCR (Bottom) analysis of E-Cadherin expression in four NSCLC cell line models with or without depleted MSI2. Graphs represent data from three independent runs. For all graphs, *P < 0.05, **P < 0.01, and ***P < 0.001 relative to SCR (scrambled shRNA) controls.
NSCLC cells have been shown to express multiple claudins with partially redundant function (26), most not represented in the RPPA panel. In direct testing, we found MSI2 depletion also induced CLDN3 and CLDN5 in all four cell lines at the protein level (3.8–22 fold for CLDN3 and 3.4–41 fold for CLDN5) (Fig. 2B and SI Appendix, Fig. S6 E and F), making restraint of claudin expression a consistent feature of MSI2 function. Studies of the MSI proteins (predominantly focused on MSI1) have defined these proteins as RNA-binding proteins that regulate mRNA translation (22, 24, 27). The induction of claudins may reflect a combination of transcriptional and posttranscriptional consequences of MSI2 depletion, as the mRNA level shows induction that is less marked than at the protein level (SI Appendix, Fig. S6 A, B, E, and F). However, the claudin mRNAs lack [(G/A)U(n)AGU (n = 1–3)] the consensus motifs for MSI2 binding described in Wang et al. (28), suggesting direct regulation of translation is not involved.
Direct MSI2 translational targets defined in other cell types that might be relevant to the invasiveness of NSCLC cells and tumors include the TGF-β receptor (TGFβR1) and its effector, the small mothers against decapentaplegic homolog 3 (SMAD3) (24), which promote EMT by down-regulating E-cadherin (CDH1) and inducing other transcriptional changes (29). We found that stable or transient MSI2 knockdown caused strong down-regulation of TGFβR1 and SMAD3, predominantly at the protein level, in all four models (Fig. 2 C and D and SI Appendix, Fig. S6 G and H). Reciprocally, exogenous overexpression of MSI2 induced TGFβR1 and SMAD3 expression and caused loss of CLDN3, CLDN5, and CLDN7 expression in the 393P cell line (SI Appendix, Fig. S6 I and J).
Depletion of MSI2 Affects the Composition of Cell–Cell Junctions and Causes Partial EMT.
Based on the action of MSI2 in supporting the expression of TGFβR1 and SMAD3, while repressing CLDN3, CLDN5, CLDN7, and FN1, we hypothesized that the reduced invasiveness of MSI2-depleted cells might reflect changes involving TJs and reduced EMT, associated with elevated E-cadherin (CDH1). Unexpectedly, immunofluorescence analysis demonstrated that expression of epithelial protein E-cadherin at cell–cell junctions was much reduced by MSI2 depletion (Fig. 2E), as was total E-cadherin protein expression, whereas mRNA levels were not consistently affected (Fig. 2F). In contrast, there was a significant increase in CLDN3 and CLDN7 staining at cell–cell contact points, whereas TJP1 (ZO-1), which localizes to the cytoplasmic surface of TJs, was unaffected (SI Appendix, Fig. S7). We examined expression of additional proteins associated with mesenchymal identity (SI Appendix, Fig. S8 A and B). MSI2 depletion up-regulated the pro-EMT factors ZEB1, ZEB2, and FOXC2 but down-regulated VIM (vimentin) and the zinc finger proteins SNAI1 (SNAIL) and SNAI2 (SLUG). Conversely, MSI2 overexpression induced CDH1, VIM, SNAIL, and SLUG (SI Appendix, Fig. S8 C and D). Collectively, these data indicated a mixed effect of MSI2 depletion on EMT.
MSI2 Regulation of Invasion via TGFβR1, SMAD3, and CLDN7.
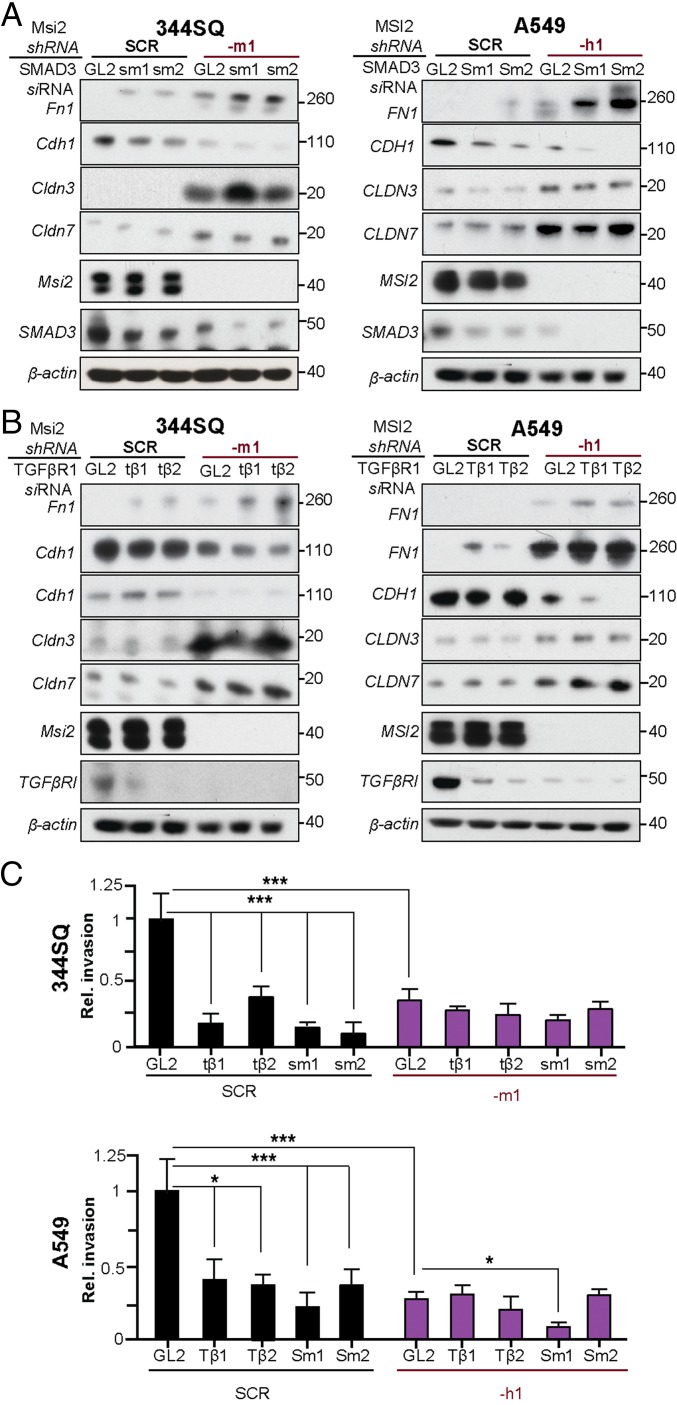
To assess the functional interaction between MSI2, its direct targets TGFβR1 and SMAD3, and claudins, we depleted SMAD3 or TGFβR1 in MSI2-depleted versus control cell lines. SMAD3 knockdown reduced CDH1 expression levels in both parental and MSI2-depleted lines (Fig. 3A). By contrast, the relationship between TGFβR1 and CDH1 expression was modulated by MSI2 status, with the TGFβR1 knockdown elevating CDH1 expression in the parental cell lines but reducing it in MSI2-depleted cell lines (Fig. 3B). MSI2 also influenced the response of cells growing as spheroids to treatment with TGF-β (SI Appendix, Fig. S2), with control scrambled shRNA (SCR)-depleted cells responding by increasing sphere size but MSI2-depleted cells showing little proliferative response and instead responding by showing a greater phenotype of collective migration. Importantly, depletion of TGFβR1 or SMAD3 caused a statistically significant decrease in invasion in SCR-depleted NSCLC cell lines but not in those with depleted MSI2 (Fig. 3C). Conversely, overexpression of TGFβR1 partially but incompletely rescued the decrease in invasion seen in MSI2-depleted cells (SI Appendix, Fig. S9), suggesting other contributing factors.
Fig. 3.
Functional interaction of MSI2 with TGFβR1, SMAD3, FN1, E-Cadherin, and CLDN3 and CLDN7. (A) Western analysis for expression of indicated proteins in the 344SQ and A549 cell lines with (–m1 and –h1) or without (SCR) shRNA depletion of MSI2, and with (–sm1, –sm2, and –Sm1, –Sm2) or without (GL2) siRNA depletion of SMAD3. (B) Western analysis for expression of indicated proteins in the 344SQ and A549 cell lines with (m1 and h1) or without (SCR) shRNA depletion of MSI2, and with TGFβR1 (–tβ1, –tβ2 and –Tβ1, –Tβ2) or without (GL2) siRNA depletion of TGFβR1. (C) Quantification of results from three independent Matrigel invasion assays for 344SQ and A549 with (–m1 and –h1) or without (SCR) shRNA depletion of MSI2, in the context of additional siRNA depletion of TGFβR1 (–tβ1, –tβ2 and –Tβ1, –Tβ2) or SMAD3 (–sm1, –sm2 and –Sm1, –Sm2) versus siRNA negative control (GL2). *P < 0.05 and ***P < 0.001 relative to controls.
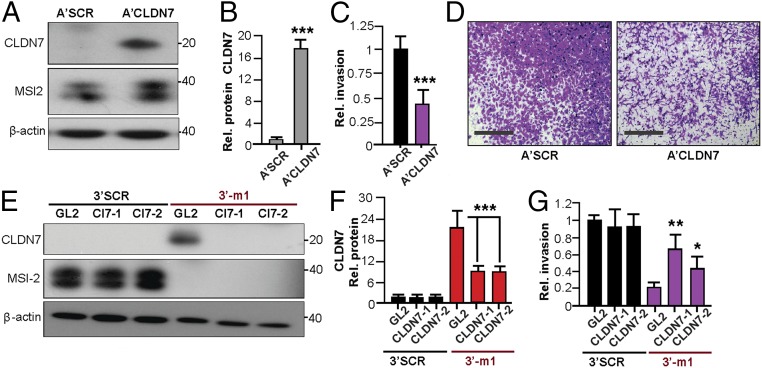
The profile of mixed pro- and anti-EMT changes, and incomplete rescue by TGFβR1 overexpression, suggested a possible important role for claudin-associated TJs in limiting NSCLC invasion induced by MSI2-dependent TGFβR1/SMAD3 signaling. Exploring the relationship between these proteins, we found that siRNA depletion of TGFβR1 or SMAD3 did not significantly affect the expression of CLDN3, CLDN7, or MSI2. This indicated that MSI2 regulates CLDN3/CLDN7 expression independently of TGFβR1 and SMAD3 (Fig. 3 A and B and SI Appendix, Fig. S10 A and B). Functionally, overexpression of CLDN7 significantly decreased invasion in cells expressing high levels of endogenous MSI2 (Fig. 4 A–D). Conversely, siRNA depletion of CDLN7 significantly increased invasion in 344SQ cells with depleted MSI2 but had no effect in cells with significant endogenous MSI2 (Fig. 4 E–G). Taken together, we conclude that MSI2 stimulates invasion in lung cancer in part by sustaining TGFβR1 signaling and suppressing the expression of CLDN7 and potentially other claudins.
Fig. 4.
CLDN7 regulation of invasion. (A and B) Representative Western blot analysis of indicated proteins (A) and quantification of CLDN7 expression (B) in negative control A’SCR and A’CLDN7 stably transfected A549 cells. (C and D) Quantification (C) and representative images (D) of Matrigel invasion for A’SCR and A’CLDN7 stably transfected A549 cells. (E and F) Western blot analysis (E) and quantification of CLDN7 expression (F) in 344SQ cells expressing MSI2 (SCR) or stably MSI2-depleted (–m1) and transiently transfected with GL2 control or Cl7 siRNAs (Cl7-1, Cl7-2). (G) Quantification of Matrigel invasion assay for cells in E and F, based on three experiments. For all graphs, *P ≤ 0.05, *P ≤ 0.01, and ***P ≤ 0.001 relative to controls.
Discussion
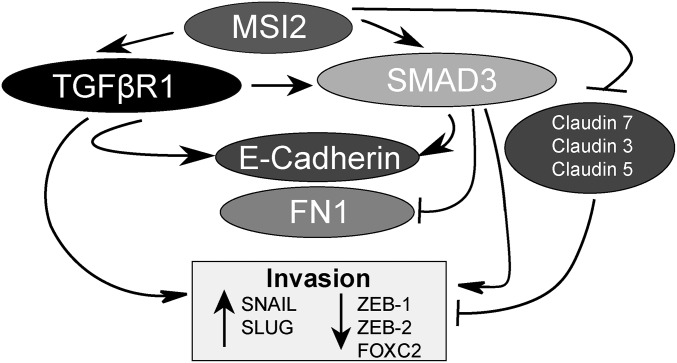
In summary, our study for the first time, to our knowledge, shows that elevation of MSI2 expression progressing NSCLC supports tumor cell invasion and metastasis by modulating TGF-β–dependent EMT and repressing claudin expression (Fig. 5). Although a number of studies have previously identified a role for the MSI2 paralogue MSI1 as oncogenic in a number of cancer types (30–32), MSI2 has attracted much less scrutiny. However, MSI2 has been shown to be oncogenic in a mouse model of colon cancer (28); MSI2 expression is induced by loss of the adenomatous polyposis coli (APC) gene, and overexpression of MSI2 phenocopies APC loss (28). MSI2 is also overexpressed and oncogenic in human leukemias. Elevated MSI2 expression is associated with poor survival in leukemia, and MSI2 knockdown or genetic deletion reduced engraftment and caused a defect in hematopoietic stem cell maintenance in vivo as a result of decreased proliferation (23, 24, 33), in part due to a loss of sensitivity to TGF-β–mediated expansion and compromised TGFβR1 and SMAD3 signaling. Our data confirm the relevance of TGFβR1 and SMAD3 to some MSI2 activities in NSCLC but also establish for the first time, to our knowledge, that MSI2 status conditions TGFBβR1 regulation of E-cadherin/CDH1 as well as the ability of TGFβR1 and SMAD3 to influence cell invasion. These results coupled with our evidence showing rising levels of MSI2 as tumors become increasingly metastatic in mouse models or human specimens suggest that MSI2 expression status may be relevant to the change in TGF-β signaling from prodifferentiation to proinvasive during tumor progression (29). Full understanding of the functional role of MSI2 in NSCLC metastasis will require additional studies, such as clinical correlative analysis.
Fig. 5.
Model for MSI2 action in coordinating EMT and invasion potential. See Discussion for details.
Importantly, our data suggest that, in NSCLC, proliferation is a much less important target of MSI2 regulation than control of invasion, in a marked difference from leukemia models. Analysis of MSI proteins in breast cancer has led to the suggestion that these proteins may be required to support an epithelial luminal cell identity (25). However, our findings point to a more complex mode of action, with MSI2 supporting expression of CDH1, VIM, SLUG, and SNAIL but suppressing that of Zeb1/2, FOXC2, and multiple claudins. TGF-β has previously been shown to directly support expression of SNAIL but not ZEB1/2 in an NSCLC cell model (34), suggesting these downstream effects of MSI2 include both TGF-β–dependent and –independent outputs. Together with MSI2-dependent expression of CDH1, these results are compatible with a model in which MSI2 creates conditions that favor collective migration (35), an idea bearing further investigation.
Although essentially unaddressed in lung cancer, a claudin-low phenotype has been linked to EMT, stemness, and chemotherapy resistance in breast and bladder cancer (36–38). CLDN7 is known to inhibit human lung cancer invasion (15), and low expression of CLDN7 is linked to poor prognosis in NSCLC (39). Our data for the first time, to our knowledge, indicate that MSI2 represses the expression of multiple claudins, with at least CLDN7 functionally important for MSI2-dependent invasion. Although technical issues limit simultaneous targeting of multiple claudins, it is likely that control of this suite of proteins significantly influences NSCLC metastasis, particularly given the mixed EMT phenotype of modulating MSI2 expression.
Finally, given their role as noncatalytic RNA-binding proteins, it is likely to be difficult to develop effective small molecule inhibitors targeting MSI1 or MSI2. However, several recent therapeutic strategies to improve NSCLC treatment focus on TGF-β (40–42), and activity of such compounds could be strongly influenced by MSI2 status in tumors, with invasive or metastatic NSCLC expressing higher levels of MSI2 having a differential response. In addition, the EMT process itself has been shown to influence cellular resistance to a number of drugs of relevance to NSCLC treatment (7). Further study of MSI2 function in normal lung development, cellular transformation, and NSCLC drug resistance is clearly warranted.
Methods
Also see the extended methods in SI Appendix, SI Methods.
Cell Culture.
Mouse cell lines (344SQ, 531LN2, others) from p53R172HΔg/+K-rasLA1/+ mice were derived from tumor tissues at necropsy from different mice as previously described (13). Human alveolar basal epithelial adenocarcinoma cell lines A549, H358, H322, and H226 were obtained from the American Type Culture Collection (ATCC). All cells were grown in RPMI 1640 with 10% FBS (50 mL FBS/450 mL RPMI). The 344SQ-m1/2, 531LN2-m1/2, A549-h1/h2, and H358-h1/h2 MSI2-knockdown cell lines were created by transfection with the pLKO.1 system-based shRNA lentivirus (SIGMA) (SI Appendix, SI Methods).
Cell Proliferation and Invasion Assays.
Cell growth was measured by CellTiterBlue and water-soluble tetrazolium salts assays and by direct quantitation of DAPI-stained nuclei. Invasion assays were performed using standard Matrigel invasion assays. Details of protocols and statistical analyses are provided in detail in SI Appendix, SI Methods.
In Vivo Analysis of Tumor Growth and Metastasis.
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committees at the MD Anderson Cancer Center and at the Fox Chase Cancer Center. Orthotopic and s.c. xenograft studies were performed to study the proliferation and metastatic potential of NSCLC cell lines with overexpressed or depleted MSI2. Detailed protocols are provided in SI Appendix, SI Methods.
RPPA Reverse-Phase Protein Analysis.
The 344SQ-SCR, 344SQ-m1, and 344SQ-m2 mouse cells were lysed and prepared according to MD Anderson Core Facility instructions, as previously described, and RPPA was performed at the facility (43–45). Data were visualized using the MultiExperiment Viewer program (www.tm4.org/mev.html) (46).
Protein Expression Analysis by Western Blotting.
Lysates were prepared for analysis, and Western blotting and analysis were performed using standard protocols. Quantified data were averaged from at least three independent runs for all experiments. See SI Appendix, SI Methods for the specific antibody reagents used.
siRNA Transfection.
SMARTpool siRNAs targeting human/mouse TGFβR1/TgfβR1, SMAD3/Smad3, and CLDN7/Cldn7 (SI Appendix, Table S4) and nonspecific control pool siRNA were purchased from Dharmacon. NSCLC cells at 50% confluence were transfected with siRNA at final concentrations of 50 nmol/L using the LipofectAMINE 2000 transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank Drs. Jamie Rodriguez and Ignacio Wistuba for preparing tester blocks for immunohistochemistry. We thank Cara Dubyk for technical assistance with TMA staining, Dr. Margret Einarson for assistance with high throughput analyses, Dr. Marcos Estécio for advice, Dr. Waun Ki Hong for general support, and Vladislav Korobeynikov for assistance with the Vectra Imaging System. The authors and the work were supported by the NCI Core Grant P30 CA006927 (to Fox Chase Cancer Center); NCI P30 Core Grant CA016672 (to MD Anderson Cancer Center); a Conquer Cancer Foundation (ASCO) Young Investigator Award, American Cancer Society IRG Pilot Grant, Lung Cancer Research Foundation (LCRF), AHEPA Foundation grant, and DOD Career Development Award LC140074 (to Y.B.); UNM Core Funding (C.F.M., F.A.S., and S.R.); NCI Grants CA181287 and R21CA191425 (to E.A.G.); a Ruth L. Kirschstein NRSA F30 fellowship (F30 CA180607) from the NIH (to T.N.B.); NIH Grant K08 CA151651 and the MD Anderson Physician-Scientist Award (to D.L.G.); and NIH Grant R01 CA157450 (to J.M.K.). This research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF 2010-0027945; to Y.-H.A.). Bioinformatics analysis for this work was supported by Russian Science Foundation Grant 15-15-20032.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.G.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513616113/-/DCSupplemental.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Howlader NNA, et al. SEER Cancer Statistics Review, 1975-2010. In: Cronin KA, editor. National Cancer Institute; Bethesda: 2013. [Google Scholar]
- 3.Zheng S, El-Naggar AK, Kim ES, Kurie JM, Lozano G. A genetic mouse model for metastatic lung cancer with gender differences in survival. Oncogene. 2007;26(48):6896–6904. doi: 10.1038/sj.onc.1210493. [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T, et al. Mutations of ras genes distinguish a subset of non-small-cell lung cancer cell lines from small-cell lung cancer cell lines. Oncogene. 1991;6(8):1353–1362. [PubMed] [Google Scholar]
- 5.Takahashi T, et al. p53: A frequent target for genetic abnormalities in lung cancer. Science. 1989;246(4929):491–494. doi: 10.1126/science.2554494. [DOI] [PubMed] [Google Scholar]
- 6.Bertolini G, et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci USA. 2009;106(38):16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Licciulli S, et al. Notch1 is required for Kras-induced lung adenocarcinoma and controls tumor cell survival via p53. Cancer Res. 2013;73(19):5974–5984. doi: 10.1158/0008-5472.CAN-13-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arasada RR, Amann JM, Rahman MA, Huppert SS, Carbone DP. EGFR blockade enriches for lung cancer stem-like cells through Notch3-dependent signaling. Cancer Res. 2014;74(19):5572–5584. doi: 10.1158/0008-5472.CAN-13-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan KA, et al. Notch pathway activity identifies cells with cancer stem cell-like properties and correlates with worse survival in lung adenocarcinoma. Clin Cancer Res. 2013;19(8):1972–1980. doi: 10.1158/1078-0432.CCR-12-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirino V, et al. TGF-β1 exposure induces epithelial to mesenchymal transition both in CSCs and non-CSCs of the A549 cell line, leading to an increase of migration ability in the CD133+ A549 cell fraction. Cell Death Dis. 2013;4:e620. doi: 10.1038/cddis.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck TN, Chikwem AJ, Solanki NR, Golemis EA. Bioinformatic approaches to augment study of epithelial-to-mesenchymal transition in lung cancer. Physiol Genomics. 2014;46(19):699–724. doi: 10.1152/physiolgenomics.00062.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons DL, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23(18):2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z, et al. Claudin-7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res. 2011;317(13):1935–1946. doi: 10.1016/j.yexcr.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prat A, et al. Genomic analyses across six cancer types identify basal-like breast cancer as a unique molecular entity. Sci Rep. 2013;3:3544. doi: 10.1038/srep03544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruna A, et al. TGFβ induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat Commun. 2012;3:1055. doi: 10.1038/ncomms2039. [DOI] [PubMed] [Google Scholar]
- 18.Liu W, Cheng S, Asa SL, Ezzat S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 2008;68(19):8104–8112. doi: 10.1158/0008-5472.CAN-08-2132. [DOI] [PubMed] [Google Scholar]
- 19.Urtreger AJ, et al. Fibronectin is distinctly downregulated in murine mammary adenocarcinoma cells with high metastatic potential. Oncol Rep. 2006;16(6):1403–1410. [PubMed] [Google Scholar]
- 20.Zhang X, et al. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50(2):490–499. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- 21.Yi M, Ruoslahti E. A fibronectin fragment inhibits tumor growth, angiogenesis, and metastasis. Proc Natl Acad Sci USA. 2001;98(2):620–624. doi: 10.1073/pnas.98.2.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rezza A, et al. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J Cell Sci. 2010;123(Pt 19):3256–3265. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466(7307):765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SM, et al. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J Exp Med. 2014;211(1):71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz Y, et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. eLife. 2014;3:e03915. doi: 10.7554/eLife.03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon MJ. Emerging roles of claudins in human cancer. Int J Mol Sci. 2013;14(9):18148–18180. doi: 10.3390/ijms140918148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakakibara S, Nakamura Y, Satoh H, Okano H. Rna-binding protein Musashi2: Developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001;21(20):8091–8107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat Commun. 2015;6:6517. doi: 10.1038/ncomms7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sureban SM, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134(5):1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 31.Li D, et al. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Ann Surg Oncol. 2011;18(7):2074–2083. doi: 10.1245/s10434-011-1567-9. [DOI] [PubMed] [Google Scholar]
- 32.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17(7):867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kharas MG, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16(8):903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, et al. Suppression of SCARA5 by Snail1 is essential for EMT-associated cell migration of A549 cells. Oncogenesis. 2013;2:e73. doi: 10.1038/oncsis.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shamir ER, Ewald AJ. Adhesion in mammary development: Novel roles for E-cadherin in individual and collective cell migration. Curr Top Dev Biol. 2015;112:353–382. doi: 10.1016/bs.ctdb.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69(10):4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi W, et al. Intrinsic basal and luminal subtypes of muscle-invasive bladder cancer. Nat Rev Urol. 2014;11(7):400–410. doi: 10.1038/nrurol.2014.129. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto T, et al. Reduced expression of claudin-7 is associated with poor outcome in non-small cell lung cancer. Oncol Lett. 2010;1(3):501–505. doi: 10.3892/ol_00000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhurst RJ, Hata A. Targeting the TGFβ signalling pathway in disease. Nat Rev Drug Discov. 2012;11(10):790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dituri F, et al. Differential inhibition of the TGF-β signaling pathway in HCC cells using the small molecule inhibitor LY2157299 and the D10 monoclonal antibody against TGF-β receptor type II. PLoS One. 2013;8(6):e67109. doi: 10.1371/journal.pone.0067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park CY, et al. An novel inhibitor of TGF-β type I receptor, IN-1130, blocks breast cancer lung metastasis through inhibition of epithelial-mesenchymal transition. Cancer Lett. 2014;351(1):72–80. doi: 10.1016/j.canlet.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Iadevaia S, Lu Y, Morales FC, Mills GB, Ram PT. Identification of optimal drug combinations targeting cellular networks: Integrating phospho-proteomics and computational network analysis. Cancer Res. 2010;70(17):6704–6714. doi: 10.1158/0008-5472.CAN-10-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kornblau SM, et al. Functional proteomic profiling of AML predicts response and survival. Blood. 2009;113(1):154–164. doi: 10.1182/blood-2007-10-119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tibes R, et al. Reverse phase protein array: Validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5(10):2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 46.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.