Summary
The 2A proteinase (2Apro) of human rhinoviruses cleaves the virally encoded polyprotein between the N-terminus of VP1 and its own C-terminus. Poor understanding of the 2Apro substrate specificity of this enzyme has hampered progress in developing inhibitors that may serve as anti-viral agents. We show here that the 2Apro of human rhinovirus (HRV) 1A and 2 (rhinoviruses from genetic group A) cannot self-process at the HRV14 (a genetic group B rhinovirus) cleavage site. When the amino acids in the cleavage site of HRV2 2Apro (IleIleThrThrAla*GlyProSerAsp) were singly or doubly replaced with the corresponding HRV14 residues (AspIleLysSerTyr*GlyLeuGlyPro) at positions from P3 to P2’, HRV1A and HRV2 2Apro cleavage took place at wild-type levels. However, when three or more positions of the HRV1A or 2 2Apro were substituted (e.g. at P2, P1 and P2’), cleavage in vitro was essentially eliminated. Introduction of the full HRV14 cleavage site into a full-length clone of the HRV1A and transfection of HeLa cells with a transcribed RNA did not give rise to viable virus. In contrast, revertant viruses bearing Cys at the P1 position or Pro at P2’ were obtained when an RNA bearing the three inhibitory amino acids was transfected. Reversions in the enzyme affecting substrate specificity were not found in any of the in vivo experiments. Modeling of oligopeptide substrates onto the structure of HRV2 2Apro revealed no appreciable differences in residues of HRV2 and HRV14 in the respective substrate binding sites, suggesting that the overall shape of the substrate is important in determining binding efficiency.
Introduction
Human rhinoviruses, members of the genus enterovirus within the family of picornaviruses (King et al., 2012), are the main causative agent of the common cold (Makela et al., 1998). Their single stranded, positive sense RNA genome contains only one open reading frame from which a single polyprotein is expressed. Proteolytic processing, performed by the two virally encoded proteases 2Apro and 3Cpro, is therefore essential to generate mature viral proteins. In an intramolecular manner, 2Apro initiates the cascade of cleavages by self-processing the VP1-2Apro junction (Toyoda et al., 1986). All other proteolytic reactions on the polyprotein are accomplished by 3Cpro (Hanecak et al., 1982; Palmenberg et al., 1979; Skern et al., 2002).
Enteroviral 2Apro have been shown to be cysteine proteases with a truncated chymotrypsin-like fold (Baxter et al., 2006; Gorbalenya et al., 1986; Petersen et al., 1999) and a tightly bound zinc ion. They vary in size between 142 and 147 amino acids for the over 100 known HRV serotypes (Knowles et al., 2012). HRV2 and HRV14 2Apro (members of rhinoviral Group A and Group B respectively) share only about 40% sequence identity (Skern et al., 1985; Stanway et al., 1984). Furthermore, their respective VP1-2Apro cleavage sites differ substantially (Fig. 1a). Thus, HRV2 2Apro cleaves at IleIleThrThrAla*GlyProSerAsp (Sommergruber et al., 1992) whereas HRV14 2Apro processes at AspIleLysSerTyr*GlyLeuGlyPro (Callahan et al., 1985) (the asterisk indicates the scissile bond). Indeed, clear differences in the specificities of these two 2Apro have been documented. Sousa et al. (Sousa et al., 2006) examined their abilities to self-process at cleavage sites with Arg at P1 (the nomenclature is that of Schechter and Berger (Schechter & Berger, 1967)) and found appreciable differences.
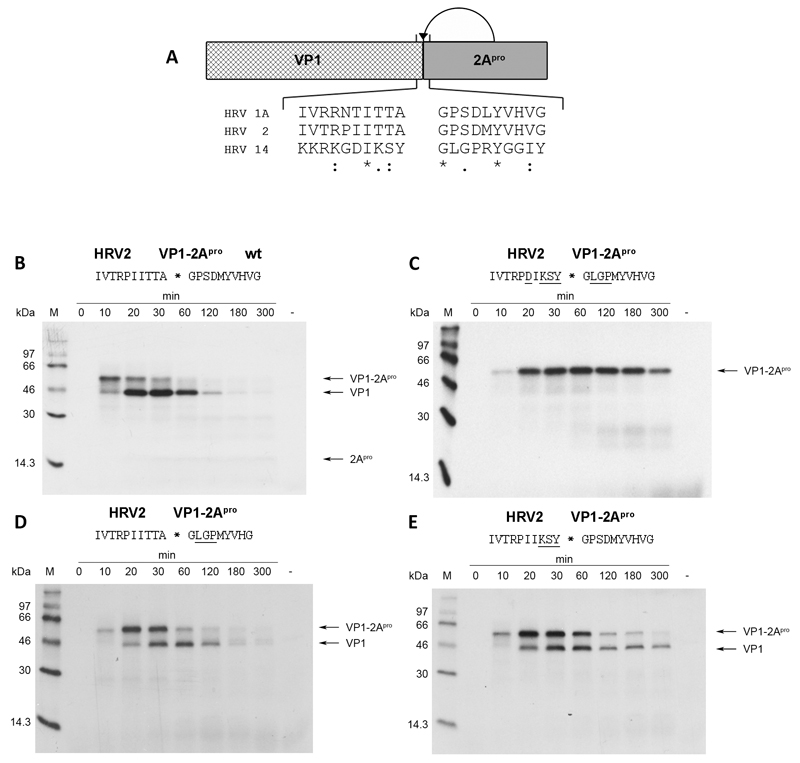
Fig. 1.
HRV2 2Apro fails to self-process on different substrates. (a) Schematic overview of in vitro translated protein precursor VP1-2Apro and self-processing sequences from the indicated HRV serotypes. Identical, very similar and similar residues are indicated by asterisks, colons and dots, respectively. (b) and (c) HRV2 2Apro self-processing at the wild-type HRV2 VP1-2Apro and HRV14 VP1-2Apro cleavage sites, respectively. (d) and (e) HRV2 2Apro self-processing at modified cleavage sites. For all reactions, RRLs were programmed with the indicated in vitro transcribed RNA (10ng/μL) and incubated at 30°C. Translated proteins were labeled with 35S-methionine. 10μl aliquots were taken at the given time-points and the reaction stopped by the addition of 25μl 2x Laemmli sample buffer, 1μl unlabeled methionine/cysteine (20mM) and 14μl H2O. Negative controls were prepared by adding water instead of RNA followed by incubation for 300 minutes. Samples were then analyzed on 17.5 % SDS PAGE (Dasso & Jackson, 1989) and proteins were visualized by fluorography. Underlined residues in the cleavage sites indicate variations from the wild-type HRV2 cleavage site.
In 2005, Crowder and Kirkegaard (2005) pinpointed the potential of 2Apro as a target for antiviral compounds. However, the absence of a high resolution structure of a HRV 2Apro bound to substrate and the considerable heterogeneity between rhinovirus group A and group B sequences have impeded an exact understanding of substrate binding. Moreover, despite extensive efforts using peptide cleavage assays (Sommergruber et al., 1992; Wang et al., 1997) and fusion proteins (Skern et al., 1991), the exact specificity of group A and group B 2Apro self-processing remains unclear, strongly hampering the specific design of compounds that inhibit this reaction.
In order to better understand the mechanism of substrate recognition better, we tested whether HRV2 2Apro would be able to self-process at the VP1-2Apro cleavage site found in HRV14. We found that this was not the case either in vitro or in vivo and set out to identify the amino acids that are responsible for this inhibitory effect. Our results show that the occupancy of both P and P’ sites is crucial for efficient processing by HRV2 and HRV1A 2Apro.
Results
HRV2 2Apro cannot cleave intramolecularly at the cleavage site of HRV14 2Apro
Fig. 1a shows a schematic overview of the in vitro cleavage assay and the residues at the VP1-2Apro junction of the three HRV serotypes examined in this study. The VP1-2Apro cleavage sites of the group A viruses HRV1A and HRV2 exhibit high conservation. In contrast, the cleavage site found in the group B virus HRV14 shows substantial differences to the group A viruses. The differences at positions P1 and P2’ attract special attention as both have been reported to be important for substrate recognition (Skern et al., 1991; Sommergruber et al., 1992).
Self-processing of HRV2 2Apro on its wild-type cleavage site as observed in rabbit reticulocyte lysate (RRL) is shown in Fig. 1b. Unprocessed VP1-2Apro precursor can be detected about 10 min after addition of mRNA to the RRLs. Processed VP1 can also be detected at this time point; the majority of precursor is cleaved after 30 min. Detection of 2Apro is impaired by the low number of methionine residues and the resulting reduced sensitivity. Nevertheless, a faint band can be detected at a molecular weight corresponding to 2Apro at about 30 min.
In order to investigate whether HRV2 2Apro can self-process the VP1-2Apro cleavage site found in HRV14, we used site directed mutagenesis to exchange the residues from P5-P4’ of the HRV2 cleavage site with those of the HRV14 site (Fig. 1c). Differences to the wild-type HRV2 cleavage site are underlined. Initial translation products can again be detected 10 min after start of translation. Nevertheless, no processed products could be observed even after 300 min of translation. This indicates that HRV2 2Apro is unable to process this cleavage site.
Next, we aimed to narrow down the residues preventing cleavage by HRV2. Fig. 1d and Fig. 1e show HRV2 2Apro processing on cleavage sites with corresponding variations in either the P-site or the P’-site. Clearly, both of these sites are cleaved, as indicated by the conversion of the VP1-2Apro precursor, even though both mutants show slightly lowered cleavage kinetics (50% cleavage of precursor occurs at about 30-60 min).
The results imply that a combination of variations on both sides of the cleavable peptide bond would be necessary to prevent HRV2 2Apro from processing at the HRV14 site. Accordingly, the variations from P3 to P1 were maintained and combined with single amino acid substitutions at P2’, P3’ and P4’ (Fig. 2). The most severe effect could be observed when proline at P2’ was exchanged for leucine (Fig. 2a). The VP1-2Apro precursor accumulated over time but no clear pattern of conversion into VP1 and 2Apro was observed during the experiment.
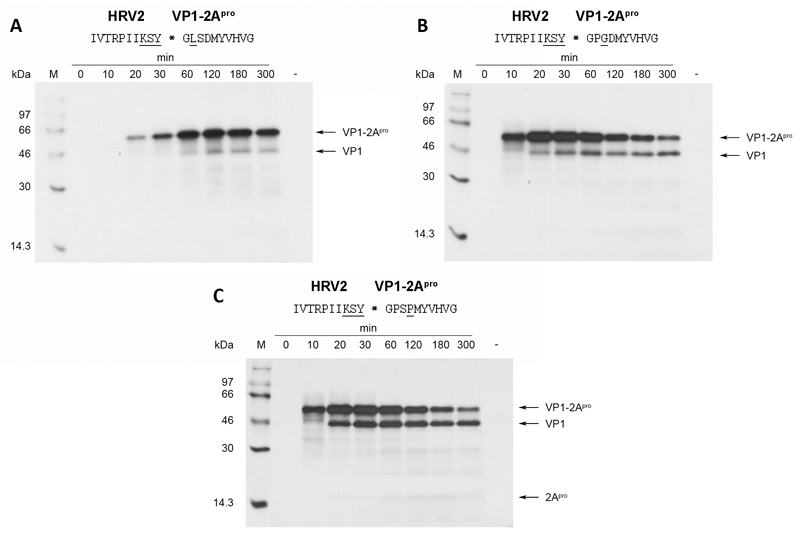
Fig. 2.
HRV2 2Apro self-processing on HRV2/HRV14-hybrid cleavage sites bearing substitutions in the P’ region. Substitutions in the wild-type HRV2 cleavage site are underlined. RRLs were programmed with the respective RNAs and the proteins labeled and analysed as described in figure 1.
Moving the single amino acid variation at the P’-site further away from the cleavable bond lead to increased activity compared to the P2’ Leu mutant (Fig. 2b and Fig. 2c). Nevertheless, a strong reduction of self-processing activity compared to the wild-type cleavage site is still observed. In Fig. 2b and 2c, the first cleavage products can be detected 20 min after start of translation. However, a time-point of 50% turnover of precursor is hard to detect as the pattern shows no clear conversion from precursor to cleavage products but rather an accumulation of cleavage products at a constant level during the course of the experiment.
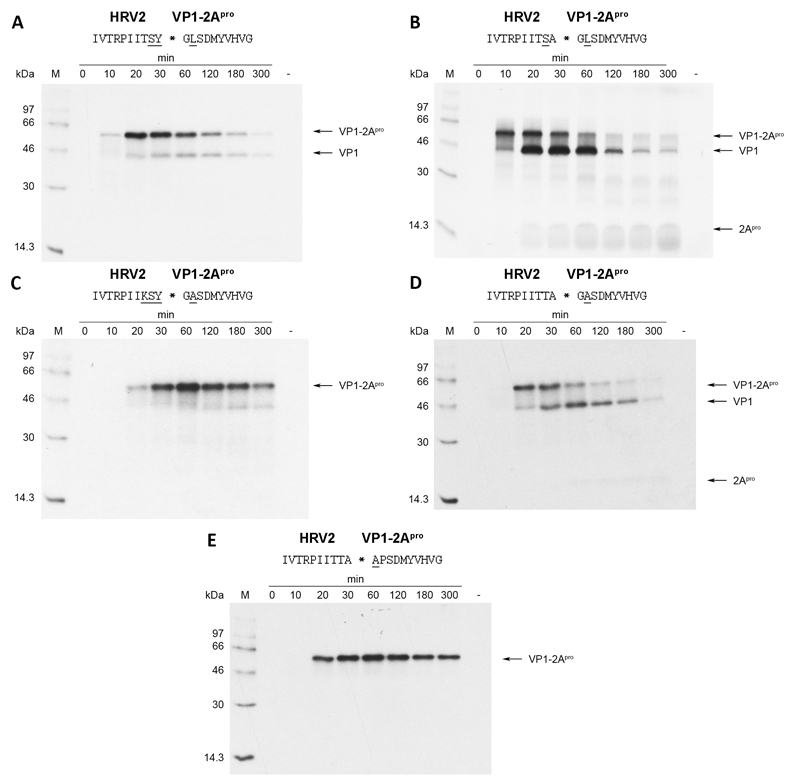
Assuming a more or less extended β-sheet conformation of the substrate in the bound form, the side-chain of the P3 residue is most likely to face the solvent. It therefore should not contribute to substrate specificity. For this reason, we decided to mutate lysine at P3 back to the wild-type threonine leaving just the variations at P2, P1 and P2’ (Fig. 3a). Interestingly, HRV2 2Apro still performed very weakly in processing this site, indicating that these three variations are sufficient to inhibit self-processing to almost 100%. Nevertheless, we aimed to investigate whether it is possible to reduce the number of substituted residues even further and still observe strong inhibition of self-processing. To this end, we mutated tyrosine at P1 back to the wild-type alanine and tested the mutant in our in vitro system (Fig. 3b). The processing kinetics of this mutant are comparable with wild-type levels (see Fig. 1b). Hence, tyrosine at P1 was an essential component of the variation that lead to inhibition. We can thus define the cleavage site with variations in P2, P1 and P2’ as the minimal substitutions required to inhibit the HRV2 2Apro.
Fig. 3.
HRV2 2Apro self-processing on substrates bearing substitutions across the entire cleavage site. Underlined substitutions in the wild-type HRV2 cleavage site are underlined. RRLs were programmed with the respective RNAs and the proteins labeled and analysed as described in figure 1.
Role of the P1’ residue
The above results showed the importance of occupancy at the positions P2, P1 and P2’. Although the P1’ residue glycine is conserved in all enteroviral 2Apro, it has recently been observed by Park et al. (Park et al., 2010) that two HRV2 2Apro cleavage sites on the nuclear pore protein (Nup) 62 show alanine at P1’. In addition, two sites also showed alanine at P2’. In order to investigate how the presence of alanine at P1’ and P2’ influence self-processing, we generated appropriate substitutions for our in vitro system (figures 3c and 3d). At P1’, the presence of a methyl group at P1’ through the introduction of alanine leads to a complete inhibition of self-processing. In contrast, the substitution of proline with alanine at P2’ did not affect cleavage, with processing occurring at essentially wild-type levels. However, the presence of alanine at P2’ in a cleavage site in which the P3-P1 residues had been substituted with those from HRV14 (i.e. P3 Lys, P2 Ser and P1 Tyr) again led to inhibition of HRV2 2Apro self-processing (Fig. 3e). Indeed, inhibition is comparable to that seen with the mutant bearing leucine at P2’ (see Fig. 2a). This again emphasizes the need for mutations at both sides of the cleavable peptide bond for inhibition.
Specificity of 2Apro in vivo
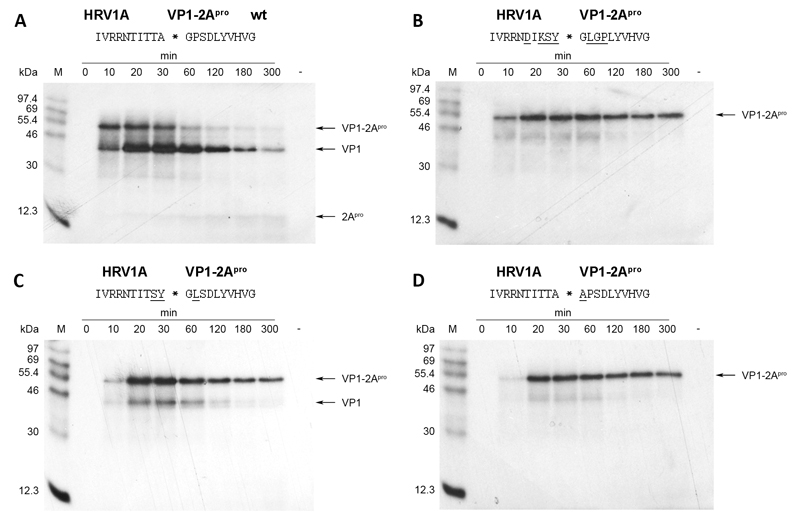
We then wished to investigate the in vivo phenotype of viruses with variations in the VP1-2Apro cleavage site that inhibited self-processing in vitro and, if possible, isolate second site revertants that had regained activity. The HRV2 full length RNA either isolated from virions or transcribed from the cDNA clone described by Duechler et al. (Duechler et al., 1989a) has a low transfection efficiency. Thus, transfection with DEAE-dextran of HeLa cells (Ohio) with in vitro transcribed HRV2 RNA yields only about 50 plaques/μg (Duechler et al., 1989b). However, the transfection efficiency of the RNA of the closely related genetic A group HRV1A has been reported to be about 2×105 plaques/μg RNA (McKnight, 2003), a figure that is in accordance with our observations from transfection efficiency studies (data not shown). In order to increase the likelihood of obtaining second-site revertants, we therefore decided to introduce the corresponding mutations into a full length cDNA clone of HRV1A (McKnight, 2003). Before starting this approach, we nevertheless first wished to confirm whether the self-processing kinetics of HRV2 and HRV1A 2Apro are comparable. Fig. 4 shows that this is indeed the case. Self-processing of HRV1A 2Apro on its wild-type cleavage site reaches 50% after 20-30 min as does the HRV2 enzyme (compare Fig. 4a with Fig. 1b). The HRV1A enzyme also fails to cleave when the HRV14 sequence is introduced (Fig. 4b); furthermore, the residues determining this property lie at P2, P1 and P2’ (Fig. 4c). Finally, the presence of alanine at P1’ leads to a complete inhibition of cleavage (Fig. 4d).
Fig. 4.
HRV1A 2Apro self-processing on substrates bearing mutations across the entire cleavage site. Substitutions in the wild-type HRV1A cleavage site are underlined. RRLs were programmed with the respective RNAs and the proteins labeled and analysed as described in figure 1.
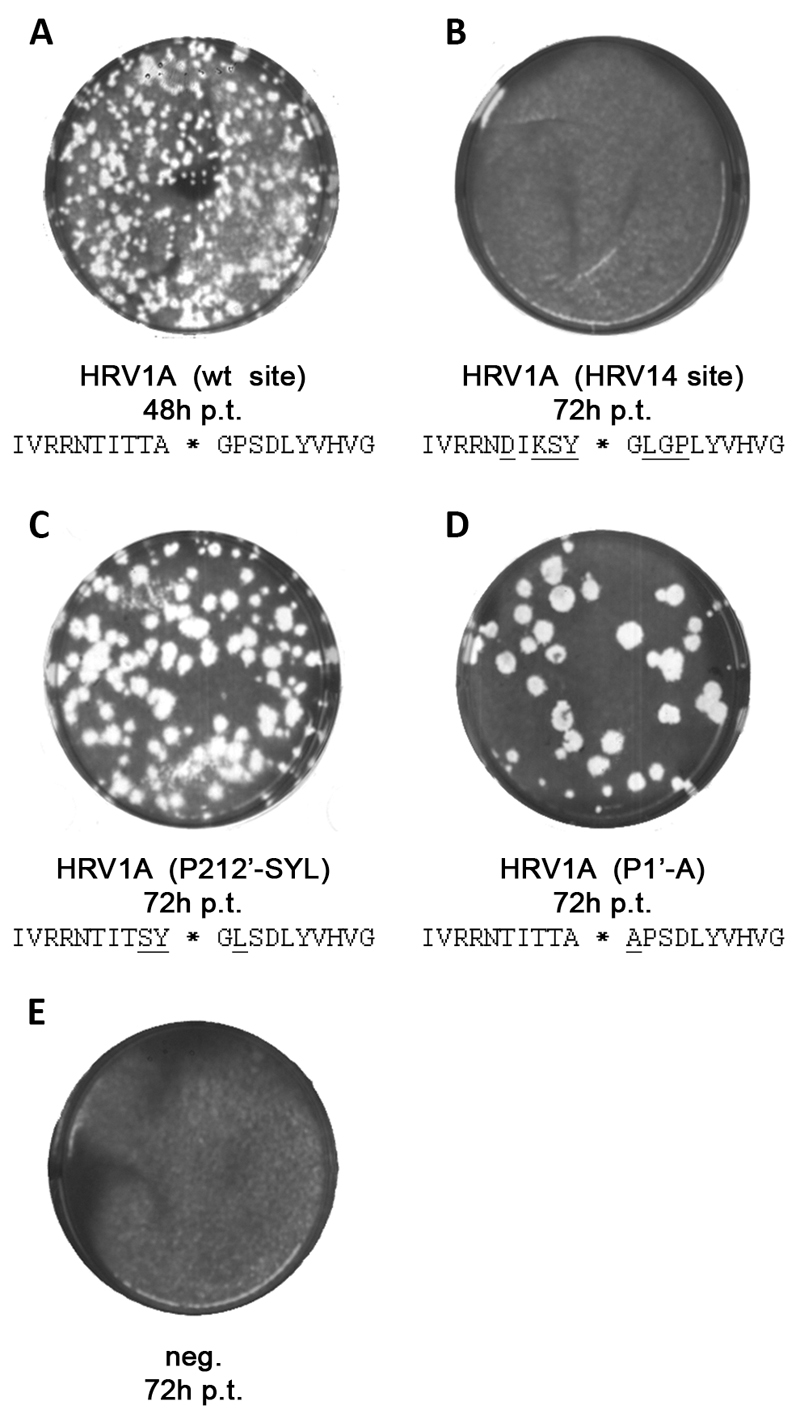
Given the very similar self-processing kinetics of HRV2 and HRV1A, we therefore introduced selected mutations into the HRV1A full length cDNA clone and generated three plasmids subsequently referred to as HRV1A-(HRV14 site: i.e. the sequence shown in Fig. 4b), HRV1A-(P212’-SYL: i.e. the sequence shown in Fig. 4c) and HRV1A-(P1’-A: i.e. the sequence shown in Fig. 4d). Then, full-length RNA was in vitro transcribed with T7 RNA polymerase and used to transfect a monolayer of HeLa cells with DEAE dextran. Cells were washed and covered with 1% low melting agarose in infection medium and incubated at 34°C for 48 or 72 hours as indicated. In order to visualize viral plaques, the agarose was carefully removed and cells were stained with crystal violet (Fig. 5). Upon transfection with wild-type HRV1A RNA, viral plaques could be detected 48 hours post-transfection (Fig. 5a); all cells were lysed 72 hours post-transfection (data not shown). In contrast, we were not able to retrieve virus after transfecting HeLa cells with infectious HRV1A-(HRV14 site) RNA (Fig. 5b). Even 72 hours after transfection, no viral plaques could be observed, indicating that the virus was not viable as a consequence of the multiple changes in this cleavage site. However, when we investigated the HRV1A-(P212’-SYL) mutant, very small plaques could be detected 48 hours post-transfection which became easily visible after 72 hours (Fig. 5c). A similar situation was observed with mutant HRV1A-(P1’-A) (Fig. 5d) 72 hours post-transfection. In short, these two mutants yielded plaques that were too small to detect at 48 hours post-transfection and lower in number compared to those observed with wild-type HRV1A.
Fig. 5.
Investigation of reversion in three HRV1A mutants bearing VP1/2Apro cleavage site mutations. A 50% confluent monolayer of HeLa cells was transfected using DEAE dextran at 34°C with 1.2 μg of in vitro transcribed HRV1A wt or the indicated mutant RNA. H2O was used instead of RNA for negative controls. 25 minutes post transfection, cells were washed with PBS and covered with 1% low melting agarose in infection medium. Plaques were visualized with crystal violet 48h or 72h post transfection to visualize viral plaques.
To determine whether the viruses obtained after transfection with the mutant HRV1A-(P212’SYL) and HRV1A-(P2’A) RNAs had maintained the original mutations, had reverted to wild-type or possessed second site revertants, viral plaques were picked and freeze-thawed in infection medium to release intracellular virus. The supernatant was used to infect a fresh monolayer of HeLa cells. Plaque purification was repeated for three rounds before cDNA covering the VP1-2A-2B region was amplified by RT-PCR and sequenced for further analysis.
Table 1 shows the results of the sequence analysis using cDNAs amplified from ten plaques of HRV1A-(P212’-SYL) and five plaques of HRV1A-(P2’-A). Generally, we were not able to identify second site revertants in the sequenced region. However, nine out of ten isolates of HRV1A-(P212’-SYL) were found to have one of two rescue mutations within the cleavage site. In six cases, we identified a Tyr to Cys mutation at P1 while in three other sequences we observed a Leu to Pro mutation at P2’. Proline at P2’ is the wild-type residue found in the HRV1A VP1-2Apro junction. In contrast, cysteine at the P1 site is not a naturally occurring amino acid in any of the introduced HRV serotypes. Given the three possible residues created by a single base transition from the tyrosine codon (cystein, histidine, tyrosine), cystein exhibits the smallest space requirements and might therefore best mimic the wild-type alanine. We also observed one viral plaque with the original transfected cleavage site, indicating that self-processing at this site is not completely inhibited.
Table 1.
Nucleotide sequences at the VP1-2Apro junction from HRV1A variants
| VP1-2Apro cleavage site encoded by transfected RNA | Recovered VP1-2Apro cleavage site from viral offspring | Nucleotide Sequence of cDNA at the VP1-2Apro junction | Frequency§ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TITSY*GLSD | ACT | ATA | ACA | TCC | TAT | * | GGC | CTA | AGC | GAT | ||
| TITSY*GLSD | ACT | ATA | ACA | TCC | TAT | * | GGC | CTA | AGC | GAT | 1/10 | |
| TITSC*GLSD | ACT | ATA | ACA | TCC | TGT | * | GGC | CTA | AGC | GAT | 6/10 | |
| TITSY*GPSD | ACT | ATA | ACA | TCC | TAT | * | GGC | CCA | AGC | GAT | 3/10 | |
| TITTA*APSD | ACT | ATA | ACA | ACA | GCT | * | GCA | CCC | AGT | GAT | ||
| TITTA*GPSD | ACT | ATA | ACA | ACA | GCT | * | GGG | CCC | AGT | GAT | 5/5 | |
Number of identical sequences per number of sequenced plaques
In contrast, analysis of all five isolates of HRV1A-(P1’-A) showed that the alanine codon at P1’ had reverted back to the codon for the wild-type residue glycine, with the same double base mutation being detected in all five cDNAs examined.
Discussion
The absence of a structure of an enteroviral 2Apro complexed to a substrate or inhibitor has hampered a clear understanding of the determinants of the substrate specificity of these enzymes. The experimental basis of the work shown here illustrates this problem as we cannot at present explain why the HRV1A or HRV2 2Apro cannot carry out self-processing on the HRV14 cleavage sequence, even though the introduction of the respective amino acids one at a time does not affect cleavage. Our in vitro mutational approach coupled with the in vivo selection of revertant viruses demonstrates that at least three residues must be modified in the HRV2 or HRV1A cleavage sites to inhibit cleavage; however, this inhibition can be relieved by the modification of just one of the three amino acids. Thus, spatial requirements may dominate over absolute sequence constraints in determining substrate specificity.
In addition, we also show that Gly at P1’ in the cis self-processing reaction cannot be replaced by Ala. This is in contrast to the trans cleavage reaction for which two sites have been determined that show Ala at P1’; both are in the cellular substrate Nup62 (Park et al., 2010). What is the reason for the requirement of Gly at P1’ in the cis reaction? Petersen et al. (Petersen et al., 1999) proposed that Leu19 is the key residue that, through its bulk, prevents the acceptance of residues other than Gly at P1’ by blocking space required for a side-chain (Petersen et al., 1999). We therefore mutated Leu19 to alanine in HRV2 2Apro and examined self-processing with the P1’ Gly to Ala mutant. This mutant should possess sufficient space for the alanine side-chain at P1’; nevertheless, almost no processing could be observed (data not shown), suggesting that Leu19 is not involved in determining P1’ specificity. Our attempts to find residues involved in P1’ discrimination by searching for second site revertants using a HRV1A 2Apro P1’ Gly to Ala mutant were unsuccessful (Table 1), possibly indicating that more than one residue is involved.
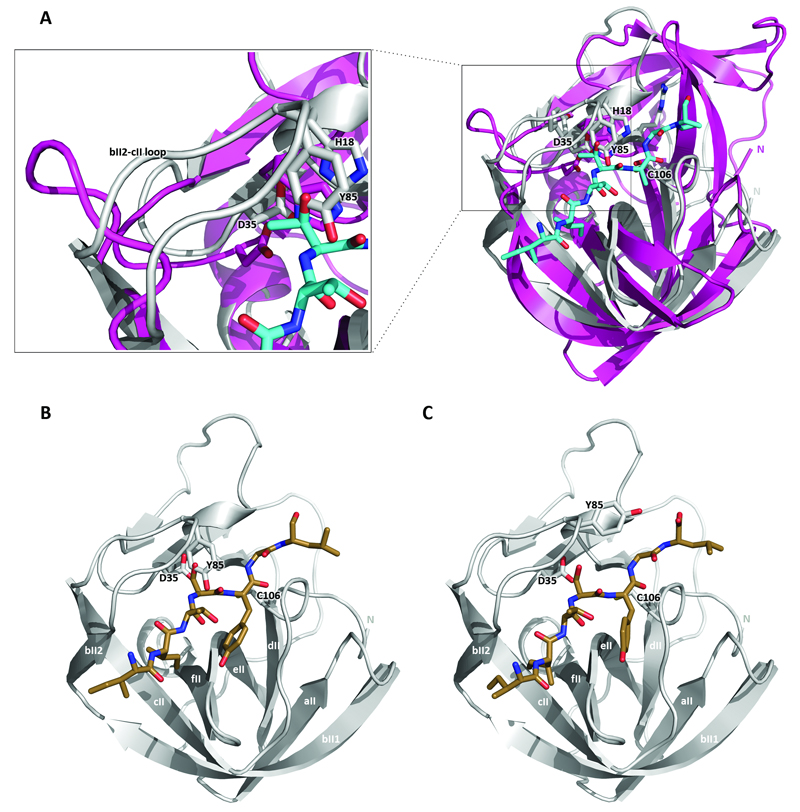
To structurally analyze spatial requirements of 2Apro substrate binding as well as P1’ discrimination in the absence of the structure of a complex of 2Apro and a substrate or inhibitor, we built a model of the binding of the HRV2 2Apro to the heptapeptide IleIleThrThrAlaGlyPro (IITTAGP) spanning the region from P5 to P2’ of the cleavage site. To this end, we used the DaliLite server (Holm & Sander, 1993a) to align HRV2 2Apro with Streptomyces griseus B protease (SGPB) complexed to the amino acid sequence ProAlaCysThrLeuGluTyr (PACTLEY) that correspond to residues P5 to P2’ of the third domain of the turkey ovomucoid inhibitor (OMTKY3). Subsequently, the amino acid side-chains of the OMTKY3 residues were mutated to the P5 to P2’ sequence IITTAGP of the HRV2 2Apro cleavage site using Pymol (Fig. 6a). Overall, the mutated peptide fits well into the substrate binding site of the HRV2 2Apro. However, a major difference between the two enzymes is the presence of the loop between strands bII2 and cII in HRV2 2Apro covering the Asp35 residue (Fig. 6a, enlargement). The tyrosine (Tyr85) residue at the end of the loop collides with the side-chain of the P2 Thr residue. This has been noted previously by Petersen et al. (Petersen et al., 1999) who used a different structural alignment approach to position the main-chain of the OMTKY3 peptide in the binding site. To examine the arrangement of the HRV14 2Apro cleavage sequence in HRV2 2Apro, we then extended the model by mutating the peptide IITTAGP to IleIleThrSerTyrGlyLeu (IITSYGL) (Fig. 6b). Again, Tyr85 still clashes with the P2 Ser residue. In addition, at P1, the side-chain of the tyrosine residue now lies very close to β-strand ell. In contrast, the leucine residue at P2’ does not appear to be causing any steric clashes.
Fig. 6.
(a) Superimposition of HRV2 2Apro onto SGBP bound to the PACTLEY peptide from OMTKY3. The PACTLEY sequence was modified to IITTAGP using Pymol. Superimposition was performed by using the DaliLite server (Krieger et al, 2009). HRV2 2Apro is in grey, SGPB is in purple. The peptide is shown as sticks, with nitrogen atoms dark blue, carbon atoms light blue and oxygen atoms red. The enlargement shows the HRV2 2Apro loop (bII2-cII) covering Asp35. (b) and (c) HRV2 2Apro model bound to a peptide corresponding to the HRV14 2Apro cleavage site before (b) and after (c) energy minimization using the YASARA energy minimization server (Krieger et al., 2009). The colour code is as in (a), except that carbon atoms are coloured brown. β-strands of the C-terminal domain of HRV2 2Apro are labeled according to SGPB nomenclature (Delbaere et al., 1979; Petersen et al., 1999). The side-chains of the indicated active site residues as well as Tyr85 are shown. Drawings were done with Pymol (DeLano, 2002).
We then minimized this model using the YASARA energy minimization server (Krieger et al., 2009) as shown in Fig. 6c. Rmsd analysis, measured with the Pymol command “rms_cur”, of positions of the Cα atoms of the non-minimized and minimized peptides shows a difference of 0.73 Å, indicating that the backbone of the substrate moves further into the binding cleft. In addition, the tyrosine at P1 shows rotation about the Cα-Cβ and Cβ-Cγ bonds placing the side-chain in the middle of the P1 pocket. Of the residues in the enzyme, there is clear movement of the residue Tyr85 in the loop covering residue Asp35. Thus, the side-chain of Tyr85 has been rotated 101° about its Cα-Cβ bond. Substrate induced movement of this residue was indeed predicted by Petersen et al. (1999).
Close examination of the minimized structure reveals that two residues come close to the Gly in the P1’ position of the substrate. These are the catalytic His18 and Tyr85 (distances 3.6 Å and 3.7 Å from P1’ Gly Cα to His18 Nε and to Tyr85 Cε, respectively). Both residues are fully conserved in all enteroviral 2Apro. The presence of any other residue at P1’ would thus cause a steric clash with His18 and Tyr85 side-chains and explain the absolute requirement for Gly at P1’ in self-processing. In addition, this would imply that the movement of the side-chain of Tyr85 is required for specificity, providing an explanation for the absolute conservation of this residue in all enteroviral 2Apro. How can Ala be accepted in trans cleavage at the P1’ position? One possibility might be that in the cis reaction, the N-terminal residues of 2Apro enter the substrate binding site before those of the C-terminus of VP1. The presence of Ala at P1’ may therefore prevent the movement of Tyr85 and the binding of the VP1 sequence. In contrast, in the trans reaction, the substrate is not covalently linked to the enzyme, so that the constraints on substrate binding may be more relaxed, giving Tyr85 greater freedom of movement.
The minimization of this model supports our experimental observations that single substitutions of the amino acids of the HRV14 cleavage site at P2, P1 and P2’ can all be accepted by the HRV2 2Apro. The model does not however explain why the presence of all three lead to inhibition of HRV2 or HRV1A 2Apro self-processing. We propose that the reason may lie in the smaller Ser residue at P2 that is less effective than Thr in driving Tyr85 out of the substrate binding pocket. In addition, the presence of the Tyr P1 and Leu P2’ side-chains may destabilise substrate binding through their bulky side-chains. In this way, substrate specificity would appear to depend on more spatial properties than on recognition of the sequence itself. Such a phenomenon has also been observed for the HIV-1 proteinase that recognizes the asymmetric shape of a group of amino acids rather than their primary sequence (Nalam et al., 2010).
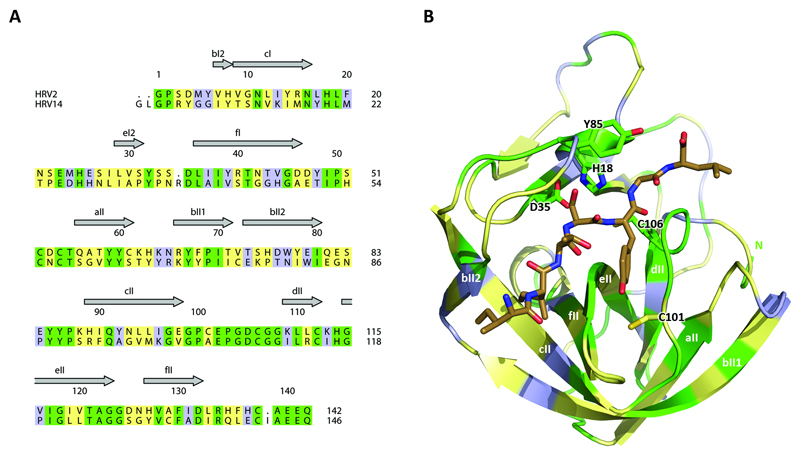
Are there differences in the residues of HRV2 and HRV14 2Apro that surround the substrate binding site or in the area of Tyr85? To investigate this point, we compared the amino acid sequences of the two enzymes and mapped the differences on the HRV2 2Apro structure (Fig. 7). Even though there is only a 40% conservation of the amino acid sequence, there is a high level of conservation of the residues flanking the P1 pocket (eII and the coil linking strands cII and dII). Indeed, only one residue differs in this region, namely at position 101 with Cys present in HRV2 and Ala in HRV14. This residue has been previously shown to be involved in preventing HRV14 from accepting Arg at P1 (Sousa et al., 2006).
Fig. 7.
Differences between the 2Apro of HRV2 and HRV14. (a) Alignment of the primary sequences of HRV2 (top) and HRV14 (bottom) 2Apro. Conserved residues are in green, similar in yellow and unrelated in light blue. (b) Mapping of the alignment (color code as in (a)) onto the HRV2 2Apro structure bound to the peptide (brown sticks) of the HRV14 2Apro (see Fig. 7B). The side-chains of the active site residues as well as Tyr85 and Cys101 are shown.
Another difference lies in the residues in the loop connecting eII with fII. However, all the side-chains in this loop point outwards, away from the S1 binding pocket. However, none of the other differences noted could be interpreted as influencing substrate specificity. This again supports the notion that spatial rather than sequence requirements are involved in determining substrate specificity.
In summary, we present experiments that substrate recognition by HRV 2Apro is not a simple matter of the recognition of certain amino acid side-chains but is rather restricted by as yet undetermined spatial considerations. Although the results are based on an energy minimized model, they suggest that the reason for the conservation of Tyr85 is its movement on substrate binding that then restricts the residues that can be accepted at P1’ to glycine. Further experiments will be performed to test this hypothesis.
Methods
Construction of plasmids. pHRV2 VP1-2Apro encoding HRV2 VP1 and 2Apro cloned downstream to the encephalomyocarditis virus IRES (CITE) and a T7 promoter has been described (Glaser et al., 2003). pHRV1A VP1-2Apro encodes the HRV1A VP1 and 2Apro and was generated from pHRV2 VP1-2Apro by replacing the VP1-2Apro coding sequence via NcoI/BamHI sites. Substitutions at the VP1-2Apro cleavage site were introduced via synthetic oligonucleotide cassettes. Restriction enzyme sites (BsiWI and BstEII) were introduced around the cleavage site without changing the amino acid sequences to facilitate the introduction of the cassettes. For transfection experiments mutated cleavage sites were introduced into a full length cDNA clone of HRV1A via cassette cloning. Again restriction enzyme sites BsiWI/BstEII were introduced and used for cloning.
In vitro transcription and translation. Following linearization with BamHI (pHRV2 VP1-2Apro and pHRV1A VP1-2Apro) or with MluI (HRV1A full length clone), DNA was purified via the Wizard® SV Gel and PCR Clean-Up System (Promega) and transcribed in vitro with T7 RNA polymerase for 2 h at 37°C. Reactions (100 μl) contained 10-15 μg of linearized DNA, 100 mM sodium phosphate pH 7.7, 50 mM DTT, 0.5 mM NTPs, 20 mM spermidine (HCl)3, 40 mM MgCl2 120 U RNasin (Promega), 1 μg T7 RNA polymerase. Template DNA was removed by addition of 150 U DNase I (Invitrogen) and incubation at 37°C for 20 min. RNA was purified by phenol/chloroform extraction and precipitated with ethanol and ammonium acetate before RNA quality was examined on 1% (w/v) agarose gels containing 0.1% (w/v) SDS. In vitro translation reactions were performed as previously described by Sousa et al. (Sousa et al., 2006).
RNA transfection and plaque isolation. Alternatively, in vitro transcribed RNA products encoding the full HRV1A genome with or without site mutations were used for HeLa cell transfection by DEAE-dextran. HeLa (Ohio) cells were cultured at 37°C with 5% CO2 in Minimal Essential Medium (MEM) containing 10% (v/v) fetal calf serum (FCS, Gibco), 1% (v/v) 200 mM L-Glutamine (Gibco), 1% (v/v) 100x penicillin/streptomycin (Gibco, 10.000 U ml-1 penicillin, 10.000 μg ml-1 streptomycin) and were prepared for transfection by seeding 0.325·106 cells per 6-well in 2ml infection medium, consisting of MEM supplemented with 2% (v/v) fetal calf serum (FCS, Gibco), 1% (v/v) 200 mM L-Glutamine (Gibco), 1% (v/v) 100x penicillin/streptomycin (Gibco) and 1% (v/v) 3M MgCl2. Cells were grown to a density of 50%. Transfection reactions (300μl/well) contained 1.2 μg in vitro transcribed RNA, 140mM LiCl2, 1mM MgCl2, 10mM HEPES pH 7.5 and 2 μM DEAE-dextran and incubated on ice for 30 min prior to incubation with cells. Prior to treatment, cells were washed twice with phosphate-buffered saline (PBS). Cells were then incubated with transfection mix for 25 min at 34°C with gentle shaking. Transfection mix was removed and cells were covered with 1% (w/v) low melting agarose (Sigma) in infection medium for 48h to 72h. Plaques were visualized by the addition of 0.05% (w/v) neutral red solution, picked, resuspended in 1 ml infection medium and freeze/thawed three times. The 1 ml supernatant was used to infect fresh HeLa cells. After the third round of plaque isolation, plaques were resuspended in 100 μl PBS instead.
Reverse transcription and PCR. Viral RNA in the supernatant from freeze/thawed plaques in PBS was reverse transcribed into cDNA using SuperScript® III Reverse Transcriptase (Invitrogen). cDNA synthesis reactions contained 11 μl supernatant, 0.1 μM antisense primer (5’-GTAACACTTGAAACCATTTGG -3’, Tim1921 binding in 2C region), 0.125 mM dNTPs (Invitrogen), 1x first strand buffer, 40 U RNasin (Promega) and 200 U SuperScript III reverse transcriptase. Reverse transcription was performed at 40°C for 60 min, followed by inactivation of the enzyme at 70°C for 15 min. Resulting cDNA was then used as template for amplification using Pfu Polymerase (Promega), following the instructions according to the standard PCR protocol from Promega using 5’-TTTTATGATGGATATGATGG-3’ (Tim1915, binding in VP1 region) as the sense and 5’-GTAACACTTGAAACCATTTGG -3’ (Tim1921, binding in 2C region) as the antisense primer. The RT-PCR products were cleaned via the Wizard® SV Gel and PCR Clean-Up System (Promega) and were sequenced by LGC Genomics using 5’-CCCTTACACACATAGTCATGTG-3’ (Tim1922, binding in VP1 region) as the sense and 5’-ATTTCACATTTGATTTTCTCTTG-3’ (Tim1923, binding in 2C region) as the antisense primer.
Modeling and energy minimization. Superimposition of SGPB in complex with the inhibitor OMTKY3 (PDB 3SGB) and chain B of the HRV2 2Apro crystal structure (PDB 2HRV) was performed using the DaliLite server at EMBL-EBI (Holm & Sander, 1993b) placing the OMTKY3 within the substrate binding cleft of HRV2 2Apro. OMTKY3 was trimmed to residues 14-20 (ProAlaCysThrLeuGluTyr, corresponding to P5-P2’) and side chains were exchanged to fit the self processing site of HRV2 2Apro (IleIleThrThrAlaGlyPro) or the sequence IleIleThrSerTyrGlyLeu using PyMol. A PDB file containing the protease as well as the peptide was submitted to the YASARA energy minimization server which uses the self-parameterizing and knowledge-based YASARA force filed (derived from AMBER) (Krieger et al., 2009). Energy minimized structures were then inspected using PyMol (DeLano, 2002).
Acknowledgements
We thank Dan Pavear for the HRV1A clone. This project was funded by the Austrian Science Fund, grants P20889 and P24038 to Tim Skern.
References
- Baxter NJ, Roetzer A, Liebig HD, Sedelnikova SE, Hounslow AM, Skern T, Waltho JP. Structure and dynamics of coxsackievirus B4 2A proteinase, an enyzme involved in the etiology of heart disease. J Virol. 2006;80:1451–1462. doi: 10.1128/JVI.80.3.1451-1462.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan PL, Mizutani S, Colonno RJ. Molecular cloning and complete sequence determination of RNA genome of human rhinovirus type 14. Proc Natl Acad Sci U S A. 1985;82:732–736. doi: 10.1073/pnas.82.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso MC, Jackson RJ. Efficient initiation of mammalian mRNA translation at a CUG codon. Nucleic Acids Res. 1989;17:6485–6497. doi: 10.1093/nar/17.16.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano DL. The PyMOL molecular graphics system. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Delbaere LT, Brayer GD, James MN. The 2.8 A resolution structure of Streptomyces griseus protease B and its homology with alpha-chymotrypsin and Streptomyces griseus protease A. Can J Biochem. 1979;57:135–144. doi: 10.1139/o79-017. [DOI] [PubMed] [Google Scholar]
- Duechler M, Skern T, Blaas D, Berger B, Sommergruber W, Kuechler E. Human rhinovirus serotype 2: in vitro synthesis of an infectious RNA. Virology. 1989a;168:159–161. doi: 10.1016/0042-6822(89)90414-5. [DOI] [PubMed] [Google Scholar]
- Duechler M, Skern T, Blaas D, Berger B, Sommergruber W, Kuechler E. Human Rhinovirus Serotype 2: In Vitro Synthesis of an Infectious RNA. Virology. 1989b;168:159–161. doi: 10.1016/0042-6822(89)90414-5. [DOI] [PubMed] [Google Scholar]
- Glaser W, Triendl A, Skern T. The Processing of eIF4GI by Human Rhinovirus Type 2 2Apro: Relationship to Self-Cleavage and Role of Zinc. J Virol. 2003;77:5021–5025. doi: 10.1128/JVI.77.8.5021-5025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Blinov VM, Donchenko AP. Poliovirus-encoded proteinase 3C: a possible evolutionary link between cellular serine and cysteine proteinase families. FEBS Lett. 1986;194:253–257. doi: 10.1016/0014-5793(86)80095-3. [DOI] [PubMed] [Google Scholar]
- Hanecak R, Semler BL, Anderson CW, Wimmer E. Proteolytic processing of poliovirus polypeptides: antibodies to polypeptide P3-7c inhibit cleavage at glutamine-glycine pairs. Proc Natl Acad Sci U S A. 1982;79:3973–3977. doi: 10.1073/pnas.79.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993a;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Structural alignment of globins, phycocyanins and colicin A. FEBS Lett. 1993b;315:301–306. doi: 10.1016/0014-5793(93)81183-z. [DOI] [PubMed] [Google Scholar]
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy, Ninth report of the international committee on taxonomy of viruses. Elsevier Inc; 2012. [Google Scholar]
- Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, Palmenberg AC, Simmonds P, Skern T, Stanway G, Yamashita T, et al. Picornaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier; 2012. pp. 855–880. [Google Scholar]
- Krieger E, Joo K, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins. 2009;77(Suppl 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimaki M, Blomqvist S, Hyypia T, Arstila P. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight KL. The human rhinovirus internal cis-acting replication element (cre)exhibits disparate properties among serotypes. Arch Virol. 2003;148:2397–2418. doi: 10.1007/s00705-003-0177-7. [DOI] [PubMed] [Google Scholar]
- Nalam MN, Ali A, Altman MD, Reddy GS, Chellappan S, Kairys V, Ozen A, Cao H, Gilson MK, Tidor B, Rana TM, et al. Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J Virol. 2010;84:5368–5378. doi: 10.1128/JVI.02531-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg AC, Pallansch MA, Rueckert RR. Protease required for processing picornaviral coat protein resides in the viral replicase gene. J Virol. 1979;32:770–778. doi: 10.1128/jvi.32.3.770-778.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park N, Skern T, Gustin KE. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J Biol Chem. 2010;285:28796–28805. doi: 10.1074/jbc.M110.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JF, Cherney MM, Liebig HD, Skern T, Kuechler E, James MN. The structure of the 2A proteinase from a common cold virus: a proteinase responsible for the shut-off of host-cell protein synthesis. EMBO J. 1999;18:5463–5475. doi: 10.1093/emboj/18.20.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Skern T, Hampoelz B, Guarné A, Fita I, Bergmann E, Petersen J, James MNG. Structure and function of picornavirus proteinases. In: Semler BL, Wimmer E, editors. Molecular Biology of Picornaviruses. Washington, DC: ASM Press; 2002. pp. 199–212. [Google Scholar]
- Skern T, Sommergruber W, Auer H, Volkmann P, Zorn M, Liebig HD, Fessl F, Blaas D, Kuechler E. Substrate requirements of a human rhinoviral 2A proteinase. Virology. 1991;181:46–54. doi: 10.1016/0042-6822(91)90468-q. [DOI] [PubMed] [Google Scholar]
- Skern T, Sommergruber W, Blaas D, Gruendler P, Fraundorfer F, Pieler C, Fogy I, Kuechler E. Human rhinovirus 2: complete nucleotide sequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985;13:2111–2126. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommergruber W, Ahorn H, Zophel A, Maurer-Fogy I, Fessl F, Schnorrenberg G, Liebig HD, Blaas D, Kuechler E, Skern T. Cleavage specificity on synthetic peptide substrates of human rhinovirus 2 proteinase 2A. J Biol Chem. 1992;267:22639–22644. [PubMed] [Google Scholar]
- Sousa C, Schmid EM, Skern T. Defining residues involved in human rhinovirus 2A proteinase substrate recognition. FEBS Lett. 2006;580:5713–5717. doi: 10.1016/j.febslet.2006.09.023. [DOI] [PubMed] [Google Scholar]
- Stanway G, Hughes PJ, Mountford RC, Minor PD, Almond JW. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 1984;12:7859–7875. doi: 10.1093/nar/12.20.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H, Nicklin MJ, Murray MG, Anderson CW, Dunn JJ, Studier FW, Wimmer E. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell. 1986;45:761–770. doi: 10.1016/0092-8674(86)90790-7. [DOI] [PubMed] [Google Scholar]
- Wang QM, Sommergruber W, Johnson RB. Cleavage specificity of human rhinovirus-2 2A protease for peptide substrates. Biochem Biophys Res Commun. 1997;235:562–566. doi: 10.1006/bbrc.1997.6830. [DOI] [PubMed] [Google Scholar]