Abstract
Antimicrobials are widely used, not only for treating human infections, but also for treatment of livestock and in fish farms. Human habitats in Southeastern Asian countries are located in close proximity to aquatic environments. As such, the human populations within these regions are at risk of exposure to antimicrobial resistant bacteria, and thereby disseminating antimicrobial resistance genes (ARGs). In this study, we collected water samples from 15 sites (5 sites in Chao Phraya River, 2 sites at the mouth of Chao Phraya River, 3 sites in Ta Chin River, and 5 sites at city canals) and 12 sites (6 sites at city canals; 2 sites at chicken farms; 2 sites at pig farms; and 2 samples from sites at pig farms, which were subsequently treated at a biogas plant) in Thailand in 2013 and 2014, respectively. In total, 117 Aeromonas spp. were isolated from the water samples, and these organisms exhibited various antimicrobial susceptibility profiles. Notably, there was a significant correlation between the environmental concentration of tetracyclines and the rates of tetracycline resistance in the isolated Aeromonas spp.; however, both the concentration and rates of tetracycline resistance in samples derived from pig farms were higher than those of samples harvested from other aquatic environments. These findings suggest that the high concentrations of antimicrobials observed in these aquatic environments likely select for ARGs. Furthermore, they indicate that Aeromonas spp. comprise an effective marker for monitoring antimicrobial resistance in aquatic environments.
Keywords: Aeromonas spp., antimicrobial resistance, aquatic environments, tetracycline resistance, indicator bacteria
Introduction
The use of antimicrobials for treating/preventing bacterial infections is essential for promoting the health and welfare of humans and animals. As such, the emergence and spread of antimicrobial resistance is of global concern. To address these issues, the World Health Organization (WHO) and other international organizations require the development of programs for monitoring antimicrobial resistance in zoonotic bacteria, animal pathogens, and indicator bacteria derived from food-producing animals (Dehaumont, 2004; FAO/OIE/WHO, 2004).
Antimicrobials are widely used, not only for treating human infections, but also in livestock and fish farms. As a consequence, large quantities of these antimicrobials are released into aquatic environments via sewage or directly from livestock and fish farms, thereby providing a selective pressure for the development or acquisition of antimicrobial-resistant bacteria (ARB) and genes (ARGs) (Esteve et al., 2015). Indeed, aquatic environments have been shown to function as reservoirs of ARB and ARGs (Marti et al., 2014), and the transfer of plasmid-encoded ARGs from aquatic environments to humans comprises the greatest potential risk to human health (Smith et al., 1994). To date, however, there have been few reports regarding the monitoring of antimicrobial resistance in aquatic environments.
To efficiently monitor antimicrobial resistance, it is necessary to identify an effective general indicator bacterium within aquatic environments. For example, Escherichia coli is commonly used to monitor antimicrobial resistance in clinical settings and in the veterinary field (NARMS, 2012; National Veterinary Assay Laboratory Forestry and Fisheries, 2012); however, this organism is not always isolated from aquatic environments. In contrast, Aeromonas spp. are Gram-negative, waterborne organisms that are ubiquitous in most aquatic environments (Rhodes et al., 2000). We therefore selected Aeromonas spp. as a general model organism to inform on antimicrobial resistance in aquatic environments.
The spread and maintenance of ARB and ARGs in aquatic environments comprises a potential human health risk. In Southeastern Asian countries, including Thailand, human habitats are often located in close proximity to aquatic environments. Indeed, contamination of both humans and livestock animals with water-derived ARB has been demonstrated in tropical Asian countries (Shimizu et al., 2013). Likewise, a previous study detected contamination of individuals in Indochina with ARGs derived from aquatic environments (Suzuki and Hoa, 2012). Thailand is one of the most agriculturally productive countries in Southeast Asia, and livestock animals in this country are frequently administered antimicrobials to treat and prevent infections (Daniel et al., 2015). As a result, individuals in Thailand are likely at a greater risk of exposure to ARB and ARGs than those in other countries in this region. In this study, we first demonstrated the usefulness of Aeromonas spp. as general indicator bacteria for monitoring antimicrobial resistance. We subsequently utilized this organism to evaluate the current prevalence of resistance at multiple aquatic sites in Thailand. Finally, we examined putative correlations between the prevalence of ARB and ARGs and drug concentrations in rivers and livestock wastewater.
Materials and Methods
Sample Collection
Grab water samples were collected in sterile 1-L bottles in Bangkok and in the province of Ratchaburi in September 2013 and 2014 (Figure 1). The characteristics of the sampling sites are summarized in Supplementary Table S1. Specifically, in 2013, surface water samples were taken at 5 sites (RC 1, 2, 3, 4, and 5) on the Chao Phraya River and at 2 sites (RCM 1 and 2) at the mouth of Chao Phraya River. In addition, 3 sites (RT 1, 2, and 3) were sampled on the Ta Chin River and 5 samples were harvested at city canals (C 1, 2, 3, 4, and 5) in Bangkok. In 2014, 6 samples were taken at city canals (C 6, 7, 8, 9, 10, and 11) in Bangkok, and effluent was collected from chicken farms (FC 1 and 2) and pig farms (FP 1 and 2) in the province of Ratchaburi. The pig farm effluent was also treated at a biogas plant. Treated effluent is indicated as post-treatment FP 1 and 2 (FFP1 and FFP2). All samples were stored at 4°C after collection and analyzed within 12 h.
FIGURE 1.
Map of sampling sites in Thailand. RC 1–5 indicate Chao Phraya River sites; RCM 1 and 2 indicate sites at the mouth of Chao Phraya River; RT 1–3 indicate Ta Chin River sites; C 1–5 indicate the city canal sites sampled in 2013; C 6–11 indicate the city canal sites sampled in 2014; FC 1 and 2 and FP 1 and 2 indicate chicken and pig farm sites, respectively. Sources: http://www.savgis.org/cv_EN.htm.
Analysis of Antimicrobial Concentrations
To measure aquatic concentrations of antimicrobials, including sulfonamides and tetracyclines, water samples were analyzed using a liquid chromatograph (Accela, Thermo Scientific, Waltham, MA, USA) equipped with a tandem mass spectrometer (LC-MS/MS; Quantum Access, Thermo Scientific) after extraction using a solid-phase cartridge (Oasis HLB resin; Waters Corp., Milford, MA, USA). The analytical process was the same as that described by Segura et al. (2015).
Bacterial Isolation
For isolation of Aeromonas spp. from water samples, deoxycholate-hydrogen sulfate-lactose (DHL) agar medium (Nissui Pharmaceutical, Tokyo, Japan) was inoculated with 100 μL of each respective sample and incubated at 37°C overnight. For each sample, a maximum of six colonies were identified as likely Aeromonas spp. based on colony morphology on DHL agar (pink colony) and were subjected to further analyses. Specifically, the isolates were screened for oxidase production and subjected to a variety of biochemical tests (API20E; BioMérieux, Marcy l’Etoile, France). Furthermore, the identity of each strain was verified by 16S rRNA gene sequencing (Marchesi et al., 1998).
Antimicrobial Susceptibility Testing
The minimal inhibitory concentrations (MICs) of the following antimicrobials for each isolate were determined using the agar dilution method, according to the Clinical Laboratory Standards Institute (CLSI) guidelines (Clinical Laboratory Standards Institute, 2008): ampicillin, cefazolin, cefotaxime, kanamycin, tetracycline, sulfamethoxazole, nalidixic acid, and ciprofloxacin (all obtained from Sigma-Aldrich, St. Louis, MO, USA). The breakpoints for each antimicrobial were in accordance with CLSI guidelines (Clinical Laboratory Standards Institute, 2008), except for those of sulfamethoxazole, which are not defined by the CLSI guidelines. As such, we utilized the break points for sulfonamide for this antimicrobial. E. coli ATCC25922, Staphylococcus aureus ATCC29213, Enterococcus faecalis ATCC29212, and Pseudomonas aeruginosa ATCC27853 were used as quality control strains.
Characterization of ARGs
Each strain was evaluated for the presence of the tetA, tetB, tetC, tetD, tetE, and tetG genes via multiplex PCR, as previously described (Jin Jun et al., 2004). Meanwhile, PCR analysis was utilized to detect the tetM and tetS genes (Kim et al., 2004) and the sul1, sul2, and sul3 genes (Hoa et al., 2008), respectively.
Statistical Analysis
Correlations between the environmental concentration of each antimicrobial and both the rate of antimicrobial resistance and the prevalence of resistance genes among Aeromonas spp., respectively, were evaluated using Pearson’s test. Differences were considered significant at p < 0.05.
Results
Isolation of Aeromonas spp. from Water Samples
In total, 117 Aeromonas strains were isolated from 27 water samples. Specifically, 24, 9, 13, 13, 30, 9, 12, and 7 strains were isolated from riverine of Chao Phraya River, the mouth of Chao Phraya River, the riverine of Ta Chin River, the city canal, chicken farms, pig farms, and post-treatment pig farms, respectively (Table 1).
Table 1.
Prevalence of antimicrobial resistance genes (ARGs) among Aeromonas spp. isolated from aquatic environments in Thailand.
| Tetracycline resistance genes | Sulfonamide resistance genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sampling sites | n | tetA (%) | tetB (%) | tetC (%) | tetE (%) | tetM (%) | sul1 (%) | sul2 (%) | sul3 (%) | |
| Chao Phraya River | 24 | 4.2 | 0 | 8.3 | 29.1 | 0 | 37.5 | 0 | 0 | |
| Mouth of the Chao Phraya River | 9 | 11.1 | 0 | 11.1 | 33.3 | 0 | 44.4 | 0 | 0 | |
| Ta Chin River | 13 | 0 | 0 | 15.4 | 0 | 0 | 15.4 | 0 | 0 | |
| City Canal, 2013 | 13 | 7.7 | 0 | 7.7 | 7.7 | 0 | 30.8 | 0 | 0 | |
| City Canal, 2014 | 30 | 0 | 3.3 | 20 | 23.3 | 0 | 26.7 | 3.3 | 0 | |
| Chicken Farm | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Pig Farm | 12 | 75 | 0 | 0 | 0 | 16.7 | 25 | 33.3 | 25 | |
| Pig Farm, Post-treatment | 7 | 28.5 | 0 | 0 | 0 | 14.3 | 28.6 | 42.9 | 14.3 | |
Antimicrobial Resistance
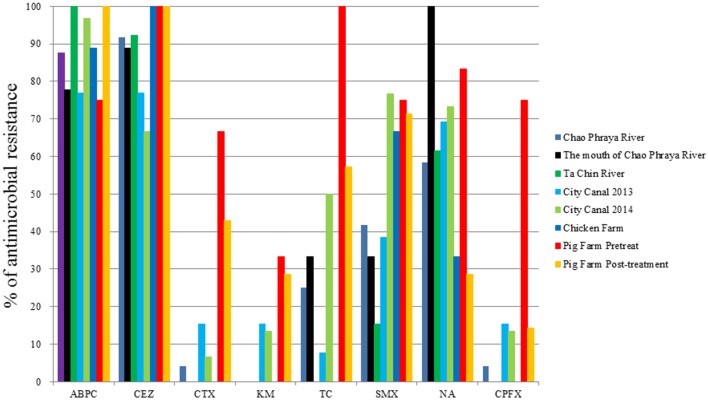
The proportions of antimicrobial-resistant Aeromonas spp. are summarized in Figure 2. Nearly all isolates were resistant to ampicillin and cefazolin. Likewise, we frequently detected sulfamethoxazole- and nalidixic acid-resistant isolates. Notably, however, the rates of cefotaxime, kanamycin, tetracycline, and ciprofloxacin resistance were markedly higher among isolates derived from pig farms than among those obtained from other sites.
FIGURE 2.
Antimicrobial resistance rates of Aeromonas spp. obtained from several aquatic environments. ABPC, ampicillin; CEZ, cefazolin; CTX, cefotaxime; KM, kanamycin; TC, tetracycline; SMX, sulfamethoxazole; NA, nalidixic acid; CPFX, ciprofloxacin.
Antimicrobial Resistance Genes (ARGs)
While the tetA gene was detected in multiple river and canal samples, the prevalence of this gene was highest among the pig farm isolates (75.0%; Table 1). Moreover, both the tetM and sul3 genes (16.7 and 25%, respectively; Table 1) were detected only in isolates harvested from pig farms, while the sul2 gene was detected in pig farm and canal (2014) isolates only (33.3 and 3.3%, respectively; Table 1). The tetC and tetE genes were the most frequently detected resistance genes among the river isolates (Table 1), and the sul1 gene was detected in all water environments except for chicken farms (15.4 to 44.4%). Notably, the tetC, tetG, and tetS genes were not detected in any of the isolates examined in this study, while tetB was detected in only 1 of the 30 canal isolates from 2014 (3.3%).
Relationship between Rates of Antimicrobial Resistance and Concentrations of Antimicrobials
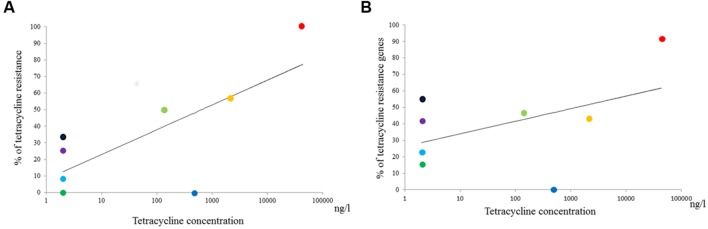
The average concentrations of tetracyclines and sulfonamides at each sample site are summarized in Table 2. Subsequent statistical analyses detected significant correlations between the environmental concentration of tetracyclines and both the rate of tetracycline resistance and the prevalence of tetracycline resistance genes among Aeromonas spp. (R2 = 0.63 and R2 = 0.57, p = 0.019 and p = 0.030, respectively; Figure 3). In contrast, there was no statistical correlation between the environmental concentration of sulfonamides and the rate of sulfamethoxazole resistance or the prevalence of sulfonamide resistance genes among Aeromonas spp.
Table 2.
The average concentrations of tetracyclines and sulfonamides at the aquatic environments examined in this study.
| Average concentrations (ng/L) | |||
|---|---|---|---|
| Sampling sites | n | Tetracyclines | Sulfonamides |
| Chao Phraya River | 5 | <2 | 12.6 ± 5.0 |
| Mouth of the Chao Phraya River | 2 | <2 | 13.7 ± 2.4 |
| Ta Chin River | 3 | <2 | 33.7 ± 1.9 |
| City Canal, 2013 | 5 | <2 | 74.8 ± 54.8 |
| City Canal, 2014 | 6 | 138.8 ± 123.2 | 111.5 ± 119.2 |
| Chicken Farm | 2 | 482.0 ± 503.5 | ND |
| Pig Farm | 2 | 43,500.0 ± 4,0527.1 | 48.0 ± 9.9 |
| Pig Farm, Post-treatment | 2 | 2,135.5 ± 365.6 | 12.5 ± 16.3 |
ND, not detected.
FIGURE 3.
Relationship between the tetracycline concentration at individual aquatic environments and (A) rates of tetracycline resistance or (B) the prevalence of tetracycline resistance genes among Aeromonas spp, respectively. Purple, Chao Phraya River; Black, The mouth of Chao Phraya River; Green, Ta Chin River; Sky Blue, City canal 2013; Yellow-Green, City canal 2014; Blue, Chicken farm; Red, Pig farm; Orange, Pig farm post-treatment.
Discussion
In this study, Aeromonas spp. were frequently isolated from water samples. Notably, these isolates exhibited various resistance patterns against the antimicrobials tested, with the exception of ampicillin and cefazolin, to which nearly all strains were resistant. These findings are therefore consistent with those of a previous study demonstrating that Aeromonas spp. are endogenously resistant to these two antimicrobials (Cisar et al., 2014). Meanwhile, the cephalosporin-resistant Aeromonas spp. exhibited resistance against one of the latest generation of cephalosporins, cefotaxime. As such, monitoring of cefotaxime resistance in Aeromonas spp. would provide information regarding the resistance rates to latest generation cephalosporins among aquatic isolates. Furthermore, Yano et al. (2015) previously reported that Aeromonas spp. exhibited a greater variety of antimicrobial susceptibility profiles than other aquatic bacteria (Yano et al., 2015). Together, these results suggest that Aeromonas spp. comprise an effective general indicator organism for monitoring antimicrobial resistance in aquatic environments.
Aeromonas spp. were previously found to be as prevalent in humans as in environmental sources, indicating that these organisms could disseminate antimicrobial resistance from environmental to clinical environments (Sanchez-Cespedes et al., 2009). Meanwhile, yet-to-be cultured bacteria in aquatic environments can act as non-visible ARG reservoirs (Suzuki et al., 2015), and may be capable of transferring these ARGs to both Aeromonas spp. and other pathogenic aquatic bacteria. As such, monitoring the prevalence and dissemination of antimicrobial resistance in aquatic environments using Aeromonas spp. as an indicator organism is essential to minimize potential risks to human health.
In this study, we detected 40-fold higher concentrations of tetracyclines in pig farm wastewater than in river water samples. Indeed, in the two pig farms tested, chlortetracycline was used as a feed additive for 15- to 20-week-old piglets (personal communications). In contrast, the concentrations of sulfonamides detected in this study were markedly lower than those previously detected on pig farms in Vietnam and South Africa (Le and Munekage, 2004; Hoa et al., 2011; Shimizu et al., 2013; Suzuki et al., 2015). These results suggest that while tetracyclines are currently used intensively, sulfonamides have begun to be phased out for agricultural use in Thailand.
We detected a significant correlation between the environmental concentration of tetracyclines and the rate of tetracycline resistance, but not between the concentration of sulfonamides and the rate of sulfonamide resistance among the Aeromonas strains isolated in this study. Our findings are therefore partially consistent with those of a previous study, which reported that high concentrations of antimicrobials (ciprofloxacin, ofloxacin, cefazolin, cefotaxime, sulfamethoxazole, and clarithromycin) were associated with high copy numbers of ARGs in hospital wastewater samples (Rodriguez-Mozaz et al., 2015). Indeed, these high concentrations of antimicrobials played an important role in the selection and maintenance of ARB and ARGs. These findings therefore suggest that aquatic environments, particularly those containing high concentrations of antimicrobials, act as reservoirs of ARB and ARGs. However, our sampling data were limited, especially in regard to sampling points associated with high concentrations of antimicrobials. As such, in future studies, a greater number of such sampling points is needed to obtain more reliable findings.
The tetA gene was the most frequently detected ARG among tetracycline-resistant Aeromonas isolates derived from pig farms. In a previous report, tetA was the most prevalent tetracycline-resistant gene among E. coli strains isolated from pig feces (Schwaiger et al., 2010), suggesting that the tetA gene of Aeromonas spp. isolated from pig farm wastewater originated from pig feces. Meanwhile, the tetE gene was the mostly frequently observed ARG among isolates derived from river and canal water. Notably, tetE has also been detected in aquatic sediments from aqua culture farms, despite the absence of tetracycline treatments (Tamminen et al., 2011). In most aquatic environments, antimicrobial concentrations are low [e.g., sub- to low-μg/L levels, even in anthropogenically impacted waters (Shimizu et al., 2013)]. However, long-term consistent exposure to sub-threshold doses of antimicrobials can result in increased rates of bacterial resistance (Harnisz, 2013). These results suggest that antimicrobial resistance genes (ARGs), including tetE, can be maintained in aquatic environments regardless of the concentration of the corresponding antimicrobial. Consistent with this conclusion, we recently demonstrated that multidrug resistance plasmids can be retained without selective pressure in viable but non-culturable cells in oligotrophic water environments (Bien et al., 2015).
In this study, treatment of pig farm samples at a biogas plant resulted in decreased rates of resistance to six antimicrobials, as well as reduced concentrations of antimicrobials. Likewise, a previous study demonstrated that biogas plant treatments decrease the prevalence of ARB and tetracyclines in cattle feces (Ihara et al., 2012). These results suggest that biogas plant treatments comprise an effective method for decreasing the concentration of certain antimicrobials and ARB in livestock wastewater.
Author Contributions
Study conception and design: MU, CB, SS, HT, YT. Acquisition of data: MU, CT, AF, TO, KS, HT. Analysis and interpretation of data: MU, CT, SS, HT, YT. Drafting of manuscript: MU, SS, HT, YT.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by MEXT KAKENHI Grant Number 25257402.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00710
References
- Bien T. L., Sato-Takabe Y., Ogo M., Usui M., Suzuki S. (2015). Persistence of multi-drug resistance plasmids in sterile water under very low concentrations of tetracycline. Microbes Environ. 30 339–343. 10.1264/jsme2.ME15122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisar C. R., Henderson S. K., Askew M. L., Risenhoover H. G., McAndrews C. R., Kennedy S. D., et al. (2014). Antibiotic resistance in Aeromonas upstream and downstream of a water resource recovery facility. Water Environ. Res. 86 835–843. 10.2175/106143014X14062131177917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI Clinical and Laboratory Standards Institute (2008). Performance Standards for Antimicrobial Disk Dilution Susceptibility Tests for Bacteria Isolated from Animals: Approved Standard-Third Edition Documents M31-A3. Wayne. PA: CLSI. [Google Scholar]
- Daniel D. S., Lee S. M., Dykes G. A., Rahman S. (2015). Public health risks of multiple-drug-resistant Enterococcus spp. in Southeast Asia. Appl. Environ. Microbiol. 81 6090–6097. 10.1128/AEM.01741-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehaumont P. (2004). OIE International standards on antimicrobial resistance. J. Vet. Med. B Infect. Dis. Vet. Public Health 51 411–414. 10.1111/j.1439-0450.2004.00784.x [DOI] [PubMed] [Google Scholar]
- Esteve C., Alcaide E., Gimenez M. J. (2015). Multidrug-resistant (MDR) Aeromonas recovered from the metropolitan area of Valencia (Spain): diseases spectrum and prevalence in the environment. Eur. J. Clin. Microbiol. Infect. Dis. 34 137–145. 10.1007/s10096-014-2210-z [DOI] [PubMed] [Google Scholar]
- FAO/OIE/WHO (2004). Joint FAO/OIE/WHO Expert Workshop on Non-Human Antimicrobial usage and Antimicrobial Resistance: Scientific Assessment. Available at http://www.oie.int/doc/ged/D895.PDF [Google Scholar]
- Harnisz M. (2013). Total resistance of native bacteria as an indicator of changes in the water environment. Environ. Pollut. 174 85–92. 10.1016/j.envpol.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Hoa P. T., Managaki S., Nakada N., Takada H., Shimizu A., Anh D. H., et al. (2011). Antibiotic contamination and occurrence of antibiotic-resistant bacteria in aquatic environments of northern Vietnam. Sci. Total Environ. 409 2894–2901. 10.1016/j.scitotenv.2011.04.030 [DOI] [PubMed] [Google Scholar]
- Hoa P. T. P., Nonaka L., Hung Viet P., Suzuki S. (2008). Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 405 377–384. 10.1016/j.scitotenv.2008.06.023 [DOI] [PubMed] [Google Scholar]
- Ihara I., Yoshida G., Toyoda K., Iwasaki M., Umetsu K. (2012). Environmental impacts of veterinary antibiotics and reduction of related risk by anaerobic digestion. Soil Microorgan. 60 47–50. [Google Scholar]
- Jin Jun L., Bum Jeong J., Huh M. D., Chung J. K., Choi D. L., Lee C. H., et al. (2004). Detection of tetracycline-resistance determinants by multiplex polymerase chain reaction in Edwardsiella tarda isolated from fish farms in Korea. Aquaculture 240 89–100. 10.1016/j.aquaculture.2004.07.025 [DOI] [Google Scholar]
- Kim S. R., Nonaka L., Suzuki S. (2004). Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol. Lett. 237 147–156. 10.1016/j.femsle.2004.06.026 [DOI] [PubMed] [Google Scholar]
- Le T. X., Munekage Y. (2004). Residues of selected antibiotics in water and mud from shrimp ponds in mangrove areas in Viet Nam. Mar. Pollut. Bull. 49 922–929. 10.1016/j.marpolbul.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Marchesi J. R., Sato T., Weightman A. J., Martin T. A., Fry J. C., Hiom S. J., et al. (1998). Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti E., Variatza E., Balcazar J. L. (2014). The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 22 36–41. 10.1016/j.tim.2013.11.001 [DOI] [PubMed] [Google Scholar]
- NARMS (2012). NARMS Reports and Data. Available at: http://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM453398.pdf [Google Scholar]
- National Veterinary Assay Laboratory Forestry and Fisheries. (2012). A Report on the Japanese Veterinary Antimicrobial Resistance Monitoring System 2008 to 2011. Available at: http://www.maff.go.jp/nval/tyosa_kenkyu/taiseiki/pdf/jvarm2008_2011.pdf [Google Scholar]
- Rhodes G., Huys G., Swings J., McGann P., Hiney M., Smith P., et al. (2000). Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tet A. Appl. Environ. Microbiol. 66 3883–3890. 10.1128/AEM.66.9.3883-3890.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mozaz S., Chamorro S., Marti E., Huerta B., Gros M., Sànchez-Melsió A., et al. (2015). Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 69 234–242. 10.1016/j.watres.2014.11.021 [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes J., Figueras M. J., Aspiroz C., Aldea M. J., Toledo M., Alperi A., et al. (2009). Development of imipenem resistance in an Aeromonas veronii biovar sobria clinical isolate recovered from a patient with cholangitis. J. Med. Microbiol. 58 451–455. 10.1099/jmm.0.47804-0 [DOI] [PubMed] [Google Scholar]
- Schwaiger K., Holzel C., Bauer J. (2010). Resistance gene patterns of tetracycline resistant Escherichia coli of human and porcine origin. Vet. Microbiol. 142 329–336. 10.1016/j.vetmic.2009.09.066 [DOI] [PubMed] [Google Scholar]
- Segura P. A., Takada H., Correa J. A., El Saadi K., Koike T., Onwona-Agyeman S., et al. (2015). Global occurrence of anti-infectives in contaminated surface waters: impact of income inequality between countries. Environ. Int. 80 89–97. 10.1016/j.envint.2015.04.001 [DOI] [PubMed] [Google Scholar]
- Shimizu A., Takada H., Koike T., Takeshita A., Saha M., Rinawati, et al. (2013). Ubiquitous occurrence of sulfonamides in tropical Asian waters. Sci. Total Environ. 45 108–115. 10.1016/j.scitotenv.2013.02.027 [DOI] [PubMed] [Google Scholar]
- Smith P., Hiney M. P., Samuelsen O. B. (1994). Bacterial resistance to antimicrobial agents used in fish farming: a critical evaluation of method and meaning. Ann. Rev. Fish Dis. 4 273–313. 10.1016/0959-8030(94)90032-9 [DOI] [Google Scholar]
- Suzuki S., Hoa P. T. (2012). Distribution of quinolones, sulfonamides, tetracyclines in aquatic environment and antibiotic resistance in indochina. Front. Microbiol. 3:67 10.3389/fmicb.2012.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Ogo M., Koike T., Takada H., Newman B. (2015). Sulfonamide and tetracycline resistance genes in total- and culturable-bacterial assemblages in South African aquatic environments. Front. Microbiol. 6:796 10.3389/fmicb.2015.00796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminen M., Karkman A., Lohmus A., Muziasari W. I., Takasu H., Wada S., et al. (2011). Tetracycline resistance genes persist at aquaculture farms in the absence of selection pressure. Environ. Sci. Technol. 45 386–391. 10.1021/es102725n [DOI] [PubMed] [Google Scholar]
- Yano Y., Hamano K., Tsutsui I., Aue-Umneoy D., Ban M., Satomi M. (2015). Occurrence, molecular characterization, and antimicrobial susceptibility of Aeromonas spp. in marine species of shrimps cultured at inland low salinity ponds. Food Microbiol. 47 21–27. 10.1016/j.fm.2014.11.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.