ABSTRACT
Sphingolipids are bioactive molecules playing a key role as membrane components, but they are also central regulators of many intracellular processes including macroautophagy/autophagy. In particular, sphingosine-1-phosphate (S1P) is a critical mediator that controls the balance between sphingolipid-induced autophagy and cell death. S1P levels are adjusted via S1P synthesis, dephosphorylation or degradation, catalyzed by SGPL1 (sphingosine-1-phosphate lyase 1). Intracellular pathogens are able to modulate many different host cell pathways to allow their replication. We have found that infection of eukaryotic cells with the human pathogen Legionella pneumophila triggers a change in the host cell sphingolipid metabolism and specifically affects the levels of sphingosine. Indeed, L. pneumophila secretes a protein highly homologous to eukaryotic SGPL1 (named LpSPL). We solved the crystal structure of LpSPL and showed that it encodes lyase activity, targets the host's sphingolipid metabolism, and plays a role in starvation-induced autophagy during L. pneumophila infection to promote intracellular survival.
KEYWORDS: bacterial nutrition, Legionella pneumophila, lipid metabolism, sphingosine-1-phosphate lyase, xenophagy
Xenophagy is a cellular mechanism allowing an infected cell to rapidly degrade the invading bacteria by relying on the core autophagy machinery. The important role of autophagy in restricting the replication of pathogens has been demonstrated for several intracellular bacteria: the invaders are surrounded by autophagosome-like structures that allow fusion with lysosomes and consequently the degradation of the pathogen. Furthermore, it has been shown that the induction of autophagy suppresses the survival of intracellular bacteria. Thus the subversion of autophagy is a key strategy employed by pathogens to block the host response and to promote their survival.
Legionella pneumophila is one of those intracellular pathogens that interferes with the host autophagy machinery. L. pneumophila is a Gram-negative bacterium that is naturally found in aquatic environments where it replicates in protists, but that can also cause a severe pneumonia in humans called Legionnaires' disease. The high conservation of many signaling pathways in human macrophages and protists allows L. pneumophila to invade and to replicate in human cells. Once the host cell is infected, L. pneumophila is able to delay its delivery to lysosomes and to build up a specialized vacuole where it replicates until nutrients are depleted. The ability of L. pneumophila to subvert host defenses to set up its intracellular cycle relies on its uniqueness to encode more than 300 secreted effector proteins that interfere with diverse cellular pathways. Our analyses of L. pneumophila genomes showed for the first time that many of these effectors share high similarity with eukaryotic proteins, and are thus proteins never or only rarely found in prokaryotic genomes, a finding that led to the hypothesis that these proteins had been acquired through horizontal gene transfer from its hosts.
One example is the lpp2128/spl gene that encodes a protein highly similar to eukaryotic SGPL1, named LpSPL for Legionella peumophila SGPL1. Phylogenetic analyses of the LpSPL sequence show that it branches within the eukaryotic clade of SGPL1, closest to the one of Acanthamoeba castellanii, further supporting the idea that the LpSPL-encoding gene had been acquired from a protist host. In order to characterize its function and role in host-pathogen interaction, we resolved its crystal structure. This showed that LpSPL exhibits a dimeric multidomain architecture like eukaryotic SGPL1 and revealed a high level of structural conservation in its active site composition with its eukaryotic counterparts. Eukaryotic SGPL1 catalyzes the irreversible cleavage of S1P, a product of the sphingolipid degradation pathway. By using a fluorogenic homolog of S1P we showed that LpSPL exhibits lyase activity in both transfected and infected cells. Furthermore, mutations of the residues reported to be involved in eukaryotic SGPL1 activity also abrogated the lyase activity of LpSPL.
Sphingolipids are a family of metabolites essential for membrane biogenesis, but they are also important signaling molecules modulating numerous physiological processes. The central component of the sphingolipid degradation pathway is ceramide that, hydrolyzed from sphingomyelin or synthetized de novo, undergoes successive conversions to various other sphingolipid intermediates like ceramide-1-phosphate, sphingosine and S1P. S1P levels are tightly fine-tuned through synthesis and degradation because once S1P is present in the intracellular milieu it functions as a cellular mediator (Fig. 1A).
Figure 1.
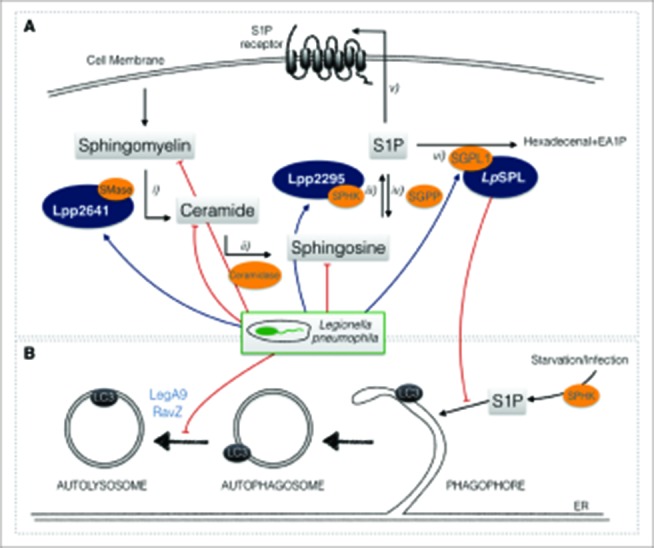
Legionella pneumophila modulates sphingolipid metabolism and autophagy during infection. (A) Simplified illustration of sphingolipid metabolism: i) sphingomyelin from plasma membranes is turned into ceramide by the enzyme sphingomyelinase (SMase). ii) Ceramide is then metabolized by ceramidase into sphingosine that iii) is phosphorylated by SPHK (sphingosine kinase) into sphingosine-1-phosphate (S1P), iv) a reaction that can be reverted by SGPP (sphingosine-1-phosphate phosphatase). v) S1P is secreted into the extracellular milieu, where it might act in a paracrine or autocrine way, or vi) may by broken down to hexadecenal and ethanolamine-1-phosphate (EA1P) by sphingosine-1-phosphate lyase (SGPL1). L. pneumophila decreases sphingolipid levels during infection (red arrows). It encodes 3 eukaryotic-like proteins that are homologs to enzymes that act in sphingolipid metabolism: Lpp2641 (putative sphingomyelinase), Lpp2295 (putative sphingosine kinase) and LpSPL (sphingosine-1 phosphate lyase) (blue arrows). LpSPL has been characterized functionally. (B) L. pneumophila restrains host autophagy during infection by secreting effectors that inhibit both autophagosome formation (LpSPL) and maturation (RavZ and LegA9).
To understand the role LpSPL plays in the host cell, we measured host cell sphingolipid levels during infection by using mass spectrometry techniques. We observed that infection of macrophages with L. pneumophila leads to an overall reduction of bioactive sphingolipids such as sphingomyelin and ceramide, showing for the first time that L. pneumophila infection affects host cell sphingolipid metabolism. Interestingly, cells infected with a L. pneumophila strain that does not express LpSPL reveal an accumulation of sphingosine compared to cells infected with wild-type L. pneumophila. Thus, the bacterium specifically secretes LpSPL to prevent an increase of sphingosine levels in the infected host cell. This observation correlates with the fact that ectopically expressed LpSPL localizes at the endoplasmic reticulum, like eukaryotic SGPL1. Given the role of sphingolipids as regulators of autophagy, we further explored whether secretion of LpSPL into the host cell affects the autophagic response to infection.
Indeed, L. pneumophila is already known to interfere with the autophagy machinery: 2 secreted effectors have been previously identified to target the autophagy machinery (RavZ and LegA9). By using multiple assays, including SQSTM1/p62 and LC3 western blots we found that LpSPL is a third effector secreted by L. pneumophila that modulates autophagy, as LpSPL restrains starvation-induced autophagy by acting on autophagosome biogenesis (Fig. 1B). Importantly, this effect depends on its enzymatic activity as a catalytically inactive mutant fails to decrease host authophagy. By using high-content image-based analyses we quantified the formation of LC3 puncta in cells infected with wild type or a mutant strain deleted for the spl gene, showing that LpSPL limits the autophagic response during L. pneumophila infection to counteract antibacterial response by the host cell.
Taken together, our work discovered that the modulation of the sphingolipid metabolism of the host cell is a sophisticated strategy used by L. pneumophila to exploit the host autophagy machinery and to evade the host-cell response.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
CB's lab was funded by the Institut Pasteur, the French Region Ile de France (DIM Malinf), ANR-10-LABX-62-IBEID and Infect-ERA project EUGENPATH (ANR-13-IFEC-0003-02); PE was funded by the Fondation pour la Recherche Médicale grant N° DEQ20120323697.


