Abstract
CD4+ T cells predominate in salivary gland (SG) inflammatory lesions in Sjögren’s syndrome (SS). However, their antigen specificity, degree of clonal expansion, and relationship to clinical disease features remain unknown. We used multiplex reverse-transcriptase PCR to amplify paired T cell receptor α (TCRα) and β transcripts of single CD4+CD45RA– T cells from SG and peripheral blood (PB) of 10 individuals with primary SS, 9 of whom shared the HLA DR3/DQ2 risk haplotype. TCRα and β sequences were obtained from a median of 91 SG and 107 PB cells per subject. The degree of clonal expansion and frequency of cells expressing two productively rearranged α genes were increased in SG versus PB. Expanded clones from SG exhibited complementary-determining region 3 (CDR3) sequence similarity both within and among subjects, suggesting antigenic selection and shared antigen recognition. CDR3 similarities were shared among expanded clones from individuals discordant for canonical Ro and La autoantibodies, suggesting recognition of alternative SG antigen(s). The extent of SG clonal expansion correlated with reduced saliva production and increased SG fibrosis, linking expanded SG T cells with glandular dysfunction. Knowledge of paired TCRα and β sequences enables further work toward identification of target antigens and development of novel therapies.
CD4+ T cell clonal expansions in Sjögren’s syndrome salivary glands correlate with impaired glandular function and share TCR sequence similarity indicative of antigen-driven selection.
Introduction
Sjögren’s syndrome (SS) is a chronic, debilitating rheumatic autoimmune disease with hallmark features of severe dry mouth, dry eyes, and autoantibodies to systemic nuclear antigens (1, 2). Criteria for disease classification include both subjective symptoms and objective measures of dry eyes and mouth, presence of Ro/SS-A and La/SS-B autoantibodies, and focal lymphocytic infiltration of biopsied minor salivary gland (SG) tissue (3). Presence of at least one cluster of ≥50 lymphocytes in 4 mm2 of labial SG tissue, defined as a “focus,” is sensitive and specific for SS (3, 4) and occurs in parallel with similar infiltrates in submandibular and parotid SGs (4). The focal lymphocytic infiltrates are dominated by CD4+ T cells (5–8) expressing αβ T cell receptors (TCRs) (9, 10) with markers of activation (6, 8) and memory (10, 11), though CD8+ T cells are invariably present. B lymphocyte and macrophage populations increase with disease severity (12).
T cells expressing αβ TCRs interact with peptide antigen in the context of HLA molecules. The amino acids responsible for peptide antigen binding are located in the third complementarity-determining regions (CDR3s) of the α and β chains. CDR3 is the most variable portion of the TCR, as recombination allows for various combinations of variable (V), diversity (D, in the case of the β chain), and joining (J) gene segments as well as for the addition of random, nontemplated nucleotides into the joints between gene segments; these are referred to as NDN-region additions in the β chain and simply N-region additions in the α chain. In development, T cells simultaneously rearrange both TCRα loci (13), resulting in a potential for mature cells containing dual functional TCRα gene rearrangements (14).
Prior studies evaluated TCR Vβ gene family usage in primary SS (pSS) SG tissue by immunostaining (15, 16), single-strand conformational polymorphism analysis (17, 18), or PCR in combination with hybridization techniques (10, 19, 20). TCR sequences derived from bulk tissue and sequenced following cloning into bacterial vectors or phage were polyclonal and exhibited some preferential Vβ gene usage that varied from patient to patient. Some studies evaluating few patients found TCR motifs in CD3+ T cells within individuals suggesting antigen-driven selection (17, 18, 21). However, whether these TCR motifs occurred in expanded clones or CD4+, CD8+, or memory subsets is unknown. There is also little knowledge of the TCRα gene usage of T cells from SG tissue of pSS patients, with two studies evaluating fewer than 20 cells each (22, 23) and a third study evaluating only a portion of the known Vα gene families (20). Knowledge of paired TCRα and β sequences from SG clonal expansions is required for discovering the antigens driving T cell activation and expansion in SG tissue. Importantly, the studies referenced above were subject to PCR amplification bias, precluding a precise evaluation of the TCR repertoire in the SG of SS patients.
The specificity of SG CD4+ T cells and their role in SS is not understood. Identification of autoantigens can uncover pathologic mechanisms and revolutionize approaches to disease prediction (24), prognosis (25), diagnosis (26), and therapy (27–29). Although dry mouth and CD4+ T cell infiltrates in SG tissue are cardinal features of SS, mechanistic connections between these elements have remained elusive. In this study, we analyzed the paired α and β TCR repertoire of pSS subjects derived from single SG CD4+CD45RA– T cells in a systematic manner, utilizing a precise single-cell approach for defining T cell clonal expansions. Our strategy is not subject to the amplification bias encountered in the study of TCR repertoires from bulk cell populations and tissues or to bias introduced during in vitro cell culture (30). From SG clonal expansions identified in 9 of 10 subjects, our study provides the first firm evidence to our knowledge for antigen-driven CD4+ T cell clonal expansion in SG, uncovers TCR CDR3 similarities among expanded clones of unrelated individuals carrying the HLA DR3/DQ2 (DRB1*0301/DQB1*0201) risk haplotype (31), and reveals correlations between the degree of SG CD4+ T cell clonal expansion and oral disease features that provide mechanistic insight into the oral pathology of SS.
Results
Subject characteristics.
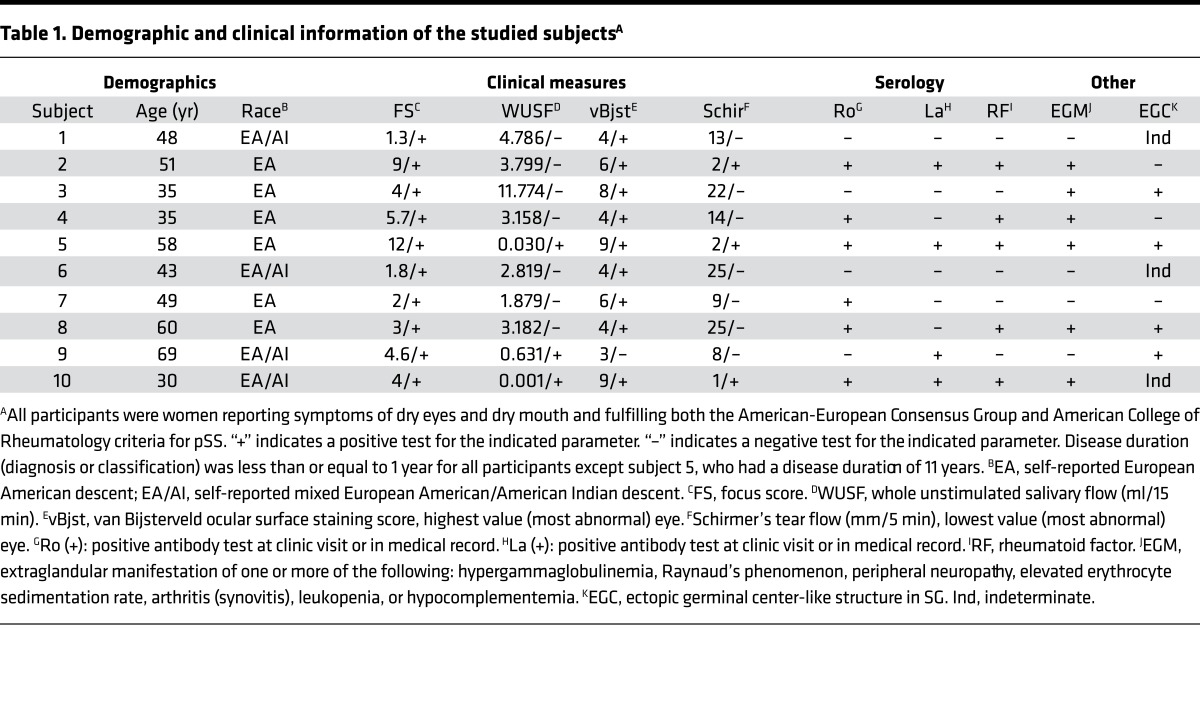
Ten individuals who met internationally accepted criteria for pSS (3), exhibited focal lymphocytic sialoadenitis with focus score ≥1, and were positive by PCR for the SS-associated DR3/DQ2 risk haplotype were chosen for analysis of their TCR repertoire in CD4+ memory T cells from SG and peripheral blood (PB). Additionally, the DR3/DQ2 haplotype was assigned to 9 of these 10 subjects by imputation of GWAS SNP data using the maximum likelihood model (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/jci.insight.85609DS1). Other demographic and clinical characteristics of the participants are presented in Table 1. The cohort was comprised entirely of women with a median age of 48 years (range 30–69 years) and a wide range of focus scores and salivary flow rates. The majority of individuals (70%) were positive for IgG Ro and/or La serum antibodies and more than half had extraglandular manifestations of disease.
Table 1. Demographic and clinical information of the studied subjectsA.
CD4+ T cells from SS SG exhibit biased TCR gene usage compared with those from PB, despite the presence of a broad TCR repertoire.
Over 3,000 CDR3 sequences from nearly 2,000 single SG and PB CD3+CD4+CD45RA– memory T cells were identified. The gating strategy for sorting and the numbers of cells recovered are shown in Supplemental Figure 1 and Supplemental Table 2. Assessment of the frequencies of utilized TCR genes among uniquely represented sequences revealed diverse TCR gene usage in both SG and PB (Supplemental Figure 2), as has been previously noted (10). Comparison of gene usage frequency by matched-pairs analysis revealed significant enrichment of a few selected TCRα genes (TRAV8-4, TRAV-24, and TRAJ22) in SG, while usage of others (TRAV38-1, TRAJ27, TRAJ44, and TRBV29-1) was reduced (Supplemental Figure 3).
CD4+ T cells from SS SG often express two functional TCRα chains.
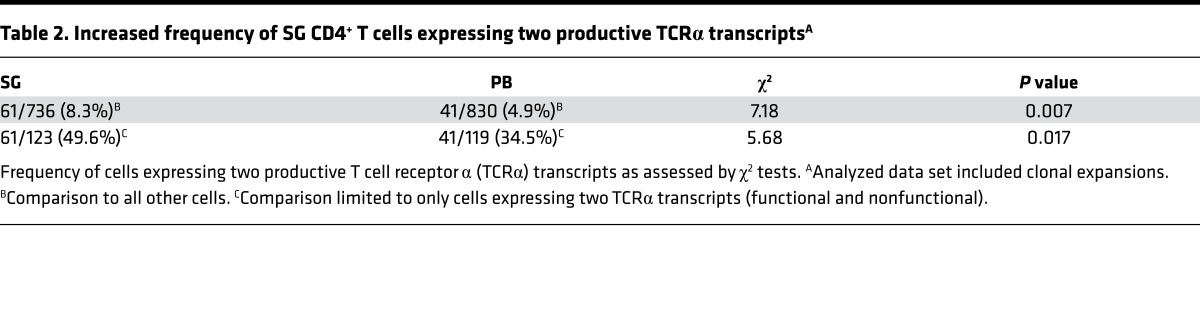
As T cells expressing dual TCRs have been proposed to contribute to autoimmunity (32–35), the prevalence of cells expressing two functional TCRα transcripts in SG versus PB was compared among all cells yielding detectable α chains. The overall frequency of cells expressing two α chains, regardless of whether the rearrangements were productive, was similar between the two tissues (13.3% of SG vs. 14.1% of PB CD4+ T cells). SG-derived cells, however, were significantly more likely to express dual productive TCRα rearrangements than those from PB (49.6% vs. 34.5%, χ2 = 5.68, P = 0.017; Table 2). This result was also found in an analysis restricted to unique sequences (48.3% vs. 33%, χ2 = 4.88, P = 0.027). This bias indicates a potential role for the surface expression of dual TCRs in the selection of CD4+ T cells found in SG tissue from individuals with SS.
Table 2. Increased frequency of SG CD4+ T cells expressing two productive TCRα transcriptsA.
Clonally expanded CD4+ T cells are frequent in SS SG tissue.
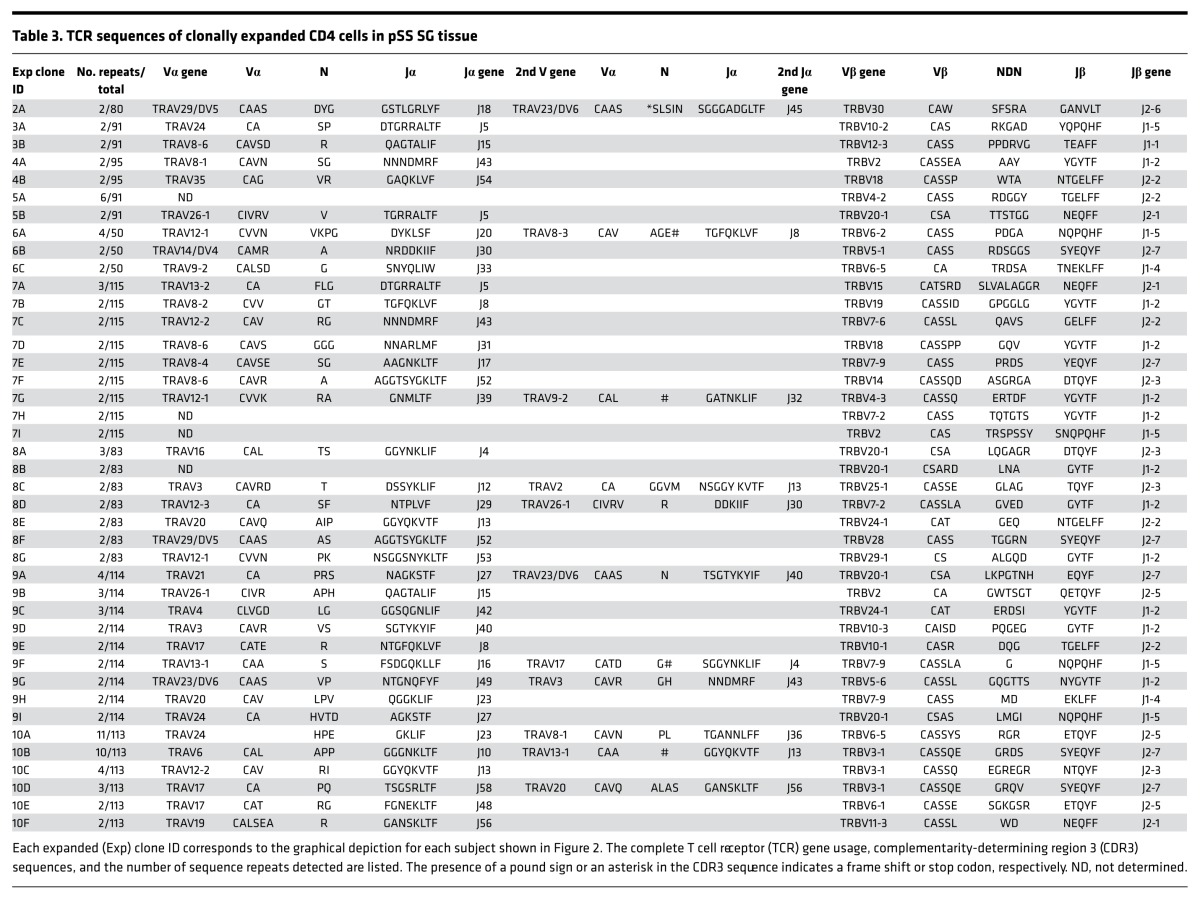
Clonal expansions were defined as two or more T cells expressing nucleotide-identical TCR CDR3 sequences in a given tissue of an individual subject. The percentages of cells comprising clonal expansions in SG and PB are shown in Supplemental Table 3 and Figure 1. Nine of the ten pSS subjects had detectable clonal expansions in SG. The TCR CDR3 sequences, TCR-V/J gene usage, and size of the expanded SG clones are shown in Table 3. Although clonal expansions were also detected in the PB of 5 individuals, the extent of clonal expansion was substantially greater in SG compared with PB (P = 0.006, median 11.95%, range 0%–28.3% vs. median 0.85%, range 0%–6.7%, Supplemental Table 3 and Figure 1). Variability in the numbers and sizes of the SG clonal expansions is shown in Figure 2. Each clone was assigned a number (corresponding to the subject number) and a letter, beginning with “A” (assigned to the largest expansion for each subject). For clones with two TCRα chains, the second α chain was designated by a prime symbol (′). The majority of subjects exhibited multiple small expansions, ranging from 2 to 6 cells per clone, with the exception of subject 10, in whom two large expansions of 11 and 10 cells each were identified.
Figure 1. Memory CD4+ T cell clonal expansions are more frequent in salivary glands than in peripheral blood of primary Sjögren’s syndrome subjects.
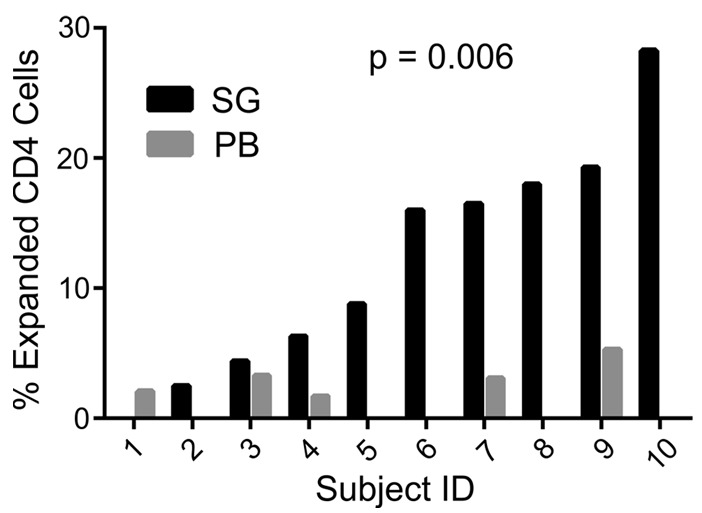
The percentages of all memory CD4+ T cells that were part of clonal expansions are shown for salivary gland (SG) and peripheral blood (PB) of each subject (P = 0.006, matched pairs Wilcoxon signed-rank test).
Table 3. TCR sequences of clonally expanded CD4 cells in pSS SG tissue.
Figure 2. Numbers and sizes of expanded clones derived from salivary gland.
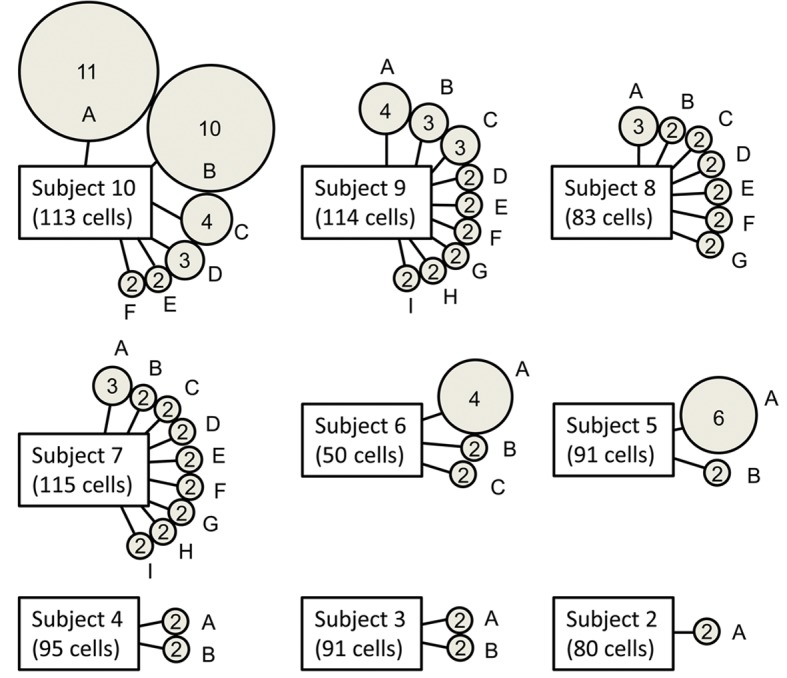
Each circle represents an expanded T cell clone. Numbers inside of circles indicate the number of cells expressing nucleotide-identical complementarity-determining region 3 (CDR3) sequences. Letters designate particular expanded clones and match designations in Table 3. The total number of single cells evaluated is shown below each subject label.
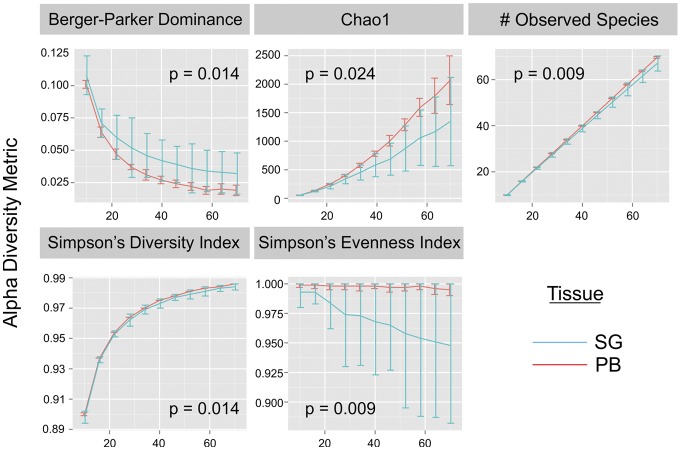
To determine whether the TCR repertoire in SG was more restricted than that in PB, we adopted statistical methods used to evaluate biodiversity. The TCR repertoire can be thought of as a collection of species and therefore a type of biodiversity. Diversity is determined by species richness (i.e., the number of unique species in a given population) and distribution evenness (i.e., how balanced the distribution of species is in an ecosystem). Differences between samples are determined by the numbers and frequencies of shared species. As methods to investigate ecological diversity have been adapted to TCR repertoire analysis (36–38), we performed diversity analyses on productive nucleotide sequences from the complete SG and PB CDR3 α and β data sets to assess the impact of clonal expansions on TCR repertoire diversity in SG. Significant reductions in richness metrics (Chao1, number of observed species), Simpson’s Diversity Index and Simpson’s Evenness, were observed at increasing depth in SG compared with PB, while a significant increase in Berger-Parker Dominance (an inverse measure of diversity) was observed in SG compared with PB (Figure 3). These results indicate reduced sequence diversity in SG compared with PB, which is likely a consequence of clonal expansions and antigenic selection of SG T cells.
Figure 3. Biodiversity metrics plotted as a function of sequence depth.
Separate salivary gland (SG) and peripheral blood (PB) data sets, each containing the complete TCRα and TCRβ CDR3 sequences, were evaluated using alpha biodiversity statistics. Under all metrics, there was a significant difference in diversity between the PB and SG samples at increasing depth. P values are from nonparametric, 2-tailed, 2-sample t tests. Error bars are SD.
Evidence of antigenic selection within individuals.
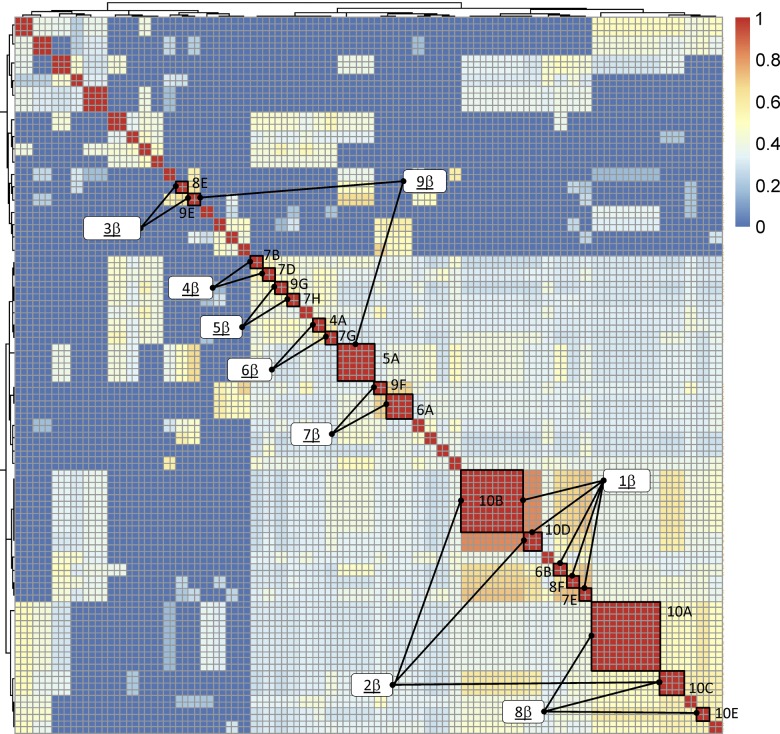
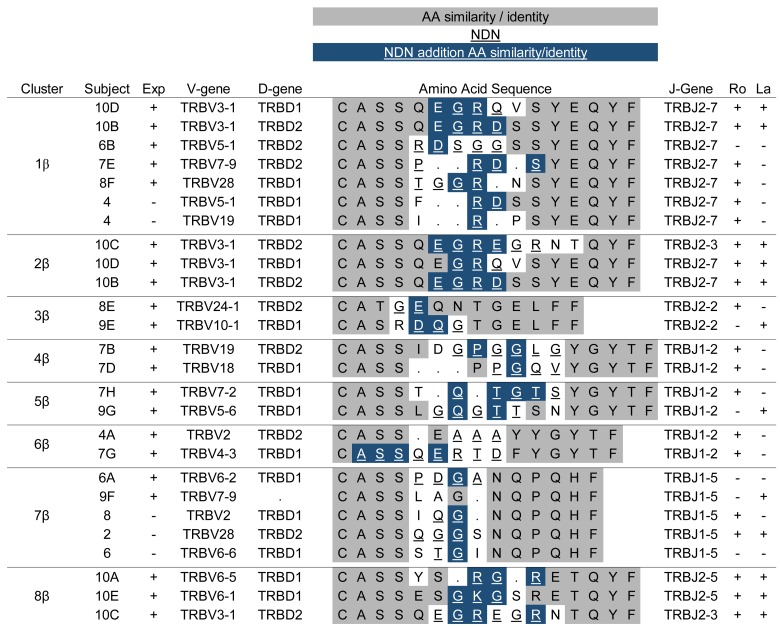
To probe for sequence similarities among expanded clones in SG, all expanded SG TCR CDR3 sequences were compared to each other using protein-protein blast matrices, evaluating TCRα and β sequences separately. The sequences were clustered by BLAST similarity scores (see Methods) and displayed as heatmaps (Figures 4 and 5). Alignment of TCRβ CDR3 sequences within related clusters revealed numerous examples of striking sequence similarity between expanded clones within individual subjects (Figure 6). For example, multiple independent clonal expansions of subject 10 align in clusters 1β and 8β, and two clonal expansions of subject 7 align in cluster 4β. In each case, these alignments included amino acids that were not germline encoded.
Figure 4. Heatmap BLAST similarity matrix of expanded salivary gland T cell receptor β complementarity-determining region 3 amino acid sequences from 10 subjects with primary Sjögren’s syndrome.
Red indicates identity. Blue indicates minimum similarity. The numbers on the color scale indicate the fraction of the maximal blast score, with 1 indicating identity and 0 indicating no similarity. Clusters selected for multiple sequence alignments are indicated by the lines connecting each to its designation, as is noted in the white boxes. Expanded clone designations are as indicated in Table 3 and Figure 2.
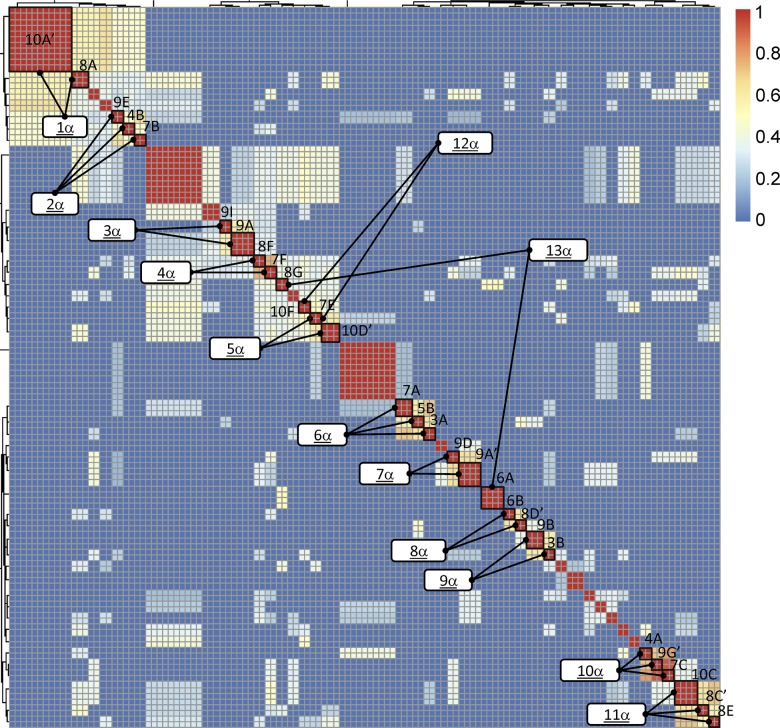
Figure 5. Heatmap BLAST similarity matrix of expanded salivary gland T cell receptor α complementarity-determining region 3 amino acid sequences from 10 subjects with primary Sjögren’s syndrome.
Red indicates maximum similarity. Blue indicates identity. The numbers on the color scale indicate the fraction of the maximal blast score, with 1 indicating identity and 0 indicating no similarity. Clusters selected for multiple sequence alignments are indicated by the lines connecting each to its designation, as is noted in the white boxes. Expanded clone designations are as indicated in Table 3 and Figure 2.
Figure 6. Alignment of T cell receptor β sequences from expanded salivary gland clones and closely related unique cells.
Cluster designations match those in Figure 4. Subject designations indicate expanded clones and match designations in Table 3 and Figure 3. Sequences that are part of clonal expansions (Exp) are indicated by “+,” and unique sequences are indicated as “–.” Underlined residues are derived from the NDN-region. Nonunderlined residues are derived from germline sequence. Gray shading indicates similar or identical residues derived from germline sequence. Blue shading indicates similar or identical residues derived from the NDN-region. Ro and La antibody status of each subject is listed as positive (+) or negative (–).
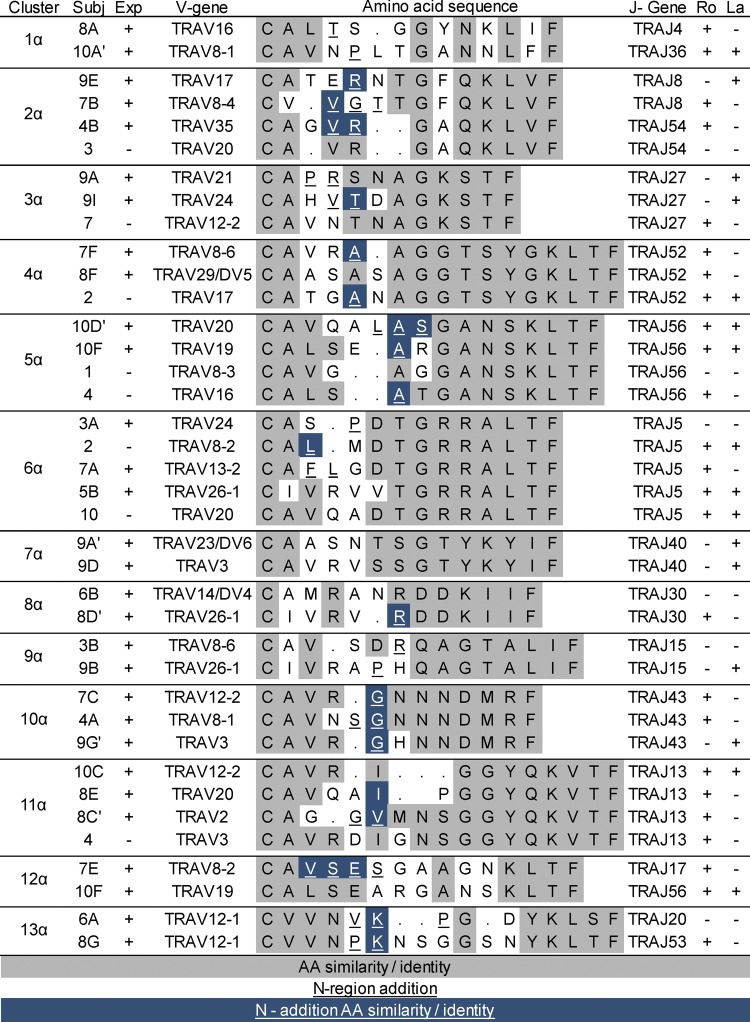
A similar analysis performed using the TCRα CDR3 sequences revealed several examples of highly similar sequences between expanded clones from the same individual (Figure 7). Because TCRα chain recombination utilizes only N-region additions without the contribution of a diversity gene, CDR3αs typically have fewer nucleotide additions at the V-J gene junction. Therefore, the extent of homology between TCRα CDR3 sequences is often driven entirely by similarity in the V and/or J gene regions. Although this was observed for sequences in clusters 7α and 11α, other TCRα CDR3 similarities involved similar or identical amino acids encoded by N-region additions (e.g., expanded clones 9A and 9I in cluster 3α and expanded clones 10D′ and 10F in cluster 5α). Taken together, the TCRα and β chain sequence similarities in clonally expanded T cells within the SG of individuals strongly suggest antigen-driven selection in the SG of these subjects.
Figure 7. Alignment of T cell receptor α sequences from expanded salivary gland clones and closely related unique cells.
Cluster designations match those indicated in Figure 5. Subject (Subj) designations indicate particular expanded clones and match designations in Table 3 and Figure 3. For cells with two α chains, the second one is designated with a prime symbol. Sequences that are part of clonal expansions (Exp) are indicated by “+,” and unique sequences are indicated as “–.” Shading and underlining are as described in the legend for Figure 6. Ro and La antibody status of each subject is listed as positive (+) or negative (–).
Evidence of recognition of common antigens among unrelated SS subjects.
More striking was our observation that amino acid sequences from expanded clones of unrelated individuals clustered together. For example, CDR3 sequences of expanded SG clones from 4 different subjects aligned in cluster 1β (Figure 6), while sequences of expanded clones from multiple pairs of subjects aligned in clusters 5β, 6β, 7β, and 9β. All of these alignments include amino acids encoded by N-region nucleotides. Similar clustering of CDR3α sequences among unrelated subjects is shown in Figure 7. As expected, some TCRα clusters contained no or few N-region-encoded amino acids in the alignments (clusters 1α, 3α, 6α, 8α, and 9α). However, multiple N-region-encoded amino acids were involved in other alignments: valine and arginine in cluster 2α; alanine in clusters 4α and 5α; glycine in cluster 10α; isoleucine and valine in cluster 11α; valine, serine, and glutamic acid in cluster 12α; and lysine in cluster 13α. These data suggest common antigen recognition among expanded SG CD4+ T cell clones.
We next examined unique sequences for similarity to those in our identified clusters. Motifs were constructed from individual clusters and used to search our database of unique SG sequences. Several unique sequences (indicated by the subject number alone) were found with similarities to those in the TCRβ alignments; these were added to the clusters displayed in Figure 6. Several of these used NDN- or N-region nucleotides to encode the amino acids conferring similarity. For example, 2 unique CDR3β sequences from subject 4 contributed to the large cluster 1β, 3 unique CDR3β sequences from subjects 2, 6, and 8 aligned with clonal expansions of subjects 6 and 9 in cluster 7β, and 1 unique sequence from subject 4 aligned with expanded sequences from subjects 5 and 9 in cluster 9β. It is of interest that this latter sequence is nucleotide-identical to expanded clone 5A. Unique TCRα CDR3 sequences from subjects 1, 2, 4, and 10 aligned with expanded clones from other individuals in clusters 4α, 5α, 6α, and 11α (Figure 7). Nongermline nucleotides contributed to the amino acid similarities in many of these cases. Due to the small numbers of cells analyzed, it is possible that the unique sequences included in these clusters are part of undetected clonal expansions.
Subjects with similar SG CDR3s are discordant for autoantibodies to canonical SS antigens.
We next determined whether unrelated subjects sharing TCR CDR3 amino acid similarities from expanded SG clones were concordant for serologic reactivity to the canonical SS antigens Ro or La (Figures 6 and 7). Surprisingly, most of the subjects with CDR3β amino acid similarities between their expanded SG clones were discordant for Ro and La antibody status. Cluster 6β was one exception, as both subjects had anti-Ro antibodies. In contrast, 4 of 10 defined TCRα clusters (clusters 1α, 4α, 11α, and 12α) contained sequences from unrelated subjects who were concordant for Ro or La autoantibody status. However, TCRα clusters discordant for Ro antibody status were more prevalent than concordant examples, and no clusters were concordant for anti-La antibody positivity. These observations suggest that many of the expanded SG T cells may not recognize the prototypic Ro and La antigens.
Convergent recombination and TCR CDR3 sequence identity among CD4+ T cells from unrelated SS subjects.
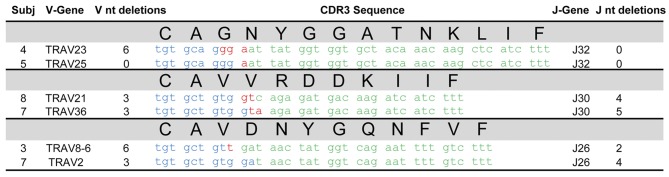
We next searched our SG database for examples of convergent recombination in which two or more CDR3s exhibit the same amino acid sequence, but through the use of different nucleotides, as convergent recombination provides evidence for antigenic selection (39). This analysis revealed 3 pairs of amino acid-identical CDR3α sequences from unrelated individuals (Figure 8). Convergent TCRs frequently arise from sequences with very few N-region additions and/or V/J gene nucleotide deletions. The 3 pairs of convergent sequences from SG had up to 8 V and J gene nucleotide deletions combined with a smaller number of N-region nucleotide additions. These data provide further strong support for recognition of common antigens by CD4+ T cells from SG tissue of unrelated individuals.
Figure 8. Convergent recombination between unrelated individuals in salivary gland CD4+ T cells.
Differing gene segments and/or N-region additions lead to identical complementarity-determining region 3 (CDR3) amino acid sequences. Nucleotide sequences deriving from the indicated V and J genes are shown in blue and green, respectively. N-region additions are indicated in red.
In addition to the examples of convergent recombination described above, two unique SG clones from subjects 8 and 9 shared the same CDR3α amino acid sequence (CAENAGGTSYGKLTF) encoded by identical nucleotides. In this case, there was 1 V gene and 5 J gene nucleotide deletions but no N-region nucleotide additions, suggesting that it may be a public CDR3.
CD4+ memory T cells using CDR3s that are identical or similar to those of clonal expansions in SG can be found in PB.
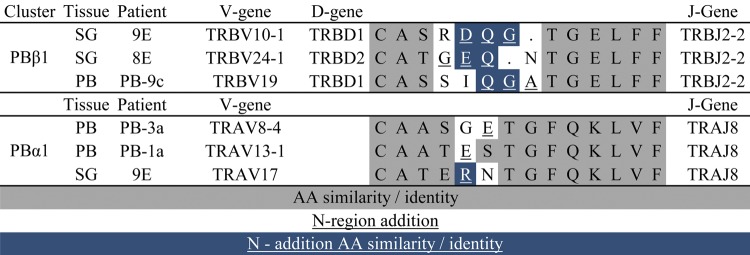
We next examined the CDR3 sequences of memory CD4+ T cells from PB to determine whether we could find evidence of circulating cells with CDR3s that were identical or similar to those in SG. Three examples of CD4+ memory T cells detected as unique sequences in PB had identical CDR3β sequences to cells in SG of the same subject. Two of these corresponded to SG expanded clones 5A and 7E. The third corresponded to a unique SG sequence (CDR3α: CVVSESSSNTGKLIF, CDR3β: CASSVDGQGIGYTF). Thus, T cell clonotypes occurring in SG can also be found in PB, though at a lower frequency.
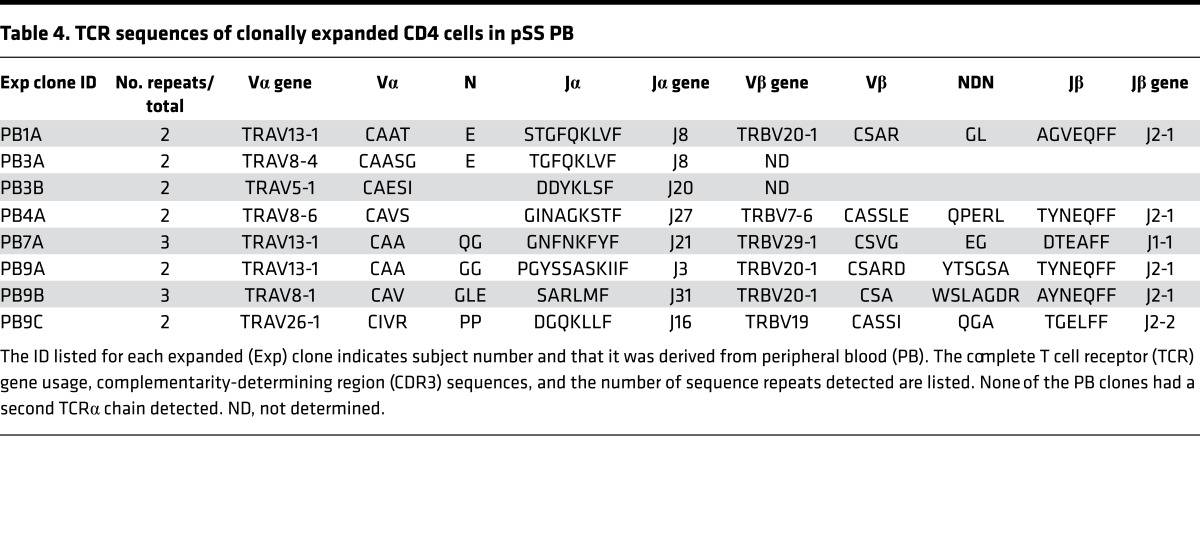
We performed a similar analysis of the CDR3s from the PB clonal expansions. Of the 8 detected clonal expansions in PB (Table 4), one (PB9C) exhibited CDR3β amino acid similarity both to SG expansions from the same individual (9E) (Figure 9) and from an unrelated individual (8E). Furthermore, two CDR3α sequences (PB1A and PB3A) from unrelated individuals displayed sequence similarity to each other and to expanded SG clone 9E. These alignments depended in part upon nongermline-encoded amino acids. Taken together, these results suggest that T cells capable of recognizing SG antigens may be present in the PB of pSS subjects. The significance of the other expanded clones in PB is unknown, but they may be the result of immune responses to common pathogens.
Table 4. TCR sequences of clonally expanded CD4 cells in pSS PB.
Figure 9. Alignment of complementarity-determining region 3 sequences from expanded clones from peripheral blood and salivary gland.
Shading and underlining are as described in the legend for Figure 6.
To determine if TCRs from expanded SG clones from subjects with pSS can also be found among the memory T cell repertoire of healthy controls, we examined the TCR repertoire of two healthy individuals without any evidence of autoimmune disease. As it is difficult to obtain SG tissue from healthy subjects, we used PB memory CD4+ T cells of two healthy controls positive for the DR3/DQ2 haplotype for this analysis. We employed a TCR deep-sequencing approach (40, 41) that minimizes amplification bias and sequence errors by applying unique molecular identifiers (UMI) to cDNA molecules prior to amplification. We found 7 of the 43 productive TCRα sequences and 1 of the 41 TCRβ sequences from the expanded SG clones shown in Table 3 in the control PB TCR deep-sequencing data. The frequency of these sequences was very low (ranging from 2.4 × 10–5 to 5.1 × 10–4). Only two pairs of α and β sequences were found. None of the PB sequences from SS subjects shown in Figure 9 or Table 4 were found in the PB of our two control subjects. So, while PB CD4+ memory T cells with the same CDR3 sequences as those in expanded clones from the SG of SS subjects can be found in healthy individuals, they are not common, and cells with the exact same TCR are extremely rare.
The TCR repertoire in SG of SS patients contains more related CDR3 sequences and is less diverse than that in PB of healthy individuals.
We hypothesized that the high degree of SG memory CD4+ T cell clonal expansions and the presence of related CDR3 sequences is an important characteristic of SS subjects. To test this hypothesis, we compared the PB TCR deep-sequencing data from the healthy controls described above to TCR deep-sequencing data generated from remaining stored SG memory CD4+ T cells from subjects 2–5 and 10 using the same deep-sequencing approach. These deep-sequencing data sets were analyzed by three different methods.
First, to assess the similarity of the TCR sequences in each tissue, we generated pairwise alignments of the TCR sequences and determined what percentage of them had pairwise maximum BLAST bit scores ≥50% of the maximum score. This threshold is the same one used to arrive at alignments shown in Figures 6 and 7. Only one copy of each unique sequence from each individual was included in the analysis. A median of 1,712 (range 638–2,948) productive sequences were recovered from deep sequencing of SGs from our SS subjects, a markedly smaller number than that recovered from PB of healthy controls (range 1,923,142–2,204,540). We therefore downsampled the PB sequences using probability proportional-to-size (PPS) sampling to match the total number of TCRα and TCRβ sequences recovered from SGs, 2,259 and 2,751, respectively. The downsampling was repeated 2,000 times to estimate a hypothetical distribution of the frequency of pairwise maximum BLAST similarity scores ≥50% among TCR sequences from PB of healthy controls. Figure 10 shows that the observed frequency of SG sequences meeting this similarity threshold (3.05% for TCRα chains and 6.5% for TCRβ chains) was far greater than what can be expected from a hypothetical sampling from healthy control PB data (P = 4.1 × 10–6 and P = 3.4 × 10–19 for the TCRα and TCRβ comparisons, respectively).
Figure 10. T cell receptor complementarity-determining region 3 sequences from salivary gland CD4+ T cells are significantly enriched in degree of sequence relatedness compared with healthy peripheral blood CD4+ T cells.
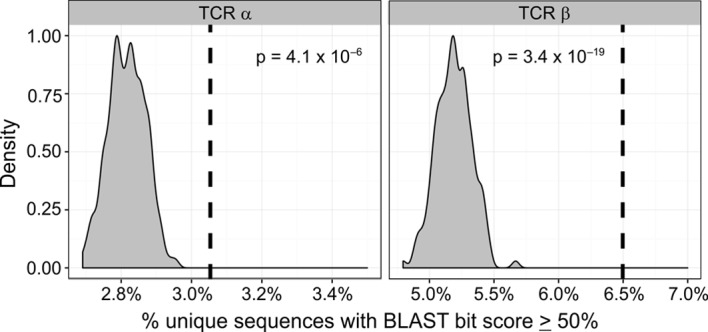
T cell receptor (TCR) deep-sequencing data from two healthy DR3/DQ2+ control subjects were sampled 2,000 times, and pairwise BLAST similarity scores were obtained. These were used to generate hypothetical distributions of the percentage of comparisons having scores ≥50% of the maximum value (gray). The dashed lines indicate the percentage of salivary gland (SG) CD4+ memory T cell TCR sequences having BLAST scores ≥50% of the maximum value in the actual SG TCR sequence data. Significance was calculated from the cumulative distribution function of the estimated hypothetical distribution (P = 4.1 × 10–6 and P = 3.4 × 10–19 goodness-of-fit tests for the TCRα and TCRβ comparisons, respectively).
Second, using the same TCR deep-sequencing data, we calculated the percentage of each individual’s repertoire, comprised by their 10 most frequent clonotypes. Repertoires that are less diverse will have a larger percentage comprised of their top 10 clonotypes. TCRα and β sequences were analyzed separately. As shown in Supplemental Figure 4, the top 10 clonotypes from the SGs of SS patients comprise a significantly larger proportion of the TCR repertoire than the top 10 clonotypes from PB of control subjects.
Finally, we conducted diversity analyses for healthy control PB versus SS SG TCR sequences similar to the ones shown for the single-cell sequencing data in Figure 3. Importantly, a UMI-based approach provides data that can be reliably compared using the Chao1 metric, as each UMI-labeled cDNA molecule approximately represents a single T cell (40, 42). As we had more sequences from deep sequencing, separate analyses were done for TCRα and β chains. PPS sampling was used to randomly select 500 UMI-based cDNA molecules from each individual’s deep-sequencing data. The selected cDNAs were clustered to define the operational taxonomic units (OTUs). Except for one metric in the β chain analysis (Berger-Parker Dominance), SG TCR sequences showed significantly less diversity for both α and β chains (Supplemental Figure 5).
Taken together, our analyses of the above TCR deep-sequencing data strongly support the concept that restricted diversity of the TCR repertoire of memory CD4+ T cells in SG of subjects with SS and their degree of relatedness are not features of the TCR repertoires of healthy individuals.
The frequency of clonally expanded CD4+ T cells in SG correlates with measures of oral disease.
SS is diagnosed using a combination of subjective symptoms, serology, SG histology, and objective measurements of oral and ocular dryness. Having established that SG CD4+ T cells are enriched for clonal expansions in individuals with SS, we investigated relationships between the degree of SG CD4+ T cell clonal expansions and clinical measures of disease. The frequency of clonally expanded SG T cells exhibited a strong inverse correlation with whole unstimulated salivary flow (r = –0.75, P = 0.015, Figure 11A). This analysis links glandular T cell expansion with decreased glandular function.
Figure 11. The frequency of detected salivary gland clonal expansions significantly correlates with measures of oral disease.
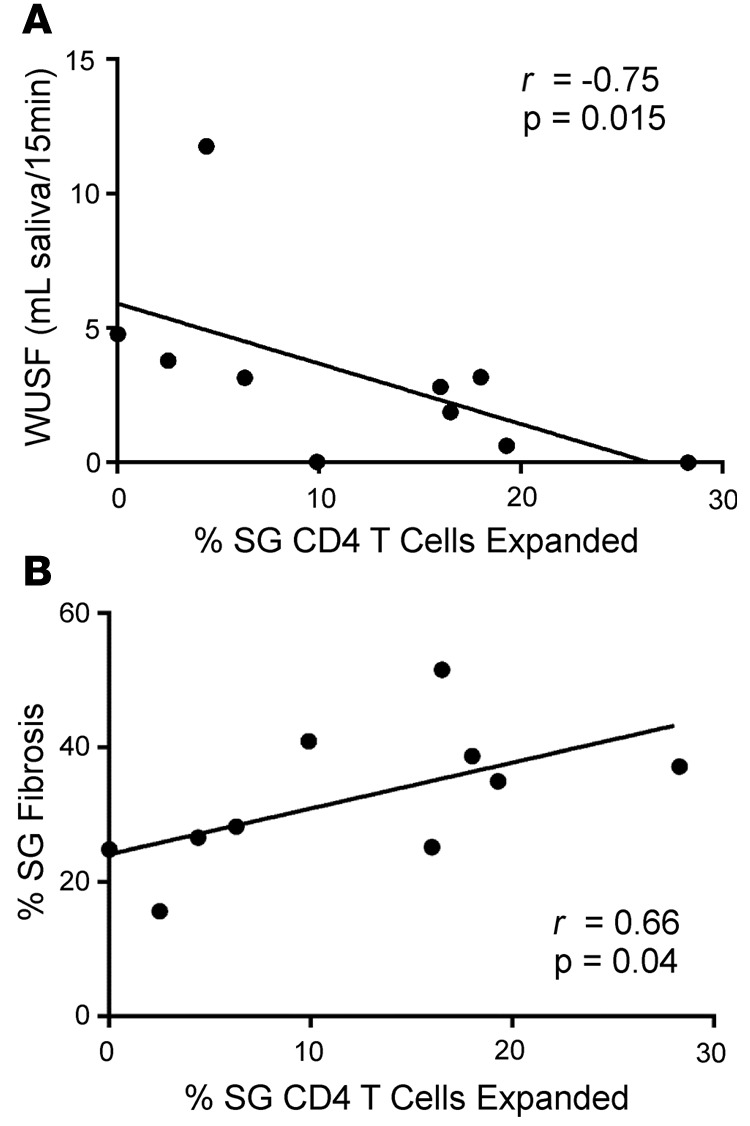
The percentages of clonally expanded salivary gland (SG) CD4 T cells (A) negatively correlate with whole unstimulated salivary flow (whole unstimulated salivary flow [WUSF]) and (B) positively correlate with the extent of SG fibrosis. Two-tailed Spearman rank correlation test.
The role of fibrosis in SG dysfunction is currently unclear. SS patients have elevated labial SG fibrosis compared with patients with sicca symptoms who do not meet SS criteria (43). To assess a possible correlation between degree of SG clonal expansion and extent of fibrotic damage in tissue, histopathological sections of SG tissue for each subject were scored for fibrotic damage, and the percentage of fibrotic tissue was calculated. As shown in Figure 11B, an increasing frequency of SG T cell expansions significantly correlated with an increasing degree of fibrosis (r = 0.66, P = 0.04, Spearman’s rank correlation, 2-tailed test). However, the correlation of the percentage of fibrotic tissue with whole unstimulated salivary flow did not reach statistical significance (P = 0.054, Spearman’s rank correlation). The frequency of SG clonal expansion did not correlate or associate with age, biopsy focus score, objective dry eye measures, Ro or La antibody status, hypergammaglobulinemia, or extraglandular manifestations (data not shown).
Discussion
We report here a comprehensive analysis of the CD4+ memory T cell TCR repertoire in pSS and a description of paired TCRα and β sequences from matched SG tissue and PB. We found clonal expansions of CD4+ memory T cells constituting 4% to 30% of the total cells analyzed in SG of 9 of 10 subjects. The extent of these expansions correlated with diminished SG function. Furthermore, amino acid sequence similarities involving nontemplated nucleotides among expanded clones both within and between individual subjects implicate antigenic selection as a primary determinant in shaping the SG repertoire in pSS.
Early studies of the TCR repertoire of SS SG T cells provided evidence of clonal expansion and preferential gene segment usage but were limited by the use of bulk tissue, the separate evaluation of either TCRα or β gene usage, and/or the small numbers of cells analyzed (17, 19, 22, 23). We believe our work is the first to report the direct ex vivo analysis of large numbers (>3,000 CDR3 sequences) of defined, SG-localized memory CD4+ T cells and matched PB samples at the single-cell level from pSS subjects with extensive clinical data. Our PCR approach captured sequences from >90% of the known TCRα and β variable genes (Supplemental Figure 2), indicating robust detection of TCR diversity. Paired TCR retrieval was more efficient from PB than from SG (median 69%, range 53%–73% vs. median 37%, range 19%–79%), possibly due to a nearly 4-fold higher surface TCR expression on cells from PB compared with SG (data not shown). However, the percentage of paired TCRs retrieved from cells with successfully amplified cDNA was similar between the two tissues (Supplemental Table 3), indicating no inherent difference in the performance of the PCR on cells from different tissues. Thus, the TCR data in this report represent an unbiased assessment of TCR repertoire from SS labial SG tissue compared with that from PB.
Numerous reports have linked dual TCR-expressing cells to autoimmunity (32, 34, 44). Thus, our observations that CD4+ T cells expressing two functional TCRα transcripts are significantly enriched in SG tissue compared with PB in pSS (Table 2) and are found in approximately 15% of the clonal expansions (Table 3) are of particular interest. During T cell development, the rearrangement of TCRβ chains is tightly regulated to ensure production of a single TCRβ chain in each T cell. However, TCRα rearrangement initiates simultaneously on both loci, sometimes resulting in the production of two in-frame rearrangements and, potentially, two distinct TCRs in a single cell (13, 14). Cells bearing two functional TCRs have been shown to directly contribute to the breadth of TCR diversity against foreign antigens (45). However, they may also inflict autoimmune tissue damage. For example, virus-specific T cells expressing dual TCRα chains were the mediators of autoimmune central nervous system damage after infection (44). Dual TCR expression may lead to increased autoreactivity by protecting autoreactive TCRs from deletion (32, 33) and by increasing the breadth of peptide recognition through enhanced positive selection (34). Our data suggest that such phenomena may contribute to the development of SS.
We present several lines of evidence supporting antigen-driven CD4+ T cell selection in SS SGs. First, we observed differential TCR gene segment usage in SG compared with PB (TRAV8-4, TRAV24, TRAV38-1, TRAJ22, TRAJ27, TRAV44, and TRBV29-1, Supplemental Figures 2 and 3). While earlier publications also showed biased usage of particular variable gene families in SG (46), our work directly assesses the gene usage of antigen-experienced CD4 T cells, not the sum of all T cells present in SG, which vary in composition by patient and include variable numbers of CD8+ cells (5, 6). Second, we found multiple clonal expansions in the SG of most subjects, which was likely the result of antigen-driven proliferation in the SG (Figure 2). Because sampling at the single-cell level imposes limitations on the number of cells that can be processed, the fact that nearly one-third of subject 10’s evaluated SG cells were part of clonal expansions is extraordinary. Third, when we assessed the diversity of the TCRs for both SG and PB at the cohort level, we found that the SG repertoires were significantly less diverse than those from PB (Figure 3). Fourth, we observed a high degree of CDR3 amino acid similarity among clonally expanded SS SG T cells as well as some single T cells within individuals and across unrelated individuals, particularly for TCRβ (Figures 6 and 7). Several of the similar or identical amino acids in our alignments were encoded by nontemplated nucleotides. This is unlikely to occur by chance, i.e., without antigenic selection. Fifth, we also observed two examples of nucleotide-identical CDR3 sequences from unrelated individuals. Finally, we noted convergent recombination yielding amino acid-identical CDR3 sequences in SG T cells from 3 pairs of unrelated subjects (Figure 8). As a whole, our data provide strong evidence for antigen-driven CD4+ T cell proliferation and selection in SG of SS subjects and further suggest that some SG T cells from unrelated individuals may recognize the same SG antigen(s).
Although previous studies examining the TCR repertoire of SG resident T cells noted a number of CDR3 sequence motifs (17, 19, 22, 23, 46), we did not find the same motifs in our data set. This may be due to a number of factors, including differing ethnicity, HLA haplotype, or T cell type, as these prior studies were conducted in Japanese populations that evaluated total T cells, including naive and CD8+ T cells. Our study selectively evaluated memory CD4+ T cells from individuals of European American descent or individuals of mixed European American/American Indian descent carrying the HLA DR3/DQ2 haplotype. The CDR3 sequences from SS SG that we describe are novel, as there were no direct BLAST matches in the public NCBI database. Although we did not identify a broad “one-size-fits-all” CDR3 motif, we did uncover strong similarities among different expanded clones within individuals and among unrelated subjects, as described above. Perhaps one reason for the heterogeneity of motifs we observed is that, although our subjects share the HLA DR3/DQ2 risk haplotype, the HLA restriction of the TCRs reported here remains unknown.
Immune-driven clonal selection typically occurs at the site of pathological insult, but this does not preclude circulation of the disease-relevant TCR clones in the periphery. A recent study examining rheumatoid arthritis patients found that circulating T cells with surface phenotypic markers matching those in the inflamed synovium shared TCR clonotypes (38), suggesting that cells expanding in the target tissue circulate systemically. In another rheumatoid arthritis study, the same clonotypes were observed in synovial tissue and blood but with differing transcriptional profiles (47). In our study, we found nucleotide-identical cells from 3 subjects in both PB and SG; in 2 cases, the CDR3s found in PB matched those of expanded SG clones of the same individual (5A and 7E). From this, we conclude that expanded T cell clones found in SG of SS subjects are also likely to be frequently found in PB of pSS subjects. However, the frequency of CD4 memory T cells bearing TCRs identical to those of the expanded SG CD4 T cell clones from our cohort was very low in PB of 2 healthy individuals. Further TCR deep-sequencing studies employing more subjects and controls will reveal the true extent of repertoire overlap between the two tissues, and single-cell whole-transcriptome RNA sequencing will give insight into the functional relationship between SG and PB cells of matching clonotypes.
Both focal lymphocytic infiltrates and autoantibodies to the hallmark Ro and La antigens are key features of SS (3). In approximately one-third of pSS patients, germinal center-like structures form in labial SG tissue (48), indicating tissue-antigen driven immune responses. Consistent with this, Ro and La autoantibodies are made in the SG in situ (48–50). Although the presence of these structures has been reported to be a useful predictor of non-Hodgkin’s lymphoma (51), no direct evidence for the role of T cells or germinal center-like structures in oral symptoms has been reported. We detected no association between presence of ectopic germinal center-like structures or presence of extraglandular disease with degree of SG T cell clonal expansion (data not shown). Rather, we found direct correlations between the frequency of clonally expanded CD4 T cells and measures of SG dysfunction and tissue damage. Increased clonal expansions correlated with reduced salivary flow and increased SG fibrosis (Figure 10), indicating that these cells may be direct pathogenic effectors in the SG. In earlier studies with much larger sample sizes, reduced whole unstimulated salivary flow correlated with increased age (52), focus score ≥1.0 (53), rheumatoid factor positivity (53, 54), anti-nuclear autoantibody titer >1:320 (53, 54), elevated serum IgM (54), and duration of disease (55). These relationships were not observed in our study, perhaps because of patient heterogeneity. Bookman et al. observed “definite association” between reduced whole stimulated saliva production, increased SG focus score, increased grade of SG fibrosis, duration of dry mouth symptoms, and tooth loss/damage (56). However, that study found no relationship between whole unstimulated salivary flow and SG fibrosis. Unlike in prior reports, the subject cohort in our study was selected for the presence of the HLA DR3/DQ2 risk haplotype as well as a focus score ≥1. We speculate that patients in this selected subgroup may be more likely to harbor SG T cells that promote oral dryness and fibrosis.
One of the long-standing questions regarding SS is what drives the exocrine tissue selectivity of disease? We hypothesize that exocrine gland-specific antigens are recognized by self-reactive immune cells, leading to tissue-specific dysfunction and damage. Identifying these antigens will provide a crucial piece of the puzzle and is critical for understanding the tissue selectivity and pathogenesis of SS. To this end, the identification of paired TCRα and β gene sequences of clonally expanded CD4+ T cells is a crucial first step toward this objective. These TCRs can be expressed in a cell line that is responsive to antigen stimulation in vitro, such as the murine T cell hybridoma, 58α–β–, which has been previously used to express human TCRs (57). Then, we can screen for reactivity to Ro and La, viral antigens (e.g., EBV), candidate SG antigens, a SG expression library, or fractions of SG or acinar cell extracts. This latter approach has recently been used to successfully identify autoantigens in the NOD diabetic mouse (58). Interestingly, the observation that several of our clustered expansions contain CDR3s from unrelated subjects with discordant Ro/La autoantibody status (Figures 6 and 7) provides evidence that some of the clonally expanded T cells may recognize antigens that are not Ro or La proteins. Some of them may recognize antigens from infectious agents or other environmental sources. Our demonstration that the frequency of CD4+ clonal expansions correlates with oral disease features documents the importance of identifying these antigens, as knowledge of disease-relevant SG antigens may lead to the development of new therapeutic strategies for SS.
Methods
Subjects and sample collection.
Samples and clinical data were obtained from subjects evaluated at the OMRF Sjögren’s Syndrome Research Clinic. All participants with SS evaluated in this study met the American-European Consensus Group Criteria for pSS (3) and the 2012 provisional American College of Rheumatology classification criteria for pSS (59). Rheumatologic, ophthalmologic, and oral examinations were performed and samples were collected during clinic visits. Laboratory tests and data from medical records were collected as previously described (60). Additional cohort selection criteria were based on (a) positive PCR screening tests for HLA DRB1*0301 (DR3) and DQB1*0201 (DQ2) alleles using previously described primers (61) and (b) minor labial SG focus score ≥1. Healthy individuals donating PB were positive by PCR screening tests for HLA DRB1*0301 (DR3) and DQB1*0201 (DQ2) alleles, were ages 42 and 58, and had no known autoimmune disorders.
HLA imputation.
To confirm the presence of HLA DR3 and DQ2 alleles, classical HLA alleles were imputed from SNP data by the HLA imputation software that applies an attribute BAGging technique with European-specific models of 4-digit resolution (human genome reference 19), as described previously (62). Genotype data were obtained using Illumina Omni-Express arrays (31) employing the 4,189 MHC region SNPs on chromosome 6 (27–34 Mb, human genome reference 19) that were common across both arrays. Maximum likelihood models are reported.
SG tissue processing and cell sorting.
Labial SG and PB samples were collected from subjects and processed for single-cell sorting. Biopsy tissue was mechanically separated and enzymatically digested as previously described (49). PB mononuclear cells were isolated from fresh blood samples using density-gradient centrifugation (Ficoll-Paque; GE Healthcare Life Sciences). Cell suspensions were filtered through a 40-μm cell strainer and counted by trypan blue exclusion. SG and PB mononuclear cell suspensions were stained with a cocktail of antibodies, phycoerythrin (PE) anti-CD3 (clone SK7, BD Biosciences), PE-Cy5 anti-CD4 (RPA-T4, BD Biosciences), Alexa Fluor 488 anti-CD8 (RPA-T8, BD Biosciences and eBiosciences), and V450 anti-CD45RA (HI100, BD Biosciences and eBiosciences), followed by bulk sorting of the CD3+CD4+CD8–CD45RA– population on a FACSAria (BD Biosciences) (Supplemental Figure 1). Single cells were sorted into 96-well PCR plates containing 10 μl/well of catch buffer (RNase-free water containing 10 mM Tris, pH 8.0, and 750 units/ml of RNasin [Promega]) using a MoFlo XDP (Beckman Coulter), leaving the bottom row of each plate free of cells as negative controls. Plates were immediately sealed (Microseal F, Bio-Rad) and placed on dry ice until transfer to a –80°C freezer where they were stored until processing for single-cell RT-PCR. Numbers of SG processed (median 6 SG per biopsy, range 3–8) and numbers of memory CD4+ T cells isolated (median 4,756, range 408–48,628) from SG single-cell suspensions varied considerably among participants (Supplemental Table 1). A maximum of 378 viable single cells (range 168–378) were sorted per subject.
Single-cell RT-PCR.
The single-cell RT-PCR protocol for TCRs was adapted from Wang et al. (63) with minor modifications. In the first step, cDNA was generated using the iScript cDNA Synthesis Kit (Bio-Rad) by adding 2 μl of 5X iScript reaction mix and 0.5 μl of iScript enzyme mix/well to each 10 μl volume whole-cell lysate in catch buffer. Plates were incubated at 25°C for 5 minutes, 42°C for 30 minutes, and 85°C for 5 minutes. In the second step, all cDNA material was added to a first-round multiplex PCR containing Advantage 2 Polymerase (Clontech) and external primers for both TCRα and β V- and C-regions (63) in a total volume of 50 μl. Reactions were cycled at 94°C for 20 seconds, 52°C for 20 seconds, and 72°C for 45 seconds for a total of 40 cycles. In the third step, two separate nested second-round PCRs were performed under the same cycling conditions with 1.5 μl of first-round PCR product as template to amplify the α and β chains. These reactions utilized NEB Taq polymerase (New England Biolabs) with 2 mM MgCl2, 1.25 pmol of each nested (internal) V-region primer (for α or β) (63), and 5 pmol of nested C-region primer (63). Amplified products were detected by agarose gel electrophoresis, purified by spin column (Qiagen or Enzymax), and Sanger-sequenced using the following primers: UpCβ: 5′-GGTGTGGGAGATCTCTGCTTCTGA-3′ for TCRβ amplicons and CaNV: 5′-TTTAGAGTCTCTCAGCTGGTACACGG-3′ for TCRα amplicons. When dual α sequences were evident, the second-round PCR products were cloned into a TOPO-TA vector (Invitrogen) and both transcripts were retrieved by sequencing multiple inserts.
Single-cell TCR sequence and repertoire analysis.
TCR nucleotide sequences from single-cell RT-PCR were deposited in GenBank (accession numbers KX075774-KX077178 and KX074222-KX0757733 for SG and PB sequences, respectively). Nucleotide sequences were compared to the reference sequences from the international ImMunoGeneTics (IMGT) information system (http://www.imgt.org; ref 64), using IMGT/HighV-QUEST to assign gene segments (65). VDJviz (42) was used to annotate nongermline-encoded nucleotides and amino acids. Clonal expansions were defined as two or more cells sharing nucleotide-identical CDR3 sequences within a given tissue of one individual.
The percentages of SG or PB cells that were part of clonal expansions were calculated on a per subject basis as follows: (number of SG or PB cells in all clonal expansions/number of SG or PB cells with retrieved TCR sequences) × 100 = percentage clonal expansion.
TCR diversity analysis.
Diversity analysis of the TCR repertoire was conducted with QIIME, v1.8 (66) and USEARCH, v8.0 (67). Nucleotide sequences were clustered into OTUs at 97% identity, using the “uclust” algorithm with default parameters. Sequences for each individual were mapped to these OTUs with the “usearch_global” algorithm at 97% identity and default parameters. A reference taxonomy was constructed for TCR sequences using their IMGT-assigned gene segments. Alpha diversity metrics evaluating richness (observed OTUs, Chao1) and evenness (Berger-Parker Dominance, Simpson’s Evenness) and Simpson’s Diversity Index were calculated in QIIME.
TCR multiple sequence alignment.
CDR3 amino acid sequences from expanded SG clones of all subjects were evaluated for within- and between-subject similarities using protein-protein BLAST, v2.2.29+ (68). Alignments were scored using the PAM30 substitution matrix with the following parameters: word_size = 2, window_size = 40, gapopen = 9, gapextend = 1, threshold = 11, comp_based_stats = 0. Similarity between two sequences was calculated as the percentage maximal BLAST bit score (i.e., the BLAST bit score divided by the bit score of the Query Sequence’s alignment to itself × 100). The higher of two scores between each pairwise comparison was retained. Sequences were clustered by similarity using Euclidean distance and Ward linkage metrics and visualized as heatmaps. Clusters containing BLAST bit scores equal to or exceeding a threshold of 50% of the maximum possible bit scores were selected and aligned using CLC Sequence Viewer (Qiagen) followed by manual adjustment to maximize similarities.
Sequence motifs derived from visual inspection of multiple alignments were used to query databases containing all SG or PB sequences from all participants using the “protein pattern find” function of the Sequence Manipulation Suite (69). Return of the parental expanded sequences used to create the motifs served to validate the motifs used in the search strategy. Unique CDR3 sequences sharing similarity with aligned clusters of clonally expanded sequences were added to the alignments.
TCR deep sequencing and analysis.
Next-generation TCR sequencing was performed on bulk populations of sorted CD4+CD45RA– memory T cells from SG of SS subjects with sufficient sample available after single-cell sorts (subjects 2, 3, 4, 5, and 10) and from PB of two healthy control subjects according to the method of Egorov et al. (40, 41). The number of SG cells ranged from 6.8 × 103 to 47 × 103, and the number of healthy control PB cells was 2 × 106 cells/subject. Cells were stored frozen in TriZol until they were processed for RNA. Briefly, cDNA was synthesized and UMI-labeled (70) using the SmartScribe RT kit (Clontech) followed by use of all the cDNA in the first-round PCR reactions using primers as specified by Egorov et al. (41) and Q5 Polymerase (NEB). First-round PCR products were purified (Qiagen) to remove excess primers, and a second-round PCR was performed with nested primers. This product was gel purified (Qiagen) and immediately quantified and used for library preparation using the NEBNext Library kit for Illumina (NEB). Library integrity was checked by TapeStation (Agilent Technologies) analysis followed by quantification using qPCR. The healthy control PB samples and the pSS-SG samples were sequenced separately on Illumina MiSeq paired-end 150-bp runs using separate indexing for α and β TCRs by the OMRF Next Generation Sequencing Core. UMI-tagged data sets were processed using MIGEC software to correct for PCR amplification biases and remove PCR/sequencing errors as described previously (41). V-D-J mapping and clonotype assembly was performed using the MIGEC/CdrBlast routine. TCR deep-sequencing data sets were submitted to the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP073308.
Measurement of SG fibrosis.
Degree of SG fibrosis was quantitatively assessed from imaged, H&E-stained SG cross sections (4–6 SG cross sections per subject) using morphometric grids (43). Whole-gland digital reconstructions were assembled (in Zeiss ZenBlue software) from overlapping ×200 high-resolution images taken on a Zeiss 710 confocal microscope. The total area of each SG cross section was measured using the Automated Measurement program feature of Axiovision 4.8 software (Zeiss) with manual adjustments. Morphometric grids were applied at a standardized scaling to all images using ImageJ (NIH). Each grid square ≥50% occupied by tissue was scored. Scored squares in which available tissue displayed ≥50% fibrotic change were marked as positive. The number of fibrosis-positive squares in each SG cross section was multiplied by 2,500 μm2 (the area of each grid square). This value was divided by the total section area and then multiplied by 100 to generate the percentage of fibrosis area for each SG cross section. Percentages of fibrosis area for all available cross sections from a given subject were averaged to obtain the mean percentage of SG fibrosis area.
Statistics.
Differences in frequency of dual TCRα usage in SG versus PB were evaluated using the χ2 test, in which cells having only a nonfunctional TCRα chain detected were counted as cells bearing two TCRα transcripts. Preferential usage of particular gene segments in SG compared to PB was evaluated by the matched-pairs Wilcoxon rank-sum test using only unique sequences. A nonparametric, 2-tailed, 2-sample t test was used to compare alpha diversity metrics between PB and SG samples. Hypothetical distributions of similarity score percentages from repeated PPS samples of PB data were modeled as gamma distributions to assess extremeness of similarity scores observed in SG samples. Relationships with clinical values were evaluated by 2-tailed Spearman rank correlations. P values of less than 0.05 were considered to be significant.
Study approval.
The study was approved by the OMRF Institutional Review Board and was conducted according to the principles of the Declaration of Helsinki. All samples and data were collected following written informed consent.
Author contributions
ADF, JSM, LFT, and MLJ conceived of and designed the study. AR, LR, DML, DUS, RHS, and KLS evaluated the subjects and/or collected the clinical data. MLJ, KML, CL, ZP, GBW, and ADF collected the experimental data. MLJ, KML, RCP, ZP, KMG, JAK, GBW, MS, DMC, CJL, CGM, and LFT analyzed the data. MLJ, KML, LFT, and ADF wrote the paper. All authors reviewed and approved the final version.
Supplementary Material
Acknowledgments
The authors are indebted to Glen D. Houston and Kimberly S. Hefner for contributing to the evaluation of oral and eye pathology for this study. The authors thank Beverly Hurt for figure preparation and Louise Williamson for clerical assistance. This study was supported by NIH Centers of Research Translation grant 1P50AR060804. The contents are the sole responsibility of the authors and do not necessarily represent the official views of the NIH. M. Shugay is supported by a fellowship grant from the Russian Foundation for Basic Research (16-34-60179). D.M. Chudakov is supported by the Molecular and Cell Biology program of the Russian Academy of Sciences.
Footnotes
Jacen S. Moore’s present address is: Department of Clinical Laboratory Sciences, University of Texas at El Paso, El Paso, Texas, USA.
Donald U. Stone’s present address is: Department of Ophthalmology, Johns Hopkins University, Baltimore, Maryland, USA, and King Khaled Eye Specialist Hospital, Riyadh, Saudi Arabia.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2016;1(8):e85609. doi:10.1172/jci.insight.85609.
Contributor Information
Michelle L. Joachims, Email: Michelle-Joachims@omrf.org.
Kerry M. Leehan, Email: kerry-leehan@omrf.org.
Christina Lawrence, Email: christina-lawrence@omrf.org.
Richard C. Pelikan, Email: richard-pelikan@omrf.org.
Jacen S. Moore, Email: jsmaiermoore@utep.edu.
Zijian Pan, Email: zijian-pan@omrf.org.
Astrid Rasmussen, Email: astrid-rasmussen@omrf.org.
Lida Radfar, Email: lida-radfar@ouhsc.edu.
Kiely M. Grundahl, Email: kiely-grundahl@omrf.org.
Mikhail Shugay, Email: mikhail.shugay@gmail.com.
Donald U. Stone, Email: donstone13@gmail.com.
R. Hal Scofield, Email: Hal-Scofield@omrf.org.
Courtney G. Montgomery, Email: courtney-montgomery@omrf.org.
References
- 1.Jonsson R, Vogelsang P, Volchenkov R, Espinosa A, Wahren-Herlenius M, Appel S. The complexity of Sjogren’s syndrome: novel aspects on pathogenesis. Immunol Lett. 2011;141(1):1–9. doi: 10.1016/j.imlet.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Mavragani CP, Moutsopoulos HM. Sjogren’s syndrome. Annu Rev Pathol. 2014;9:273–285. doi: 10.1146/annurev-pathol-012513-104728. [DOI] [PubMed] [Google Scholar]
- 3.Vitali C, et al. Classification criteria for Sjogren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61(6):554–558. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daniels TE. Labial salivary gland biopsy in Sjogren’s syndrome. Assessment as a diagnostic criterion in 362 suspected cases. Arthritis Rheum. 1984;27(2):147–156. doi: 10.1002/art.1780270205. [DOI] [PubMed] [Google Scholar]
- 5.Fox RI, Carstens SA, Fong S, Robinson CA, Howell F, Vaughan JH. Use of monoclonal antibodies to analyze peripheral blood and salivary gland lymphocyte subsets in Sjogren’s syndrome. Arthritis Rheum. 1982;25(4):419–426. doi: 10.1002/art.1780250410. [DOI] [PubMed] [Google Scholar]
- 6.Adamson TC, 3rd, Fox RI, Frisman DM, Howell FV. Immunohistologic analysis of lymphoid infiltrates in primary Sjogren’s syndrome using monoclonal antibodies. J Immunol. 1983;130(1):203–208. [PubMed] [Google Scholar]
- 7.Talal N, Sylvester RA, Daniels TE, Greenspan JS, Williams RC., Jr T and B lymphocytes in peripheral blood and tissue lesions in Sjogren’s syndrome. J Clin Invest. 1974;53(1):180–189. doi: 10.1172/JCI107536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moutsopoulos HM, Hooks JJ, Chan CC, Dalavanga YA, Skopouli FN, Detrick B. HLA-DR expression by labial minor salivary gland tissues in Sjogren’s syndrome. Ann Rheum Dis. 1986;45(8):677–683. doi: 10.1136/ard.45.8.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zumla A, Mathur M, Stewart J, Wilkinson L, Isenberg D. T cell receptor expression in Sjogren’s syndrome. Ann Rheum Dis. 1991;50(10):691–693. doi: 10.1136/ard.50.10.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohyama Y, et al. T-cell receptor Vα and Vβ gene use by infiltrating T cells in labial glands of patients with Sjogren’s syndrome. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1995;79(6):730–737. doi: 10.1016/S1079-2104(05)80308-7. [DOI] [PubMed] [Google Scholar]
- 11.Matthews JB, et al. Primed and naive helper T cells in labial glands from patients with Sjogren’s syndrome. Virchows Arch A Pathol Anat Histopathol. 1991;419(3):191–197. doi: 10.1007/BF01626347. [DOI] [PubMed] [Google Scholar]
- 12.Christodoulou MI, Kapsogeorgou EK, Moutsopoulos HM. Characteristics of the minor salivary gland infiltrates in Sjogren’s syndrome. J Autoimmun. 2010;34(4):400–407. doi: 10.1016/j.jaut.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Petrie HT, Livak F, Schatz DG, Strasser A, Crispe IN, Shortman K. Multiple rearrangements in T cell receptor alpha chain genes maximize the production of useful thymocytes. J Exp Med. 1993;178(2):615–622. doi: 10.1084/jem.178.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor alpha chains: dual receptor T cells. Science. 1993;262(5132):422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 15.Smith MD, et al. Selective expression of V beta families by T cells in the blood and salivary gland infiltrate of patients with primary Sjogren’s syndrome. J Rheumatol. 1994;21(10):1832–1837. [PubMed] [Google Scholar]
- 16.Kay RA, et al. An abnormal T cell repertoire in hypergammaglobulinaemic primary Sjogren’s syndrome. Clin Exp Immunol. 1991;85(2):262–264. doi: 10.1111/j.1365-2249.1991.tb05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto I, et al. Common T cell receptor clonotype in lacrimal glands and labial salivary glands from patients with Sjogren’s syndrome. J Clin Invest. 1996;97(8):1969–1977. doi: 10.1172/JCI118629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki M, et al. Accumulation of common T cell clonotypes in the salivary glands of patients with human T lymphotropic virus type I-associated and idiopathic Sjogren’s syndrome. J Immunol. 2000;164(5):2823–2831. doi: 10.4049/jimmunol.164.5.2823. [DOI] [PubMed] [Google Scholar]
- 19.Sumida T, et al. T cell receptor repertoire of infiltrating T cells in lips of Sjogren’s syndrome patients. J Clin Invest. 1992;89(2):681–685. doi: 10.1172/JCI115635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ajjan RA, et al. Analysis of the T-cell receptor Valpha repertoire and cytokine gene expression in Sjogren’s syndrome. Br J Rheumatol. 1998;37(2):179–185. doi: 10.1093/rheumatology/37.2.179. [DOI] [PubMed] [Google Scholar]
- 21.Sumida T, et al. TCR in Fas-sensitive T cells from labial salivary glands of patients with Sjogren’s syndrome. J Immunol. 1997;158(2):1020–1025. [PubMed] [Google Scholar]
- 22.Matsumoto I, et al. Single cell analysis of T cells infiltrating labial salivary glands from patients with Sjogren’s syndrome. Int J Mol Med. 1999;4(5):519–527. doi: 10.3892/ijmm.4.5.519. [DOI] [PubMed] [Google Scholar]
- 23.Sumida T, Kita Y, Yonaha F, Maeda T, Iwamoto I, Yoshida S. T cell receptor Vα repertoire of infiltrating T cells in labial salivary glands from patients with Sjogren’s syndrome. J Rheumatol. 1994;21(9):1655–1661. [PubMed] [Google Scholar]
- 24.Krischer JP, et al. Screening strategies for the identification of multiple antibody-positive relatives of individuals with type 1 diabetes. J Clin Endocrinol Metab. 2003;88(1):103–108. doi: 10.1210/jc.2002-020760. [DOI] [PubMed] [Google Scholar]
- 25.Weiner ES, et al. Prognostic significance of anticentromere antibodies and anti-topoisomerase I antibodies in Raynaud’s disease. A prospective study. Arthritis Rheum. 1991;34(1):68–77. doi: 10.1002/art.1780340111. [DOI] [PubMed] [Google Scholar]
- 26.van Venrooij WJ, van Beers JJ, Pruijn GJ. Anti-CCP antibodies: the past, the present and the future. Nat Rev Rheumatol. 2011;7(7):391–398. doi: 10.1038/nrrheum.2011.76. [DOI] [PubMed] [Google Scholar]
- 27.Michels AW, von Herrath M. 2011 Update: antigen-specific therapy in type 1 diabetes. Curr Opin Endocrinol Diabetes Obes. 2011;18(4):235–240. doi: 10.1097/MED.0b013e32834803ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinman L. Inverse vaccination, the opposite of Jenner’s concept, for therapy of autoimmunity. J Intern Med. 2010;267(5):441–451. doi: 10.1111/j.1365-2796.2010.02224.x. [DOI] [PubMed] [Google Scholar]
- 29.Walczak A, Siger M, Ciach A, Szczepanik M, Selmaj K. Transdermal application of myelin peptides in multiple sclerosis treatment. JAMA Neurol. 2013;70(9):1105–1109. doi: 10.1001/jamaneurol.2013.3022. [DOI] [PubMed] [Google Scholar]
- 30.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev. 2006;6(12):883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 31.Lessard CJ, et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren’s syndrome. Nat Genet. 2013;45(11):1284–1292. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zal T, Weiss S, Mellor A, Stockinger B. Expression of a second receptor rescues self-specific T cells from thymic deletion and allows activation of autoreactive effector function. Proc Natl Acad Sci U S A. 1996;93(17):9102–9107. doi: 10.1073/pnas.93.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarukhan A, Garcia C, Lanoue A, von Boehmer H. Allelic inclusion of T cell receptor alpha genes poses an autoimmune hazard due to low-level expression of autospecific receptors. Immunity. 1998;8(5):563–570. doi: 10.1016/S1074-7613(00)80561-0. [DOI] [PubMed] [Google Scholar]
- 34.Ni PP, Solomon B, Hsieh CS, Allen PM, Morris GP. The ability to rearrange dual TCRs enhances positive selection, leading to increased Allo- and Autoreactive T cell repertoires. J Immunol. 2014;193(4):1778–1786. doi: 10.4049/jimmunol.1400532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris GP, Uy GL, Donermeyer D, Dipersio JF, Allen PM. Dual receptor T cells mediate pathologic alloreactivity in patients with acute graft-versus-host disease. Sci Transl Med. 2013;5(188):e85609. doi: 10.1126/scitranslmed.3005452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321(1-2):182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 38.Spreafico R, et al. A circulating reservoir of pathogenic-like CD4+ T cells shares a genetic phenotypic signature with the inflamed synovial micro-environment. Ann Rheum Dis. 2016;75(2):459–465. doi: 10.1136/annrheumdis-2014-206226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venturi V, et al. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci U S A. 2006;103(49):18691–18696. doi: 10.1073/pnas.0608907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Britanova OV, et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J Immunol. 2014;192(6):2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 41.Egorov ES, et al. Quantitative profiling of immune repertoires for minor lymphocyte counts using unique molecular identifiers. J Immunol. 2015;194(12):6155–6163. doi: 10.4049/jimmunol.1500215. [DOI] [PubMed] [Google Scholar]
- 42.Shugay M, et al. VDJtools: unifying post-analysis of T cell receptor repertoires. PLoS Comput Biol. 2015;11(11):e85609. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leehan KM, et al. Precisely quantified fibrosis in labial salivary glands predicts Sjögren’s syndrome classification in a multiple regression model. Arthritis Rheumatol. 2014;66:e85609 [Google Scholar]
- 44.Ji Q, Perchellet A, Goverman JM. Viral infection triggers central nervous system autoimmunity via activation of CD8+ T cells expressing dual TCRs. Nat Immunol. 2010;11(7):628–634. doi: 10.1038/ni.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He X, et al. Dual receptor T cells extend the immune repertoire for foreign antigens. Nat Immunol. 2002;3(2):127–134. doi: 10.1038/ni751. [DOI] [PubMed] [Google Scholar]
- 46.Sumida T, Matsumoto I, Maeda T, Nishioka K. T-cell receptor in Sjogren’s syndrome. Br J Rheumatol. 1997;36(6):622–629. doi: 10.1093/rheumatology/36.6.622. [DOI] [PubMed] [Google Scholar]
- 47.Ishigaki K, et al. Quantitative and qualitative characterization of expanded CD4+ T cell clones in rheumatoid arthritis patients. Sci Rep. 2015;5:e85609. doi: 10.1038/srep12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jonsson MV, Skarstein K, Jonsson R, Brun JG. Serological implications of germinal center-like structures in primary Sjogren’s syndrome. J Rheumatol. 2007;34(10):2044–2049. [PubMed] [Google Scholar]
- 49.Maier-Moore JS, et al. Antibody-secreting cell specificity in labial salivary glands reflects the clinical presentation and serology in patients with Sjogren’s syndrome. Arthritis Rheumatol. 2014;66(12):3445–3456. doi: 10.1002/art.38872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tengner P, Halse AK, Haga HJ, Jonsson R, Wahren-Herlenius M. Detection of anti-Ro/SSA and anti-La/SSB autoantibody-producing cells in salivary glands from patients with Sjogren’s syndrome. Arthritis Rheum. 1998;41(12):2238–2248. doi: 10.1002/1529-0131(199812)41:12<2238::AID-ART20>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 51.Theander E, et al. Lymphoid organisation in labial salivary gland biopsies is a possible predictor for the development of malignant lymphoma in primary Sjogren’s syndrome. Ann Rheum Dis. 2011;70(8):1363–1368. doi: 10.1136/ard.2010.144782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vissink A, Bootsma H, Spijkervet FK, Hu S, Wong DT, Kallenberg CG. Current and future challenges in primary Sjogren’s syndrome. Curr Pharm Biotechnol. 2012;13(10):2026–2045. doi: 10.2174/138920112802273254. [DOI] [PubMed] [Google Scholar]
- 53.Daniels TE, et al. Associations between salivary gland histopathologic diagnoses and phenotypic features of Sjogren’s syndrome among 1,726 registry participants. Arthritis Rheum. 2011;63(7):2021–2030. doi: 10.1002/art.30381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malladi AS, et al. Primary Sjogren’s syndrome as a systemic disease: a study of participants enrolled in an international Sjogren’s syndrome registry. Arthritis Care Res (Hoboken) 2012;64(6):911–918. doi: 10.1002/acr.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jensen SB, Vissink A. Salivary gland dysfunction and xerostomia in Sjogren’s syndrome. Oral Maxillofac Surg Clin North Am. 2014;26(1):35–53. doi: 10.1016/j.coms.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Bookman AA, et al. Whole stimulated salivary flow: correlation with the pathology of inflammation and damage in minor salivary gland biopsy specimens from patients with primary Sjogren’s syndrome but not patients with sicca. Arthritis Rheum. 2011;63(7):2014–2020. doi: 10.1002/art.30295. [DOI] [PubMed] [Google Scholar]
- 57.Blank U, Boitel B, Mege D, Ermonval M, Acuto O. Analysis of tetanus toxin peptide/DR recognition by human T cell receptors reconstituted into a murine T cell hybridoma. Eur J Immunol. 1993;23(12):3057–3065. doi: 10.1002/eji.1830231203. [DOI] [PubMed] [Google Scholar]
- 58.Delong T, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351(6274):711–714. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shiboski SC, et al. American College of Rheumatology classification criteria for Sjogren’s syndrome: a data-driven, expert consensus approach in the Sjogren’s International Collaborative Clinical Alliance cohort. Arthritis Care Res (Hoboken) 2012;64(4):475–487. doi: 10.1002/acr.21591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rasmussen A, et al. Comparison of the American-European Consensus Group Sjogren’s syndrome classification criteria to newly proposed American College of Rheumatology criteria in a large, carefully characterised sicca cohort. Ann Rheum Dis. 2014;73(1):31–38. doi: 10.1136/annrheumdis-2013-203845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bunce M, et al. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;46(5):355–367. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 62.Zheng X, et al. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14(2):192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG. T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med. 2012;4(128):e85609. doi: 10.1126/scitranslmed.3003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(Database issue):D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 68.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:e85609. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28(6):e85609. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 70.Kivioja T, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2012;9(1):72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.