Summary
Background
The objective of this study is to detect HMGA2 expression in renal carcinoma to explore its relationship with clinicopathology and its significance in prognosis.
Methods
Expressions of HMGA2 mRNA and protein were detected in 50 renal carcinoma specimens, 50 corresponding adjacent normal kidney tissue samples and 40 renal benign tumour specimens via reverse transcription polymerase chain reaction and immunohistochemical assay. Expression analysis was performed along with clinical data analysis.
Results
The relative expression levels of HMGA2 mRNA in renal carcinoma, renal benign tumour tissues and adjacent normal renal tissues were 0.84±0.23, 0.19±0.06 and 0.08±0.04, respectively. HMGA2 protein positive rates were 68.0%, 7.5% and 2.0%, with a significant difference (P<0.05). HMGA2 expression was not significantly correlated with gender, age, tumour size and histological type (P>0.05), but was significantly correlated with TNM stages and lymph node metastasis (P<0.05).
Conclusions
The expressions of HMGA2 gene and protein in renal carcinoma were closely correlated with tumour formation, progression and metastasis. HMGA2 may become a powerful new pathological marker and prognostic factor for renal carcinoma.
Keywords: renal carcinoma, high mobility group A2, immunohistochemistry, RT-PCR
Kratak sadržaj
Uvod
Cilj studije je da se otkrije ekspresija HMGA2 u karcinomu bubrega kako bi se istražio njen odnos sa kliničkom patologijom kao i značaj u prognozi.
Metode
Ekspresije mDNK i proteina HMGA2 određene su u 50 uzoraka karcinoma bubrega, 50 odgovarajućih uzoraka okolnog normalnog tkiva bubrega i 40 uzoraka benignih tumora bubrega putem reverzne transkripcije – lančane reakcije polimeraze i imunohistohemijskim testovima. Analiza ekspresije obavljena je zajedno sa analizom kliničkih podataka.
Rezultati
Relativni nivoi ekspresije mDNK HMGA2 u karcinomu bubrega, tkivima benignih bubrežnih tumora i tkivima okolnog normalnog bubrežnog tkiva bili su redom: 0,84±0,23, 0,19±0,06 i 0,08±0,04. Pozitivne stope proteina HMGA2 bile su 68,0%, 7,5% i 2,0%, uz postojanje značajne razlike (P<0,05). Ekspresija HMGA2 nije značajno korelisala sa polom, starošću, veličinom tumora i histološkim tipom (P>0,05), ali je bila u značajnoj korelaciji sa stupnjevima TNM i metastazama limfnih čvorova (P<0,05).
Zaključak
Ekspresije gena i proteina HMGA2 u karcinomu bubrega bile su u bliskoj korelaciji sa formiranjem, progresijom i metastazama tumora. HMGA2 bi mogao da postane moćan novi patološki marker i prognostički faktor za karcinom bubrega.
Introduction
Renal carcinoma (RC) is a common malignancy of the urinary system. Survey results show that the incidence of RC has increased significantly in the past decade. RC is the second most frequent among the malignancies registered in the Department of Urology of our hospital (1). RC treatment has progressed significantly but the prognosis of RC patients remains pessimistic, with approximately 45% of RC patients exhibiting metastasis during initial diagnosis, and 50% exhibiting recurrence (2). The prognosis of RC patients under different pathological staging and grading systems differs significantly mainly because the biological behaviour of RC is extremely variable. Traditional histopathological staging and grading systems have certain limitations in predicting the biological behaviour of RC, but molecular biological changes that clearly indicate tumour formation and progression can provide a reasonable basis in choosing a clinical treatment. Molecular markers that can be used to evaluate RC prognosis and patient response to new anti-tumour treatments remain lacking. RC biomarker research has recently become popular, and numerous studies have focused on this area. Franciso et al. (3) summarised the results of a determination study that involved serum markers in 117 RC cases and reported that only 11% of AFP, 10% of 8-HCG, 19% of C-PTH, 2% of LDH and 1% of CEA increased, and thus, these compounds were not used as RC biomarkers. The high mobility group A2 (HMGA2) has recently become a focus in oncogene research. HMGA2 protein is a non-histone chromosomal protein that lacks a transcriptional activity, but that can change its structure by binding with chromatin (4). Its role in cancer occurrence and development may be related with the mediation of tumour-related gene transcription and the destruction of the DNA repair system (5). The present study detected HMGA2 expression in RC tissues and aimed to explore the relationship of HMGA2 with clinicopathology and its significance in prognosis.
Materials and Methods
Subjects
A total of 50 RC tissue samples were obtained from surgical treatments in the Department of Urology, Affiliated Zhongshan Hospital of Dalian University from January 2007 to December 2012. Samples from patients aged 40 years old to 75 years old, with a median age of 58 years, were included. Among the patients, 31 were males and 19 were females. RC staging followed the TNM Classification of Malignant Tumours (TNM), which was established by the Union for International Cancer Control, and was divided into 13 cases of stage I, 15 cases of stage II, 14 cases of stage III and 8 cases of stage IV. The pathological types of RC included 36 cases of clear cell carcinoma, 7 cases of chromophilic cell carcinoma, 4 cases of chromophobic cell carcinoma and 3 cases of collecting duct carcinoma. Among the cases, 24 did not exhibit lymph node metastases, whereas 26 exhibited lymph node metastases. For tumour size, 20 cases were <7 cm, whereas 30 cases were ≥7 cm. All patients had been diagnosed histopathologically and had no history of other cancers. The specimens in the control group included 1.50 samples of the corresponding adjacent normal renal tissues. From the same group of patients, renal tissues >2 cm from the tumour margin were obtained surgically and confirmed as normal renal tissues via histological examination. Benign renal tumour specimens (2.40 cases) were included. In addition, 28 cases of renal angiomyolipoma, 6 cases of renal adenoma, 4 cases of renal oncocytoma and 2 cases of renal adenoma were also included. Nephron-reserving tumour resection was performed. This study was conducted in accordance with the Declaration of Helsinki. This study was conducted with approval from the Ethics Committee of Dalian University. Written informed consent was obtained from all participants.
Specimens
All specimens were obtained at appropriate sizes within 30 min after surgical resection, and each specimen was divided into two parts under a sterile environment before being preserved. The entire process was strictly controlled to prevent contamination of RNA enzymes, and the specimens were stored in liquid nitrogen for reverse transcription polymerase chain reaction (RT-PCR) analysis. The specimens were also prepared as paraffin sections for immunohistochemical analysis.
RT-PCR
The tissue samples were soaked in liquid nitrogen for 10 min in an Eppendorf tube and subsequently stored at −80 °C in a refrigerator. RT-PCR was performed by using a quantitative PCR instrument (Bio-Rad Laboratories, Inc., California, USA). Total RNA was extracted via the Trizol one-step method (Invitrogen™, California, USA) and a spectrophotometer was used to detect the concentration of RNA samples. RNA sample (5 μL) was used to perform integrity identification by 1.0% agarose gel electrophoresis (which contained 0.5 μg/mL ethidium bromide). Then, RNA (3 μg) was reverse transcribed into cDNA, and the specific steps were performed according to the instructions of the reverse transcription kit and the 2×Taq PCR Master Mix (Tiangen Biotech (Beijing) Co., Ltd., Beijing, China). The sequences of HMGA2 specific primers were designed for PCR analysis as previously described (6). The upstream primer was 5′-CGAAAGGTGCTGGGCAGCTCCGG-3′, the downstream primer was 5′-CCATTTCCTAGGTCTGCCTCTTG-3′ and the amplified fragment length was 323 bp. For the primer sequences of the human internal reference β-actin, the upstream primer was 5′-GAAGGTGAAGGTCGGAGTC-3′, the downstream primer was 5′-GAAGATGGTGATGGGATTTC-3′ and the amplified fragment length was 228 bp (the primers were synthesised by Beijing Aoke Biotechnology Limited). The PCR reaction conditions of cDNA amplification were as follows: predenatured at 95 °C for 3 min, denatured at 93 °C for 30 s, annealed at 58 °C for 30 s and extended at 72 °C for 30 s, for a total of 30 cycles. Extension was conducted at 72 °C for 10 min, and the samples were stored at 4 °C. The reaction system was 25 μL, and the PCR products were confirmed by 1.5% agarose gel electrophoresis, which contained 0.5 μg/mL ethidium bromide. The results were displayed by an Alpha gel imaging analysis system.
Immunohistochemistry
The SP method was used for immunohistochemical detection. The immunohistochemistry kit was purchased from Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd. (Beijing, China). After fixation by formalin, the sample tissue was embedded in paraffin and cut into 4 μm-thick slices by using a Leica RM2235 rotary microtome (Leica Microsystems, Wetzlar, Germany). The paraffin slices were dewaxed until water status, and then, high-temperature antigen retrieval was performed with EDTA. The slices were washed once with phosphate buffered saline (PBS) for 1 min, incubated with 0.882 mol/L H2O2 for 10 min to eliminate the activities of the internal endogenous peroxidase and washed thrice with PBS for 1 min each time. Normal blocking serum was added dropwise to the samples, which were subsequently incubated for 15 min at room temperature. The upper fluid was decanted, and mouse anti-human HMGA2 monoclonal primary antibody (Santa Cruz Biotechnology Inc., Texas, USA) was added dropwise at 1:50. The samples were incubated overnight at 4 °C and washed thrice with PBS for 1 min each time. Then, goat anti-mouse secondary antibody (Santa Cruz Biotechnology Inc., Texas, USA) was added dropwise before the samples were incubated for 15 min. The samples were washed thrice with PBS for 1 min each time, and then, horseradish enzyme-labeled streptavidin working solution was added dropwise before the samples were incubated for 15 min. The samples were washed thrice with PBS for 1 min each time, stained with diaminobenzidine and restained with hematoxylin, dehydrated, hyalinised with xylene and mounted with neutral gum. PBS was used instead of the primary antibody in the negative control group. Positive HMGA2 expression in the immunohistochemical results was exhibited as brown and tan granular formations, mainly in the nucleus. A total of 300 tumour cells were counted within the 400× magnified view field. If the stained cells were less than 300, then all cells were counted (not less than 150 tumour cells). Tumour nuclear staining was used to count positive cells, and cells exhibiting higher than 10% expression were considered positive for immunohistochemical staining. Cells exhibiting no expression and positive cells with less than 10% expression were considered negative for immunohistochemical staining.
Statistical analysis
The SPSS 13.0.1 software (SPSS Inc., Chicago, USA) was used for the statistical analysis of the aforementioned data. All data were expressed as χ̄±S. Data counting was performed via the X2 test and data measurement was performed via t-test or ANOVA. Analysis of related factors of the clinicopathological features of RC patients was performed by Cox multivariate regression analysis, which incorporated the variables of age, gender, tumour size, histological type, TNM staging, lymph node metastasis and HMGA2 protein expression. P<0.05 was considered as statistically significant.
Results
RT-PCR
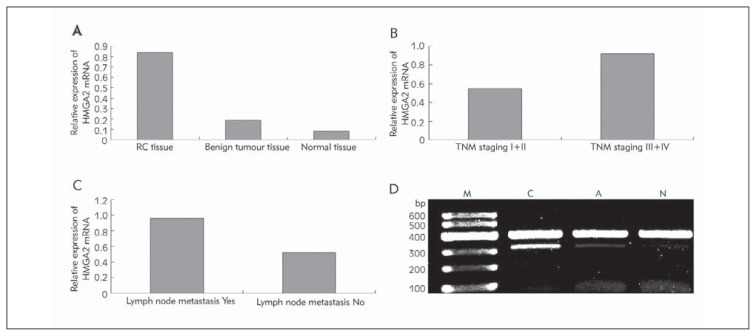
The relative expression levels of HMGA2 mRNA in RC, benign renal tumour tissues and adjacent normal renal tissues were 0.84±0.23, 0.19±0.06 and 0.08±0.04, respectively. The expression in RC was significantly higher than those in the other two groups (F=2.36, P<0.05, Figure 1A). The expression of HMGA2 mRNA was closely related with TNM staging, and the relative expressions in stages I + II and III + IV were 0.55±0.18 and 0.92±0.21 (t=8.39, P<0.05), respectively. In addition, the expression of HMGA2 mRNA was observed in the presence or absence of lymph node metastasis. The relative expression of HMGA2 mRNA in RC patients with lymph node metastasis was 0.96±0.24, whereas that in RC patients without lymph node metastasis was 0.52±0.16. The comparison between the aforementioned expressions exhibited a significant difference (t=9.87, P<0.05, Figures 1B, 1C and 1D).
Figure 1.
Expression of HMGA2 mRNA in kidney tissue by RT-PCR. A: Relative expression of HMGA2 mRNA; B: Relative expression of HMGA2 mRNA in TNM staging; C: Relative expression of HMGA2 mRNA in lymph node metastasis; D: Electrophoresis of HMGA2 mRNA RT-PCR products.
Immunohistochemical results
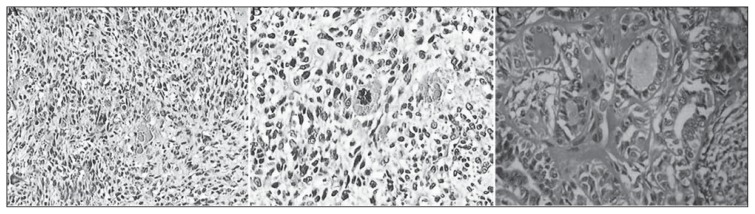
The expression of HMGA2 protein was exhibited by brown and tan granular formations, which were mainly distributed in the nucleus, and by a certain weak staining in the cytoplasm of some tumours (Figures 2A, 2B and 2C). Among the 50 cases of normal renal tissues, only 1 case exhibited a positive expression of HMGA2 protein (2.0% positive rate). Among the 40 cases of benign tumour tissues, 3 cases presented a positive expression (7.5% positive rate). Among the 50 cases of RC, 34 cases exhibited a positive expression (68.0% positive rate), which was significantly higher than those of the other two groups (X2=13.39, P<0.05).
Figure 2.
Expression of HMGA2 protein in kidney tissue by immunohistochemistry. A: No expression of HMGA2 protein in the normal renal tissues (×400); B: Low expression of HMGA2 protein in the benign renal tumour tissues (×400); C: High expression of HMGA2 protein in RC tissues (× 400).
Relationship of HMGA2 and clinicopathological features
Cox multivariate regression analysis showed that age, sex, tumour size and histological type did not exhibit statistically significant differences in HMGA2 protein expression (P>0.05), whereas significant differences were observed in different TNM staging statuses and in lymph node metastasis (P<0.05). Expression in stage I + II was 50.0%, whereas 90.9% expression was observed in stage III + IV, and intergroup comparison exhibited a significant difference (P=0.013). Thus, HMGA2 protein expression was shown to be related with TNM staging, and an advanced RC stage showed a high expression rate. The expression rate of HMGA2 protein without lymph node metastasis was 41.6%, whereas 92.3% expression was observed with lymph node metastasis, with a significant difference (P=0.007, Table I).
Table I.
Correlation analysis of HMGA2 protein expression and general clinical information of RC patients (immunohistochemistry).
| Item | Cases | HMGA2 expression | Rate | X2 | P | |
|---|---|---|---|---|---|---|
|
| ||||||
| Gender | Male | 31 | 22 | 70.9% | 2.322 | 0.313 |
| Female | 19 | 12 | 74.2% | |||
|
| ||||||
| Age | <60 | 21 | 15 | 71.4% | 1.326 | 0.25 |
| ≥60 | 29 | 19 | 65.5% | |||
|
| ||||||
| Tumor size | ≥7cm | 30 | 19 | 1.326 | 0.25 | |
| <7cm | 20 | 15 | 71.4% | |||
| Clear renal cell | 36 | 24 | 65.5% | |||
|
| ||||||
| Pathological type | Chromophilie cell | 7 | 5 | 71.4% | 4.094 | 0.44 |
| Chromophobe cell | 4 | 3 | 75.0% | |||
| Collecting duct | 3 | 2 | 66.7% | |||
|
| ||||||
| TNM staging | Stage I+II | 28 | 14 | 50.0% | 8.717 | 0.013 |
| Stage III+IV | 22 | 20 | 90.9% | |||
|
| ||||||
| Lymph node metastasis | Yes | 26 | 24 | 92.3% | 9.275 | 0.007 |
| No | 24 | 10 | 41.6% | |||
Discussion
HMGA2 is a non-histone protein with 109 amino acid residues. This protein combines with the chromosome. HMGA2 is generally located within the nucleus. It contains three AT-hook structural domains and an acid C-terminal. HMGA2 is not involved in transcription, but it can bind to DNA through the AT-hook and change the structure of chromatin, thereby regulating the transcription of target genes; thus, HMGA2 is a constructional transcription factor (7, 8). HMGA2 protein is generally involved in a wide range of biological processes, including embryonic development, cell cycle regulation, cell differentiation and tumourigenesis. HMGA2 protein is highly expressed during embryonic development, whereas its expression is difficult to detect in most differentiated adult tissues (9). Recently, scholars have detected a high expression of HMGA2 in different types of adult epithelial tumours, such as lung cancer (10), pancreatic cancer (11), oral squamous cell carcinoma (12) and thyroid cancer (13). Studies have shown that the incidence and development of these tumours were associated with the abnormal expression of the HMGA2 gene, and its increased expression was related to prognosis (14).
As a proto-oncogene, the overexpression of the HMGA2 gene in cells promotes tumour growth and metastasis. The roles of this gene in cancer occurrence and development are currently being considered to be related to its mediation of the transcription of tumour-related genes and the destruction of the DNA repair system (4). HMGA2 induces and increases the activity of the transcription factor E2FI as well as promotes the abnormal proliferation of cells, thus promoting the transformation of the G2/M phase of the cell cycle and inducing tumour growth (14). Moreover, HMGA2 induces the expression of cyclin A, inhibits p120E4F through the cAMP response element and increases cyclin A expression, thus interfering with the cell cycle and promoting tumourigenesis (16). HMGA2 overexpression damages the DNA repair system (17). In addition, HMGA2 promotes epithelial–mesenchymal transition (EMT), which is a common physiological and pathological phenomenon that is closely related with embryonic development, wound healing, tumour invasion and metastasis. EMT mainly occurs when epithelial cells lose their polarity and when tight and adhesive junctions among cells are reduced; thus, cells acquire invasion and migration abilities and finally evolve into cells with stromal morphology and characteristics. EMT is involved in most cases of cancer invasion and metastasis and thus has become a focus of studies on tumour invasion and metastasis (16). Recent studies have found that HMGA2 may be involved in EMT (18, 19). HMGA2 is expressed in stem cells, as demonstrated by its important functions in embryonic development. The roles of HMGA2 in stem cells, particularly its function in cancer stem cells, have been studied. Nishino (20) found that HMGA2 is highly expressed in embryonic neural stem cells, but the expression gradually decreases with increasing age. As of this writing, the molecular mechanisms of HMGA2 differentiation potential and self-renewal, as well as its maintenance in cells, remain unclear; these processes may be related to the ability of HMGA2 to regulate transcription by altering the structure of chromatin (21, 22).
In the present study, we found that HMGA2 expression in RC was significantly higher than in benign and normal renal tissues, with a significant difference (P<0.01). This finding indicates that HMGA2 expression is actually related with the benign or malignant behaviour of renal tissues. HMGA2 expression was 66.7% in clear renal cell carcinoma, 71.4% in chromophilic cell carcinoma, 75.0% in chromophobic cell carcinoma and 66.7% in the collecting duct carcinoma, with no statistically significant difference among these four tumours. In addition, HMGA2 expression rates among patients with stages I + II and III + IV RC were 50.0% and 90.9%, respectively, and the difference was significant (P=0.013). An advanced RC stage led to a high expression rate. The expression rate in RC with lymph node metastasis (92.3%) was higher than that in RC without lymph node metastasis (41.6%), thus suggesting that HMGA2 has an important role in the formation, progression and lymph node metastasis of RC. The mechanism might be related with EMT. In the TGF-β signal transduction pathway, HMGA2 increases the expressions of Snail, Slug and Twist proteins and reduces that of Id2 protein; changes in the expression of these proteins result in EMT, thereby promoting tumour invasion and metastasis (8).
In summary, the expressions of HMGA2 gene and protein significantly increased in RC, and were closely related with clinical staging and lymph node metastasis, thus indicating that HMGA2 has an important role in the formation and progression of RC. Based on these findings, HMGA2 detection will be significant in the diagnosis, differential diagnosis and prognosis of RC. HMGA2 may become a powerful new renal pathological marker that can provide a specific target for RC treatment. Further studies on HMGA2 will be significant in RC gene therapy.
Acknowledgements
This study was supported by Dalian Municipal Science and Technology Bureau.
Footnotes
Conflict of interest statement
The authors stated that have no conflicts of interest regarding the publication of this article.
References
- 1.Mankoff DA, Thompson JA, Gold P, Eary JF, Guinee DG, Jr, Samlowski WE. Identification of IL-2 induced complete response in metastatic RCC by FDG PET despite radiographic evidence suggesting persistent tumor. AJR. 1997;169:1049–50. doi: 10.2214/ajr.169.4.9308463. [DOI] [PubMed] [Google Scholar]
- 2.Cohen HT, Mccocern FJ. Renal–cell carcinoma. N Engl J Med. 2005;353:2477–90. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 3.Franciso HD, Kim HL, Seligson D, Liu X. Using protein expressions to predict survival in clear cell renal carcinoma. Clin Cancer Res. 2004;10:5464–71. doi: 10.1158/1078-0432.CCR-04-0488. [DOI] [PubMed] [Google Scholar]
- 4.Medeiros F, Araujo AR, Erickson-Johnson MR, Kashyap PC, Dal Cin P, Nucci M, et al. HMGA1 and HMGA2 rearrangements in mass-forming endometriosis. Genes Chromosomes Cancer. 2010;49:630–4. doi: 10.1002/gcc.20772. [DOI] [PubMed] [Google Scholar]
- 5.Lia Y, Boo LM, Wang SY, Lin HH, Wang CC, Yen Y, et al. Suppression of nonhomologous end joining repair by overexpression of HMGA2. Cancer Res. 2009;69:5699–706. doi: 10.1158/0008-5472.CAN-08-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Liu X, Li AY, Chen L, Lai L, Lin HH, et al. Overexpression of HMGA2 promotes metastasis and impacts survival of colorectal cancers. Clin Cancer Res. 2011;17:2570–80. doi: 10.1158/1078-0432.CCR-10-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves R, Beckerbauer L. HMGL/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 8.Nestorov J, Matić G, Elaković I, Tanić N. Gene expression studies: How to obtain accurate and reliable data by quantitative real-time RT PCR. J Med Biochem. 2013;32:325–38. [Google Scholar]
- 9.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and HMGA2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beige G, Meyer A, Klemke M, Burchardt K, Stern C, Wosniok W, et al. Upregulation of HMGA2 in thyroid carcinomas: a novel molecular marker to distinguish between benign and malignant follicular neoplasias. Genes Chromosomes Cancer. 2008;47:56–63. doi: 10.1002/gcc.20505. [DOI] [PubMed] [Google Scholar]
- 11.Huang ML, Chen CC, Chang LC. Gene expressions of HMGl-C and HMGI(Y) are associated with stage and metastasis in colorectal cancer. Int Colorectal Dis. 2009;24:1281–6. doi: 10.1007/s00384-009-0770-7. [DOI] [PubMed] [Google Scholar]
- 12.Meyer B, Loeschke S, Schultze A, Weigel T, Sandkamp M, Goldmann T, et al. HMGA2 overexpression in non-small cell lung cancer. Mol Carcinog. 2007;46:503–11. doi: 10.1002/mc.20235. [DOI] [PubMed] [Google Scholar]
- 13.Miyazawa J, Mitoro A, Kawashiri S, Chada KK, Imai K. Expression of mesenchyme-specific gene HMGA2 in squamous cell carcinomas of the oral cavity. Cancer Res. 2004;64:2024–9. doi: 10.1158/0008-5472.can-03-1855. [DOI] [PubMed] [Google Scholar]
- 14.Fehr A, Meyer A, Heldorn K, Röser K, Löning T, Bullerdiek J. A link between the expression of the stem cell marker HMGh2, grading, and the fusion CRTCl-MAML2 in mucoepidermoid carcinoma. Genes Chrom Cancer. 2009;48:777– 85. doi: 10.1002/gcc.20682. [DOI] [PubMed] [Google Scholar]
- 15.De Martino I, Visone R, Wierinckx A, Palmieri D, Ferraro A, Cappabianca P, et al. HMGA proteins up-regulate CCNB2 gene in mouse and human pituitary adenomas. Cancer Res. 2009;69:1844–50. doi: 10.1158/0008-5472.CAN-08-4133. [DOI] [PubMed] [Google Scholar]
- 16.Tessari MA, Gostissa M, Altamura S, Sgarra R, Rustighi A, Salvagno C, et al. Transcriptional activation of the eyelin A gene by the architectural transcription factor HMGA2. Mol Cell Biol. 2003;23:9104–16. doi: 10.1128/MCB.23.24.9104-9116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borrmann L, Schwanbeck R, Heyduk T, Seebeck B, Rogalla P, Bullerdiek J, et al. High mobility group A2 protein and its derivatives bind a specific region of the promoter of DNA repair gene ERCCI and modulate its activity. Nucleic Acids Res. 2003;31:6841–51. doi: 10.1093/nar/gkg884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Zhou BH. New insights of epithelial-mesenchymal transition in cancer metastasis. Acts Biochim Biophys Sin(Shanghai) 2008;40:643–50. doi: 10.1111/j.1745-7270.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond SM, Sharpless NE. HMGA2, microRNAs, and stem cell aging. Cell. 2008;135:1013–6. doi: 10.1016/j.cell.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16 and p19 expression. Cell. 2008;135:227–39. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young AR, Narita M. Oncogenic HMGA2: short or small. Genes Dev. 2007;21:1005–9. doi: 10.1101/gad.1554707. [DOI] [PubMed] [Google Scholar]
- 22.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. Let-7 regulates self-renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]