Summary
Background
DNA repair mechanisms are essential for maintaining genome stability, and genetic variability in DNA repair genes may contribute to cancer susceptibility. Our aim was to evaluate the influence of polymorphisms in the homologous recombination repair genes XRCC3, RAD51, and NBN on the risk for osteosarcoma.
Methods
In total, 79 osteosarcoma cases and 373 controls were genotyped for eight single nucleotide polymorphisms (SNPs) in XRCC3, RAD51, and NBN. Logistic regression was used to determine the association of these SNPs with risk for osteosarcoma.
Results
None of the investigated SNPs was associated with risk for osteosarcoma in the whole cohort of patients, however, in patients diagnosed before the age of thirty years XRCC3 rs861539 C>T and NBN rs1805794 G>C were associated with significantly decreased risk for osteosarcoma (P=0.047, OR=0.54, 95% CI=0.30–0.99 and P=0.036, OR=0.42, 95% CI=0.19–0.94, respectively). Moreover, in the carriers of a combination of polymorphic alleles in both SNPs risk for osteosarcoma was decreased even more significantly (Ptrend=0.007). The risk for developing osteosarcoma was the lowest in patients with no wild-type alleles for both SNPs (P=0.039, OR=0.31, 95% CI=0.10–0.94).
Conclusions
Our results suggest that polymorphisms in homologous recombination repair genes might contribute to risk for osteosarcoma in patients diagnosed below the age of thirty years.
Keywords: osteosarcoma, risk, DNA repair, polymorphism
Kratak sadržaj
Uvod
Mehanizmi reparacije DNK su neophodni za održavanje stabilnosti genoma i genetska varijabilnost u genima koji učestvuju u ovim procesima može doprineti riziku za pojavu kancera. Naš cilj je bio da procenimo uticaj polimorfizama u genima odgovornim za reparaciju DNK mehanizmom homologne rekombinacije, XRCC3, RAD51 i NBN, na rizik za pojavu osteosarkoma.
Metode
Ukupno 79 uzoraka bolesnika sa osteosarkomom i 373 kontrola bilo je genotipizirano na osam pojedinačnih polimorfizama (SNP-ova) u genima XRCC3, RAD51 i NBN. Logistička regresija korišćena je za određivanje povezanosti ovih SNP-ova sa rizikom za osteosarkom.
Rezultati
Nijedan od istraživanih SNP-ova nije bio povezan s rizikom za osteosarkom u čitavoj kohorti pacijenata, međutim, kod bolesnika mlađih od trideset godina kod kojih je dijagnostikovan osteosarkom, polimorfizmi XRCC3 rs861539 C>T i NBN rs1805794 G>C su bili povezani sa znatno smanjenim rizikom za osteosarkom (P=0,047, OR=0,54, 95% CI=0,30–0,99 i P=0,036, OR=0,42, 95% CI= 0,19–0,94, redom). Osim toga, nosioci kombinacije polimorfnih alela za ove SNP-ove imali su još značajnije smanjen rizik za osteosarkom (Ptrend=0,007). Rizik za razvoj osteosarkoma bio je najniži kod bolesnika koji su bili homozigoti za normalne (wt) alele za oba SNP-a (p=0,039, OR=0,31, 95% CI=0,10 do 0,94).
Zaključak
Naši rezultati pokazuju da bi polimorfizam gena odgovornih za reparaciju DNK mehanizmom homologne rekombinacije mogao doprineti riziku za pojavu osteosarkoma kod bolesnika kod kojih je bolest dijagnostikovana pre tridesete godine života.
Introduction
Osteosarcoma is the most common bone malignancy, but is still a relatively rare tumor, with the incidence of around 3 patients per 1 000 000 people per year (1). Osteosarcoma occurs primarily among adolescents and young adults, with a peak of the incidence around 16 years, however, there is also a second peak of the incidence in elderly patients (2). It often occurs at the sites of rapid bone growth and studies have shown that osteosarcoma risk increases in taller children (3, 4). The etiology of the disease is not well explained. Osteosarcoma might be associated with rapid bone proliferation, but exposure to environmental agents can also contribute to susceptibility to osteosarcoma, especially in adults (1). The incidence of osteosarcoma is also higher in some hereditary disorders with germline alterations of tumor suppressor genes and DNA repair pathways (1, 3).
DNA repair mechanisms are essential for maintaining genome stability. Double strand breaks (DSBs) are one of the most detrimental forms of DNA damage because both strands of DNA are damaged (5). DSBs can form as a result of exposure to radiation or various chemotherapeutic agents, but are also produced endogenously, for example, during DNA replication (6). Homologous recombination repair (HRR) is one of the mechanisms involved in DSB repair (7, 8) and genetic variability of HRR genes has already been associated with cancer risk (9, 10). Several enzymes participate in HRR, among them also NBN, RAD51, and X-ray repair cross-complementing protein 3 (XRCC3). NBN is involved in DSB recognition, while RAD51, with help from mediator proteins like XRCC3, catalyses the strand transfer between the damaged region and the undamaged homologous chromatid and therefore enables resynthesis of the damaged region (11).
Single nucleotide polymorphisms (SNPs) in HRR genes have been associated with modified cancer susceptibility in various cancer types (9, 10), but no study reported on association with osteosarcoma. Nevertheless, evidence that variability in DNA repair mechanisms plays a role also in this cancer came from studies reporting that some SNPs in other DNA repair genes, especially MDM2, influenced risk for osteosarcoma (3, 12–14).
We have previously shown that selected tag SNPs in NBN, RAD51, and XRCC3 genes influence the levels of DNA damage in healthy individuals (15). Our aim was to determine whether NBN, RAD51, or XRCC3 tag SNPs modify risk for developing osteosarcoma in Slovenian patients.
Materials and Methods
Subjects
We obtained the data on all the patients with histologically confirmed osteosarcoma, diagnosed between 1990 and 2008, from The Cancer Registry of Slovenia. Only patients that were treated at the Department of Hematology and Oncology, University Children’s Hospital, Ljubljana, Slovenia, or at the Institute of Oncology, Ljubljana, Slovenia, and had available sufficient formalin fixed, paraffin embedded (FFPE) material for DNA extraction were included in our study. Unrelated healthy Slovenian blood donors between the ages of 18 and 65 years were used as the control group (15). The study was approved by the Slovenian Ethics Committee for Research in Medicine and was carried out according to the Declaration of Helsinki.
DNA isolation and genotyping
Hematoxylin and eosin-stained sections of each FFPE sample were examined by an experienced pathologist to confirm the diagnosis and to select areas representative of tumor and areas representative of normal tissue. Two – three cores (1 mm in diameter) of histologically confirmed normal tissue were obtained from each specimen for DNA extraction using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Genotyping of tag SNPs in XRCC3 (rs861539 C>T, rs1799794 A>G), RAD51 (rs1801320 G>C, rs1801321 G>T, rs12593359 T>G), and NBN (rs1805794 G>C, rs709816 C>T, rs1063054 A>C) was carried out using a fluorescence-based competitive allele-specific (KASPar) assay (KBiosciences, Herts, UK) according to the manufacturer’s instructions as previously described (15).
In the control group, DNA isolation from peripheral blood leukocytes and genotyping for the selected tag SNPs were performed during the previous study (15).
Statistical analysis
Chi-squared test or Fisher’s exact test were used for the comparison of categorical data distribution, while nonparametric Mann–Whitney U-test was used for comparison of numerical data. Logistic regression was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs) to examine the associations of selected SNPs with the risk for developing osteosarcoma. Dominant genetic model was used in all analyses. All statistical tests were two-sided and the level of significance was set to 0.050. All statistical analyses were carried out by IBM SPSS Statistics, version 19.0 (IBM Corporation, Armonk, NY, USA).
Incidence rates were calculated per 1 million inhabitants for five-year age groups. Data on all osteosarcoma cases between 1990 and 2008 were obtained from The Cancer Registry of Slovenia (16) and population data of all the inhabitants in Slovenia during the same period were obtained from the Statistical Office of the Republic of Slovenia (17).
Results
Based on data from The Cancer Registry of Slovenia, 118 osteosarcoma patients were diagnosed in Slovenia between 1990 and 2008. Among them, 79 (66.9%) fulfilled all the inclusion criteria and were included in the study along with 373 healthy controls. The characteristics of the subjects are presented in Table I.
Table I.
The characteristics of controls and osteosarcoma patients.
| Characteristic | Controls (%) | All cases (%) | Cases <30 years (%) | |
|---|---|---|---|---|
| N=373 | N=79 | N=48 | ||
| Age | Median (interquartile range), years | 30 (23–53.5) | 19 (15–38) | 15.5 (12.3–18.0) |
| Gender | Male | 219 (58.9) | 40 (51.9) | 26 (54.2) |
| Female | 153 (41.1) | 37 (48.1) | 22 (45.8) | |
| Osteosarcoma histological type | Osteogenic | 70 (88.6) | 47 (97.9) | |
| Chondroblastic | 2 (2.5) | 1 (2.1) | ||
| Fibroblastic | 3 (3.8) | |||
| Teleangiectatic | 1 (1.3) | |||
| Not defined | 3 (3.8) | |||
| Primary/secondary malignancy | Primary | 75 (94.9) | 48 (100.0) | |
| Secondary | 4 (5.1) | |||
Osteosarcoma patients were significantly younger than healthy controls (P<0.001), but no differences regarding gender were observed (P=0.252).
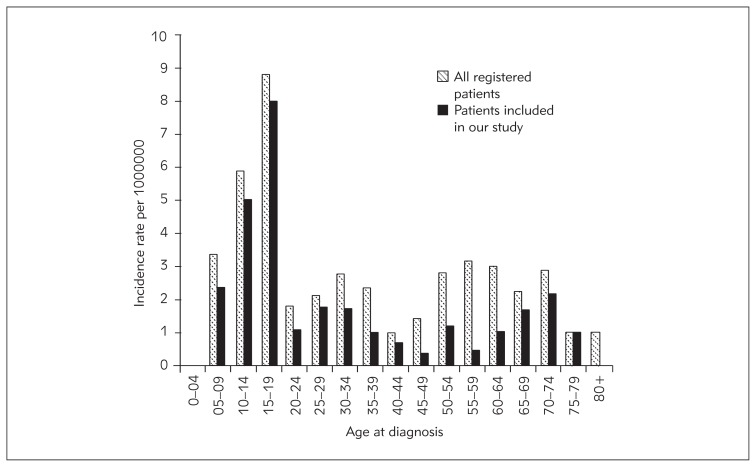
In total, 48 (60.8%) patients were younger than 30 years at the time of diagnosis. Two peaks in the average incidence rates of osteosarcoma were observed in our sample (Figure 1). The first peak occurred in the age group between 15 and 19 years and the second between 70 and 75 years. In comparison, data for all the patients diagnosed in Slovenia between 1990 and 2008 also showed the first peak in incidence rate in the age group between 15 and 19 years, however, the second increase of incidence rate started already after the age of 50 years (Figure 1).
Figure 1.
Average incidence rates of osteosarcoma in Slovenia between 1990 and 2008.
We analyzed the risk for osteosarcoma in all patients, but also separately in osteosarcoma patients younger than 30 years at the time of diagnosis. The patients younger than 30 years did not differ from the whole group regarding gender (P=0.855) or histological type (P=0.403, calculated for osteogenic osteosarcoma against other histological types). In four patients (5.1%), osteosarcoma developed as the secondary malignancy, but these patients were all older than 30 years.
No significant differences in the genotype frequency distribution of the investigated SNPs were observed when all the osteosarcoma cases were compared to healthy controls (Table II). However, patients diagnosed with osteosarcoma before the age of 30 differed significantly from healthy controls regarding the genotype distribution of XRCC3 rs861539 C>T (P=0.047) and NBN rs1805794 G>C (P=0.036) and the respective polymorphic XRCC3 rs861539 T and NBN rs1805794 C alleles were significantly associated with decreased osteosarcoma risk (OR=0.54, 95% CI=0.30–0.99 and OR=0.42, 95% CI=0.19–0.94, respectively) (Table II).
Table II.
Association of XRCC3, RAD51, and NBN SNPs with risk for osteosarcoma.
| Gene | Polymorphism | Genotype | Controls (%) | All patients | Patients <30 years | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cases (%) | OR (95% CI) | P | Cases (%) | OR (95% CI) | P | ||||
| XRCC3 | rs1799794 A>G c.-316A>G |
AA | 247 (66.2) | 47 (59.5) | 1.29 (0.78–2.14) | 0.315 | 27 (56.3) | 1.53 (0.83–2.80) | 0.175 |
| AG+GG | 126 (33.8) | 31 (39.2) | 21 (43.8) | ||||||
| rs861539 C>T p.Thr241Met |
CC | 153 (41.0) | 39 (49.4) | 0.71 (0.44–1.16) | 0.174 | 27 (56.3) | 0.54 (0.30–0.99) | 0.047 | |
| CT+TT | 220 (59.0) | 40 (50.6) | 21 (43.8) | ||||||
| RAD51 | rs1801320 G>C c.-98G>C |
GG | 304 (81.5) | 69 (87.3) | 0.58 (0.27–1.21) | 0.143 | 41 (85.4) | 0.65 (0.26–1.58) | 0.337 |
| GC+CC | 69 (18.5) | 9 (11.4) | 6 (12.5) | ||||||
| rs1801321 G>T c.-61G>T |
GG | 133 (35.7) | 25 (31.6) | 1.20 (0.71–2.01) | 0.497 | 18 (37.5) | 0.92 (0.50–1.72) | 0.802 | |
| GT+TT | 240 (64.3) | 54 (68.4) | 30 (62.5) | ||||||
| rs12593359 T>G c.*502T>G |
TT | 103 (27.6) | 22 (27.8) | 0.99 (0.58–1.70) | 0.966 | 12 (25.0) | 1.14 (0.57–2.29) | 0.702 | |
| TG+GG | 270 (72.4) | 57 (72.2) | 36 (75.0) | ||||||
| NBN | rs1805794 G>C p.Glu185Gln |
GG | 33 (8.8) | 11 (13.9) | 0.60 (0.29–1.25) | 0.170 | 9 (18.8) | 0.42 (0.19–0.94) | 0.036 |
| GC+CC | 340 (91.2) | 68 (86.1) | 39 (81.3) | ||||||
| rs709816 C>T p.Asp399= |
CC | 139 (37.3) | 29 (36.7) | 1.02 (0.62–1.69) | 0.926 | 18 (37.5) | 0.99 (0.53–1.84) | 0.975 | |
| CT+TT | 234 (62.7) | 50 (63.3) | 30 (62.5) | ||||||
| rs1063054 A>C c.*1209A>C |
AA | 163 (43.7) | 38 (48.1) | 0.78 (0.47–1.27) | 0.315 | 22 (45.8) | 0.81 (0.44–1.51) | 0.508 | |
| AC+CC | 210 (56.3) | 38 (48.1) | 23 (47.9) | ||||||
Furthermore, when the combined effect of both XRCC3 rs861539 C>T and NBN rs1805794 G>C was investigated in patients diagnosed before the age of 30 years, osteosarcoma risk was further decreased (Ptrend=0.007). The risk was the lowest in patients with no wild-type alleles for both SNPs (P=0.039, OR=0.31, 95% CI=0.10–0.94). The combination of both SNPs also showed a trend, albeit insignificant, towards decreased risk for developing osteosarcoma regardless the age at diagnosis (P=0.072; OR=0.70; 95% CI=0.47–1.03), although individual XRCC3 rs861539 C>T and NBN rs1805794 G>C SNPs were not significantly associated with osteosarcoma in the whole cohort.
Discussion
In the present study, we observed that SNPs in HRR genes influence the risk for developing osteosarcoma in adolescents and young adults. Significantly decreased risk for osteosarcoma was observed for XRCC3 rs861539 CT and TT genotypes and NBN rs1805794 GC and CC genotypes, and the risk decreased even more in the carriers of polymorphic alleles in both genes. On the other hand, none of the investigated SNPs were associated with risk for osteosarcoma in the whole cohort of patients.
We included in our study almost all the pediatric and adolescent osteosarcoma patients diagnosed in Slovenia, because most of them are treated at University Children’s Hospital, Ljubljana. On the other hand, we could not include all the adult cases, possibly because they were diagnosed and treated only surgically in other hospitals and were not admitted to the Institute of Oncology, Ljubljana.
Osteosarcoma incidence usually has a bimodal age of distribution, with the first peak among adolescents and a second peak in adults older than 60 years (2). Most patients are diagnosed before the age of 30 years, with a peak of the incidence around 16 years (18). We also observed the highest osteosarcoma incidence among adolescents between 15 and 19 years. The bimodal incidence distribution might be associated with different disease etiology in younger and older osteosarcoma patients. Environmental exposure to radiation, alkylating agents, or other factors that may contribute to osteosarcoma most likely plays an important role in older patients (1), where osteosarcoma can present as a secondary malignancy (2), which we also observed in our study. On the other hand, genetic factors probably contribute more to osteosarcoma in younger patients, where it often coincides with rapid bone growth (2). Genetic variation in DNA repair mechanisms leading to differences in DNA repair capacity and level of unrepaired DNA damage could influence risk for osteosarcoma, especially considering the fact that a higher osteosarcoma incidence was observed in hereditary disorders with alterations in DNA repair pathways. We therefore separately analyzed osteosarcoma in patients younger than 30 years at the time of diagnosis, and significantly decreased risk for osteosarcoma was observed for carriers of at least one polymorphic XRCC3 rs861539 T or NBN rs1805794 C allele.
Both SNPs, XRCC3 rs861539 T or NBN rs1805794 C allele, are non-synonymous and could therefore affect protein structure or function. Both SNPs were associated with decreased levels of DNA damage, detected using the comet assay in our previous study, suggesting that both polymorphisms may affect the DNA repair capacity of these enzymes (15). To our knowledge, no previous studies investigated SNPs in HRR genes in osteosarcoma. The results of studies investigating the associations of XRCC3 rs861539 C>T with susceptibility to other cancer types were often inconclusive, so several meta-analyses were published in the last few years, either examining susceptibility to just one or several cancer types (9, 19–22). The results differed even among meta-analyses, but most have detected increased breast or bladder cancer susceptibility in the carriers of polymorphic alleles (9, 19, 22), while lung or skin cancer susceptibility was decreased (9, 19, 20).
Fewer meta-analyses have been published regarding the influence of NBN rs1805794 G>C (10, 23–25). Increased overall cancer susceptibility (10) and increased bladder cancer susceptibility were reported (25), while the previously reported decreased breast cancer susceptibility (23) was no longer significant in a more recent meta-analysis (24). Other single studies observed no influence on acute lymphoblastic leukemia susceptibility (26), but susceptibility to acute myeloid leukemia was decreased (27). In our undergoing study, the polymorphic NBN rs1805794 C allele was also associated with decreased susceptibility to pediatric B-cell acute lymphoblastic leukemia. The most studied SNPs regarding osteosarcoma susceptibility are those in the MDM2 gene. MDM2 is a negative regulator of the tumor suppressor p53, and several studies, including a recent meta-analysis, have shown that MDM2 SNPs influence osteosarcoma susceptibility (3, 13, 14). MDM2 also contributes to the regulation of DSB repair and it was shown that it directly binds NBN (28), suggesting that our observation of NBN rs1805794 G>C modifying osteosarcoma susceptibility is biologically plausible.
The discrepancies between the results of different studies suggest that individual SNPs contribute only a small part to overall cancer susceptibility. Indeed, one of the meta-analyses concluded that while XRCC3 SNPs definitely modify cancer susceptibility, they do not represent the most important risk factor (19). Interplay of more SNPs is more likely. This is also supported by our observation that, compared to a polymorphism in a single gene, the combination of XRCC3 rs861539 C>T and NBN rs1805794 G>C SNPs has a larger protective effect on osteosarcoma susceptibility in adolescents and young adults.
As osteosarcoma is a rare form of cancer, our study was performed on a relatively small group of patients, thus our results need to be confirmed in a larger study on osteosarcoma in adolescents. However, different observations for the influence of SNPs were reported for different populations and the advantage of our study is that both patients and controls were recruited in a geographic area with an ethnically homogeneous population (29).
Although our study focused only on genes within HRR, SNPs in other DNA repair pathways have also been associated with osteosarcoma risk. The polymorphic allele of nucleotide excision repair gene XPD rs1799793 was associated with decreased risk of developing osteosarcoma (12), while various FANCM SNPs were associated with increased risk (3).
In conclusion, our results support the observations that DNA repair polymorphisms, specifically SNPs in the HRR genes XRCC3 and NBN, might contribute to risk for developing osteosarcoma in children, adolescents and young adults.
Acknowledgements
The authors thank the staff of The Cancer Registry of Slovenia for providing information on all the osteosarcoma patients in Slovenia. This work was financially supported by the Slovenian Research Agency (ARRS Grants No. P1-0170, P3-0343).
List of abbreviations
- 95% CI
95% confidence interval
- DSB
double strand break
- FFPE
formalin fixed; paraffin embedded
- HRR
homologous recombination repair
- OR
odds ratio
- SNP
single nucleotide polymorphism
- XRCC3
X-ray repair cross-complementing protein 3.
Footnotes
Conflict of interest statement
The authors stated that they have no conflicts of interest regarding the publication of this article.
References
- 1.Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–5. doi: 10.1093/annonc/mdq276. [DOI] [PubMed] [Google Scholar]
- 2.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 3.Mirabello L, Yu K, Berndt SI, Burdett L, Wang Z, Chowdhury S, et al. A comprehensive candidate gene approach identifies genetic variation associated with osteosarcoma. BMC Cancer. 2011;11:209. doi: 10.1186/1471-2407-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quartuccio N, Treglia G, Salsano M, Mattoli MV, Muoio B, Piccardo A, et al. The role of Fluorine-18-Fluoro-deoxyglucose positron emission tomography in staging and restaging of patients with osteosarcoma. Radiol Oncol. 2013;47:97–102. doi: 10.2478/raon-2013-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, Waldman AS, Wyatt MD. DNA damage and homologous recombination signaling induced by thymidylate deprivation. Biochem Pharmacol. 2008;76:987–96. doi: 10.1016/j.bcp.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger SH, Pittman DL, Wyatt MD. Uracil in DNA: consequences for carcinogenesis and chemotherapy. Biochem Pharmacol. 2008;76:697–706. doi: 10.1016/j.bcp.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He XF, Wei W, Li JL, Shen XL, Ding DP, Wang SL, et al. Association between the XRCC3 T241M polymorphism and risk of cancer: evidence from 157 case-control studies. Gene. 2013;523:10–9. doi: 10.1016/j.gene.2013.03.071. [DOI] [PubMed] [Google Scholar]
- 10.Lu M, Lu J, Yang X, Yang M, Tan H, Yun B, et al. Association between the NBS1 E185Q polymorphism and cancer risk: a meta-analysis. BMC Cancer. 2009;9:124. doi: 10.1186/1471-2407-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–35. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Biason P, Hattinger CM, Innocenti F, Talamini R, Alberghini M, Scotlandi K, et al. Nucleotide excision repair gene variants and association with survival in osteosarcoma patients treated with neoadjuvant chemotherapy. Pharmacogenomics J. 2012;12:476–83. doi: 10.1038/tpj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toffoli G, Biason P, Russo A, De Mattia E, Cecchin E, Hattinger CM, et al. Effect of TP53 Arg72Pro and MDM2 SNP309 polymorphisms on the risk of high-grade osteosarcoma development and survival. Clin Cancer Res. 2009;15:3550–6. doi: 10.1158/1078-0432.CCR-08-2249. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Liu Z, Jing P, Shao L, Chen L, He X, et al. Effects of murine double minute 2 polymorphisms on the risk and survival of osteosarcoma: a systemic review and meta-analysis. Tumour Biol. 2013 Oct 11; doi: 10.1007/s13277-013-1227-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.Goricar K, Erculj N, Zadel M, Dolzan V. Genetic polymorphisms in homologous recombination repair genes in healthy Slovenian population and their influence on DNA damage. Radiol Oncol. 2012;46:46–53. doi: 10.2478/v10019-012-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SLORA. Epidemiologija in register raka. Onkološki inštitut Ljubljana; Slovenija in rak. [database on the Internet] [cited November 11, 2013]. Available from: www.slora.si. [Google Scholar]
- 17.Statistical Office of the Republic of Slovenia. [database on the Internet] [cited November 11, 2013]. Available from: www.stat.si.
- 18.Damron TA, Ward WG, Stewart A. Osteosarcoma, chondrosarcoma, and Ewing’s sarcoma: National Cancer Data Base Report. Clin Orthop Relat Res. 2007;459:40–7. doi: 10.1097/BLO.0b013e318059b8c9. [DOI] [PubMed] [Google Scholar]
- 19.Han S, Zhang HT, Wang Z, Xie Y, Tang R, Mao Y, et al. DNA repair gene XRCC3 polymorphisms and cancer risk: a meta-analysis of 48 case-control studies. Eur J Hum Genet. 2006;14:1136–44. doi: 10.1038/sj.ejhg.5201681. [DOI] [PubMed] [Google Scholar]
- 20.Huang G, Cai S, Wang W, Zhang Q, Liu A. Association between XRCC1 and XRCC3 polymorphisms with lung cancer risk: a meta-analysis from case-control studies. PLoS One. 2013;8:e68457. doi: 10.1371/journal.pone.0068457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding G, Xu W, Hua H, Huang Q, Liang H, Ni Y, et al. Comprehensive assessment of the association between DNA repair gene XRCC3 rs861539 C/T polymorphism and lung cancer risk. Tumour Biol. 2013;34:2521–7. doi: 10.1007/s13277-013-0705-3. [DOI] [PubMed] [Google Scholar]
- 22.Ma Q, Zhao Y, Wang S, Zhang X, Zhang J, Du M, et al. Genetic polymorphisms of XRCC3 Thr241Met (C18067T, rs861539) and bladder cancer risk: a meta-analysis of 18 research studies. Tumour Biol. 2013 Oct 2; doi: 10.1007/s13277-013-1203-3. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Cui D, Lu W. NBS1 8360G > C polymorphism is associated with breast cancer risk: a meta-analysis. Breast Cancer Res Treat. 2010;123:557–61. doi: 10.1007/s10549-010-0772-8. [DOI] [PubMed] [Google Scholar]
- 24.Yao F, Fang Y, Chen B, Jin F, Wang S. Association between the NBS1 Glu185Gln polymorphism and breast cancer risk: a meta-analysis. Tumour Biol. 2013;34:1255–62. doi: 10.1007/s13277-013-0668-4. [DOI] [PubMed] [Google Scholar]
- 25.Stern MC, Lin J, Figueroa JD, Kelsey KT, Kiltie AE, Yuan JM, et al. Polymorphisms in DNA repair genes, smoking, and bladder cancer risk: findings from the international consortium of bladder cancer. Cancer Res. 2009;69:6857–64. doi: 10.1158/0008-5472.CAN-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosor M, Ziolkowska I, Januszkiewicz-Lewandowska D, Nowak J. Polymorphisms and haplotypes of the NBS1 gene in childhood acute leukaemia. Eur J Cancer. 2008;44:2226–32. doi: 10.1016/j.ejca.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Li N, Xu Y, Zheng J, Jiang L, You Y, Wu H, et al. NBS1 rs1805794G>C polymorphism is associated with decreased risk of acute myeloid leukemia in a Chinese population. Mol Biol Rep. 2013;40:3749–56. doi: 10.1007/s11033-012-2451-9. [DOI] [PubMed] [Google Scholar]
- 28.Alt JR, Bouska A, Fernandez MR, Cerny RL, Xiao H, Eischen CM. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280:18771–81. doi: 10.1074/jbc.M413387200. [DOI] [PubMed] [Google Scholar]
- 29.Vidan-Jeras B, Jurca B, Dolzan V, Jeras M, Breskvar K, Bohinjec M. Slovenian Caucasian normal. In: Gjertson D, Terasaki P, editors. HLA 1998. Kansas: American society for histocompatibility and immunogenetics, Lenexa; 1998. pp. 180–1. [Google Scholar]