Summary
Background
In this study the authors present an update to the CFTR mutation profile in Hungary, utilizing data from a selected cohort of 45 cystic fibrosis (CF) patients from different regions of the country.
Methods
Depending on the preceding analysis, four different mutation detection methods were used. A commercial assay targeting the most common CF-causing mutations was performed as the first test followed by an allele specific PCR for CFTRdele2,3(21kb), Sanger sequencing and MLPA analysis of the coding region of the CFTR gene.
Results
In our recent study 27 different mutations were detected, including 2 novel ones (c.1037_1038insA and c.1394C>T). Besides F508del (c.1521_1523delCTT), the following mutations were found at a frequency of ≥ 4.0%: W1282X (c.3846G>A), N1303K (c.3909C>G), CFTRdele2,3(21kb) (c.54-5940_273+10250del21kb) and 2184insA (c.2052_2053insA). In addition, four mutations (G542X, Y1092X, 621+1G>T, and 2143delT) were found in more than one allele.
Conclusions
The updated database of Hungarian mutations not only enables to increase the efficiency of the existing diagnostic approach, but also provides a further refined basis for the introduction of the molecular newborn screening (NBS) program in Hungary.
Keywords: cystic fibrosis, mutational spectrum, newborn screening
Kratak sadržaj
Uvod
U ovoj studiji autori predstavljaju nove podatke dobijene izučavanjem profila mutacije CFTR u Mađarskoj uz pomoć podataka izabrane grupe od 45 pacijenata sa cističnom fibrozom (CF) iz različitih krajeva zemlje.
Metode
U zavisnosti od prethodne analize, korišćene su četiri različite metode za detekciju mutacija. Prvo je rađen komercijalni test koji ciljano traga za najčešćim mutacijama koje izazivaju CF, posle čega su primenjeni PCR za CFTRdele2,3(21 kb), Sanger sekvenciranje i MLPA analiza kodirajućeg regiona gena CFTR.
Rezultati
U našoj nedavnoj studiji otkriveno je 27 različitih mutacija, uključujući dve dotad nepoznate (c.1037_1038insA i c.1394C>T). Pored F508del (c.1521 _1523delCTT), pronađene su sledeće mutacije sa učestalošću ≥ 4,0%: W1282X (c.3846G>A), N1303K (c.3909C >G), CFTRdele2,3(21kb) (c.54-5940_273+10250 del21kb) i 2184insA (c.2052_2053insA). Pored toga, četiri mutacije (G542X, Y1092X, 621+1G>T i 2143 delT) nađene su u više od jednog alela.
Zaključak
Baza podataka o mađarskim mutacijama dopunjena poslednjim informacijama ne samo da omogućava povećavanje efikasnosti postojećeg dijagnostičkog pristupa, već takođe pruža dodatno usavršen osnov za uvođenje programa molekularnog skrininga novorođenčadi (NBS) u Mađarskoj.
Introduction
Cystic fibrosis (CF) is the most common severe inherited monogenic disease in Caucasians, with an incidence of 1: 2500. Besides the F508del mutation, which accounts for the majority of CF alleles, almost 2000 different rare mutations have been described in the CFTR gene. Although no more than 20 of them occur at a frequency higher than 0.1% worldwide (1), the geographical heterogeneity of CF causing alleles demands the use of population specific mutation detection panels in order to reach reasonably high mutation detection rates.
Here, we present the mutational spectrum of CF patients in a Hungarian cohort which supplements the results of our previous study from Eastern Hungary (2).
Materials and Methods
Altogether 45 CF patients (22 males and 23 females, 10.1±8.1 years) were involved in this study. Sample collection and analysis were performed between 2010 and 2014. We selected patients with a clinical picture of classical CF, where both disease-causing mutations were identified. All patient samples were sent by Hungarian care centers (mainly from the regions of Budapest, Szeged and Debrecen). DNA was isolated from peripheral blood leukocytes with the QIAgen Blood Mini Kit (Qiagen, Hilden, Germany).
The first line molecular test – if it had not been carried out by another laboratory before – was a commercially available multiplex allele specific PCR assay, which is able to detect 29 mutations and differentiates between hetero- or homozygous forms of F508del (Elucigene™ CF29 v.2 kit, Tepnel Diagnostics Ltd, Abingdon, UK). Any mutation found other than F508del was further examined by sequence analysis of the corresponding region in order to determine if one or both alleles were affected. The first tier assay was complemented by an allele specific PCR to detect the common »Slavic« deletion, CFTRdele2,3(21kb) (3). As a second tier mutation analysis Sanger sequencing of the entire coding region was employed as described before (2) to find small-scale mutations elusive of the commercial assay. According to our previous experience (2), we started sequencing of exons 4, 6, 11, 14, 15 and 20, containing prevalent Hungarian mutations such as 2184insA (c.2052_2053insA), L101X (c.302T>G) and Y1092X (c.3276C>A); if no alteration was detected, the remaining exons were also tested. As a third tier step, MLPA analysis was used to detect large-scale rearrangements (SALSA MLPA KIT P091-B1 CFTR, MRC-Holland, Amsterdam, the Netherlands). If the sending institution had already performed first-line testing, we carried out only the second and/or third level of molecular analysis.
One gross rearrangement in the CFTR gene (CFTRdele2, c.54-5811_164+2186del273+6780_273+6961inv) which was detected by MLPA analysis was confirmed by allele specific PCR and bidirectional sequencing of the junction regions, as described in the literature (7–9).
The numbering of CFTR exons was done according to the current recommendations (Ensembl ENSG00000001626). In the mutation nomenclature both the legacy names and cDNA based HGVS names were used as proposed (4).
Results
Table I shows the distribution of detected mutations in our recent study compared to Czech (5), Polish (6) and previous Hungarian (2) papers. Altogether 27 different mutations were found. F508del was detected in 53.3%, four mutations (W1282X, N1303K, CFTRdele2,3(21 kb), and 2184insA) were detected in 4.4% of alleles, while another four mutations (G542X, Y1092X, 621+ 1G>T, and 2143delT) were found in more than one allele.
Table I.
Mutation distribution in our patient cohort compared to Czech (5), Polish (6) and previous Hungarian (2) publications.
| HGVS nomenclature | Legacy name/effect on protein level | Hungary 2011 (N=80) | Czech Republic 2013 (N=1200) | Poland 2014 (N=1476) | This study (N=90) |
|---|---|---|---|---|---|
| c.1521_1523delCTT | F508del | 70.0% | 67.4% | 54.5% | 53.3% |
| c.3846G>A | W1282X | 0.0% | 0.6% | 0.6% | 4.4% |
| c.3909C>G | N1303K | 5.0% | 2.4% | 1.2% | 4.4% |
| c.54-5940_273+10250del21kb | CFTRdele2,3(21kb) | 5.0% | 5.8% | 4.5% | 4.4% |
| c.2052_2053insA | 2184insA | 5.0% | 0.4% | 1.0% | 4.4% |
| c.1624G>T | G542X | 3.8% | 2.0% | 1.7% | 2.2% |
| c.489+1G>T | 621+1G>T | 0.0% | 0.4% | 0.3% | 2.2% |
| c.3276C>A | Y1092X | 1.3% | 0.0% | 0.1% | 2.2% |
| c.302T>G | L101X | 2.5% | 0.0% | 0.1% | 1.1% |
| c.1397C>G | S466X | 1.3% | 0.0% | 0.1% | 1.1% |
| c.2012delT | 2143delT | 0.0% | 0.9% | 1.8% | 2.2% |
| c.53+1G>T | 185+1G>T | 0.0% | 0.2% | 0.0% | 1.1% |
| c.1394C>T | p.Thr465Ile | 0.0% | 0.0% | 0.0% | 1.1% |
| c.1037_1038insA | p.Leu346Hisfs*17 | 0.0% | 0.0% | 0.0% | 1.1% |
| c.2051_2052delAAinsG | 2183AA>G | 0.0% | 0.1% | 0.7% | 1.1% |
| c.2657+5G>A | 2789+5G>A | 0.0% | 0.5% | 0.0% | 1.1% |
| c.3717+12191C>T | 3849+10kbC>T | 0.0% | 1.7% | 3.9% | 1.1% |
| c.215C>A | A72D | 0.0% | 0.0% | 0.0% | 1.1% |
| c.3454G>C | D1152H | 0.0% | 0.0% | 0.1% | 1.1% |
| c.3731G>A | G1244E | 0.0% | 0.0% | 0.0% | 1.1% |
| c.1727G>C | G576A | 0.0% | 0.0% | 0.3% | 1.1% |
| c.3302T>A | M1101K | 0.0% | 0.0% | 0.0% | 1.1% |
| c.2591_2592delTT | 2723delTT | 0.0% | 0.0% | 0.0% | 1.1% |
| c.3822G>A | W1274X | 0.0% | 0.0% | 0.0% | 1.1% |
| c.2002C>T | R668C | 0.0% | 0.0% | 0.2% | 1.1% |
| c.325_327delTATinsG | p.Tyr109Glyfs*4 | 0.0% | 0.0% | 0.0% | 1.1% |
| c.54-5811_164+2186del273+6780_273+6961inv | CFTRdele2 | 0.0% | 0.2% | 0.0% | 1.1% |
As shown in Table II, the commercially available assay was able to detect 75.9% of the mutations in our combined mutational database. The allele specific PCR for CFTRdele2,3(21kb) revealed 4.7% of disease-causing alleles. By the sequencing of exon 14 2184insA and two other rare mutations (c.2012delT and c.2002C>T) were found with a frequency of 4.7%, 1.2% and 0.6%, respectively. The remaining mutations were scattered throughout the entire coding region of the CFTR gene. MLPA analysis revealed one deletion affecting exon 2 in one patient in a heterozygous form. Allele specific PCR and sequencing analysis showed that the single exon deletion detected by MLPA was the same as previously described by others (10, 11).
Table II.
Allele frequencies and mutation detection rates in the combined mutational database. Novel mutations are in bold.
| Exon/intron | HGVS nomenclature | Legacy name/effect on protein level | No. of alleles (N=170) | Frequency of alleles | Mutation in trans | Proportion of detected mutations |
|---|---|---|---|---|---|---|
| e11 | c.1521_1523delCTT | F508del | 104 | 61.2% | various CF mutations | Elucigene CF29v.2 75.9% |
| e23 | c.3846G>A | W1282X | 4 | 2.4% | F508del, 2184insA, W1282 | |
| e24 | c.3909C>G | N1303K | 8 | 4.7% | various CF mutations | |
| e12 | c.1624G>T | G542X | 5 | 2.9% | various CF mutations | |
| i11 | c.1585-1G>A | 1717-1G>A | 1 | 0.6% | F508del | |
| e8 | c.1040G>C | R347P | 1 | 0.6% | G542X | |
| e14 | c.2051_2052delAAinsG | 2183AA>G | 1 | 0.6% | D1152H | |
| i16 | c.2657+5G>A | 2789+5G>A | 1 | 0.6% | 621+1G>T | |
| i4 | c.489+1G>T | 621+1G>T | 2 | 1.2% | F508del, 2789+5G>A | |
| e21 | c.3454G>C | D1152H | 1 | 0.6% | 2183AA>G | |
| i22 | c.3717+12191C>T | 3849+10kbC>T | 1 | 0.6% | F508del | |
| i1-i3 | c.54-5940_273+ 10250del21kb | CFTRdele2,3(21kb) | 8 | 4.7% | F508del, N1303K, Y109G | “Slavic PCR” 4.7% |
| e14 | c.2052_2053insA | 2184insA | 8 | 4.7% | F508del, G542X, W1282 | Sequencing of e14 6.5% |
| e14 | c.2012delT | 2143delT | 2 | 1.2% | F508del | |
| e14 | c.2002C>T | R668C | 1 | 0.6% | G576A | |
| e20 | c.3276C>A | Y1092X | 3 | 1.8% | F508del | Sequencing of the entire coding region 11.8% |
| e4 | c.302T>G | L101X | 3 | 1.8% | F508del, 2723delTT | |
| e11 | c.1397C>G | S466X | 2 | 1.2% | F508del | |
| i1 | c.53+1G>T | 185+1G>T | 1 | 0.6% | N1303K | |
| e11 | c.1394C>T | p.Thr465Ile | 1 | 0.6% | F508del | |
| e8 | c.1037_1038insA | p.Leu346Hisfs*17 | 1 | 0.6% | F508del | |
| e3 | c.215C>A | A72D | 1 | 0.6% | N1303K | |
| e15 | c.2491G>T | E831X | 1 | 0.6% | N1303K | |
| e6 | c.658C>T | Q220X | 1 | 0.6% | F508del | |
| e23 | c.3731G>A | G1244E | 1 | 0.6% | F508del | |
| e13 | c.1727G>C | G576A | 1 | 0.6% | R668C | |
| e20 | c.3302T>A | M1101K | 1 | 0.6% | F508del | |
| e15 | c.2591_2592delTT | 2723delTT | 1 | 0.6% | L101X | |
| e23 | c.3822G>A | W1274X | 1 | 0.6% | F508del | |
| e4 | c.325_327delTATinsG | p.Tyr109Glyfs*4 | 1 | 0.6% | CFTRdele2,3(21kb) | |
| i1-i2 | c.54-5811_164+2186del273 +6780_273+6961inv | CFTRdele2 | 1 | 0.6% | F508del | MLPA 0.6% |
Discussion
We updated our existing CF database by merging recently acquired nationwide results with our previous data from Eastern Hungary (2). Allele frequencies and proportion of detected mutations at different levels are listed in Table II. Data of our combined database are discussed below.
Altogether, 31 different mutations were identified in our previous and recent studies, two of which were novel (according to the Clinical and Functional Translation of CFTR database, cftr2.org and the Human Gene Mutation Database (HGMD)). These newly described sequence alterations are most likely pathogenic. One of them changes the reading frame, generating a premature stop codon 17 amino acids downstream (c.1037_1038insA, p.Leu346Hisfs*17). Pathogenicity of the detected novel missense mutation c.1394C>T, p.Thr465Ile is supported by the following: 1) the affected residue is located at a phylogenetically highly conserved position according to the orthologs of Bos taurus, Equus caballus, Felis catus, Mus musculus etc.; 2) another pathogenic mutation (T465N) affecting the same amino acid residue has already been described (12); 3) SIFT analysis predicts a damaging effect on the protein function.
Eleven mutations reached a frequency higher than 1%. In accordance with the literature (5, 6), the decreasing »North-to-South« gradient (13) stands for the distribution of F508del (104/170 CF alleles) compared to the Czech Republic, but not to Poland. F508del was followed by the Mediterranean mutation N1303K, the »Slavic« mutation CFTRdele2,3(21kb) and the »Galican« mutation 2184insA, each responsible for 4.7% of all CF alleles, which meets our previous observations. G542X was detected in 2.9%, the Israeli mutation W1282X in 2.4%. Other relatively frequent mutations were Y1092X, L101X found in 1.8% and 621+1G>T, S466X, 2143delT found in 1.2% of the patients. One gross rearrangement (0.6%) in the CFTR gene (CFTRdele2, c.54-5811_164+2186del273+6780_273+6961inv) was detected by MLPA analysis and confirmed by allele specific PCR and bidirectional sequencing of the junction regions, as described in the literature (7). The mutation is characterized by a deletion of 8108bp (part of intron 1, the whole exon 2 and part of intron 2) with an inverted insertion of 182 bp from intron 3 (11).
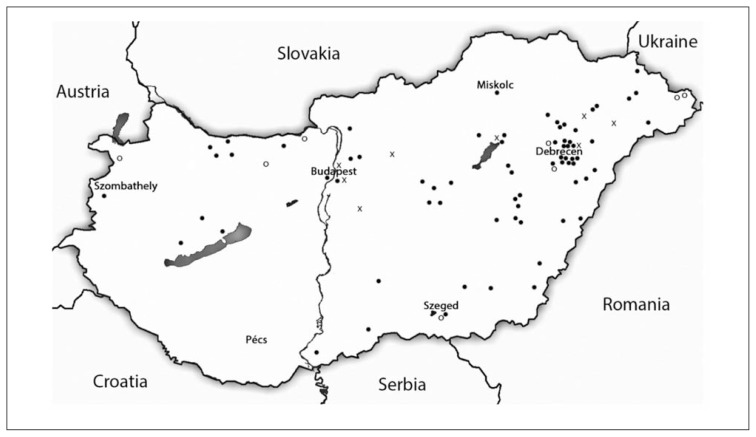
As demonstrated in Figure 1, geographical tendencies can be recognized in the distribution of both CFTRdele2,3(21kb) and 2184insA mutations. All but one CFTRdele2,3(21kb) and 2184insA positive samples originate from the northern regions of the country and 2184insA even seems to be restricted to the northeastern territories of Hungary, which is not suprising if we consider the high frequency of this mutation in Western Ukraine (14).
Figure 1.
Geographical distribution of CFTRdele2,3(21kb) and 2184insA mutations in Hungary.
Legend: o – CFTRdele2,3(21kb), x – 2184insA, ● – other mutations.
According to the mapped mutation frequencies in patients originating from different regions of Hungary using the commercially available assay 75.9% of the mutations can be identified. The allele specific PCR for the detection of the common »Slavic« deletion, CFTRdele2,3(21kb) adds 4.7%, while the sequencing of exon 14 adds 6.5% to the proportion of detected mutations. MLPA analysis revealed one rearrangement (0.6%), while direct sequencing of the entire coding region of CFTR gene identified 20 CF alleles (11.8%).
However, it should be noted that the detection of CFTRdele2,3(21kb) would have been possible by MLPA as well.
In conclusion, beside determining mutation distribution in Hungarian CF patients, our study provides a good starting point to the genetic testing following newborn screening. The commercial allele specific mutation detection method detects 75.9% of CFTR mutations. Complementing it with the detection of other two frequent population-specific alterations (i.e. CFTRdele2,3(21kb) and 2184insA) ~85% of mutations can be detected to achieve the required sensitivity (85–90%) in newborn screening (15 and Milan Macek, personal communication).
With the growing popularity of the clinical use of next generation sequencing (NGS) methods, one possible solution of the combined (i.e. all tiers) DNA analysis step of NBS programs would be the use of those tests.
However, based on our results, it also has to be emphasized that using NGS methods with a high error rate in homopolymer regions (e.g. pyrosequencing or ion semiconductor sequencing) in genetic testing will fail to detect ~5% of CF alleles (2184insA mutation). The frequent »Slavic« mutation (~5%) would be missed as well unless applying a very high coverage not usual in germline mutation detection or using commercially available kits including this mutation, but both of these solutions raise unnecessary financial burdens.
Acknowledgement
The first two authors (GI and KK) contributed equally to the work. This study was supported by the Hungarian Research Fund (K109076, I.B.)
List of abbreviations
- CFTR
cystic fibrosis transmembrane regulator
- MLPA
multiplex ligation-dependent probe amplification
- HGMD
Human Gene Mutation Database
- NGS
next generation sequencing
- NBS
newborn screening.
Footnotes
Conflict of interest statement
The authors stated that they have no conflicts of interest regarding the publication of this article.
References
- 1.Cystic Fibrosis Centre, Hospital for Sick Children, Toronto, Ontario, Canada. Cystic Fibrosis Mutation Database. http://www.genet.sickkids.on.ca/cftr. accessed April 2014.
- 2.Ivady G, Madar L, Nagy B, Gonczi F, Ajzner E, Dzsudzsak E, et al. Distribution of CFTR mutations in Eastern Hungarians: relevance to genetic testing and to the introduction of newborn screening for cystic fibrosis. J Cyst Fibros. 2011;10:217–20. doi: 10.1016/j.jcf.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Dörk T, Macek M, Jr, Mekus F, Tümmler B, Tzountzouris J, Casals T, et al. Characterization of a novel 21-kb deletion, CFTRdele2,3(21 kb), in the CFTR gene: a cystic fibrosis mutation of Slavic origin common in Central and East Europe. Hum Genet. 2000;106:259–68. doi: 10.1007/s004390000246. [DOI] [PubMed] [Google Scholar]
- 4.Dequeker E, Stuhrmann M, Morris MA, Casals T, Castellani C, Claustres M, et al. Best practice guidelines for molecular genetic diagnosis of cystic fibrosis and CFTR-related disorders – updated European recommendations. Eur J Hum Genet. 2009;17:51–65. doi: 10.1038/ejhg.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Křenková P, Piskáčková T, Holubová A, Balaščaková M, Krulišová V, Čamajová J, et al. Distribution of CFTR mutations in the Czech population: positive impact of integrated clinical and laboratory expertise, detection of novel/de novo alleles and relevance for related/derived populations. J Cyst Fibros. 2013;12:532–7. doi: 10.1016/j.jcf.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Ziętkiewicz E, Rutkiewicz E, Pogorzelski A, Klimek B, Voelkel K, Witt M. CFTR mutations spectrum and the efficiency of molecular diagnostics in Polish cystic fibrosis patients. PLoS One. 2014;9:e89094. doi: 10.1371/journal.pone.0089094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomaiuolo R, Sangiuolo F, Bombieri C, Bonizzato A, Cardillo G, Raia V, et al. Epidemiology and a novel procedure for large scale analysis of CFTR rearrangements in classic and atypical CF patients: a multicentric Italian study. J Cyst Fibros. 2008;7:347–51. doi: 10.1016/j.jcf.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Gužvić M. The history of DNA sequencing. J Med Biochem. 2013;32:301–12. [Google Scholar]
- 9.Nestorov J, Matić G, Elaković I, Tanić N. Gene expression studies: How to obtain accurate and reliable data by quantitative real-time RT PCR. J Med Biochem. 2013;32:325–38. [Google Scholar]
- 10.Férec C, Casals T, Chuzhanova N, Macek M, Jr, Bienvenu T, Holubova A, et al. Gross genomic rearrangements involving deletions in the CFTR gene: characterization of six new events from a large cohort of hitherto unidentified cystic fibrosis chromosomes and meta-analysis of the underlying mechanisms. Eur J Hum Genet. 2006;14:567–76. doi: 10.1038/sj.ejhg.5201590. [DOI] [PubMed] [Google Scholar]
- 11.Faà V, Bettoli PP, Demurtas M, Zanda M, Ferri V, Cao A, et al. A new insertion/deletion of the cystic fibrosis trans-membrane conductance regulator gene accounts for 3.4% of cystic fibrosis mutations in Sardinia: implications for population screening. J Mol Diagn. 2006;8:499–503. doi: 10.2353/jmoldx.2006.050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kammesheidt A, Kharrazi M, Graham S, Young S, Pearl M, Dunlop C, et al. Comprehensive genetic analysis of the cystic fibrosis transmembrane conductance regulator from dried blood specimens – Implications for newborn screening. Genet Med. 2006;8:557–62. doi: 10.1097/01.gim.0000237793.19868.97. [DOI] [PubMed] [Google Scholar]
- 13.Estivill X, Bancells C, Ramos C. Geographic distribution and regional origin of 272 cystic fibrosis mutations in European populations. The Biomed CF Mutation Analysis Consortium. Hum Mutat. 1997;10:135–54. doi: 10.1002/(SICI)1098-1004(1997)10:2<135::AID-HUMU6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 14.Makukh H, Krenková P, Tyrkus M, Bober L, Hancárová M, Hnateyko O, et al. A high frequency of the Cystic Fibrosis 2184insA mutation in Western Ukraine: genotype-phenotype correlations, relevance for newborn screening and genetic testing. J Cyst Fibro. 2010;9:371–5. doi: 10.1016/j.jcf.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Castellani C, Southern KW, Brownlee K, Dankert Roelse J, Duff A, Farrell M, et al. European best practice guidelines for cystic fibrosis neonatal screening. J Cyst Fibros. 2009;8:153–73. doi: 10.1016/j.jcf.2009.01.004. [DOI] [PubMed] [Google Scholar]