Abstract
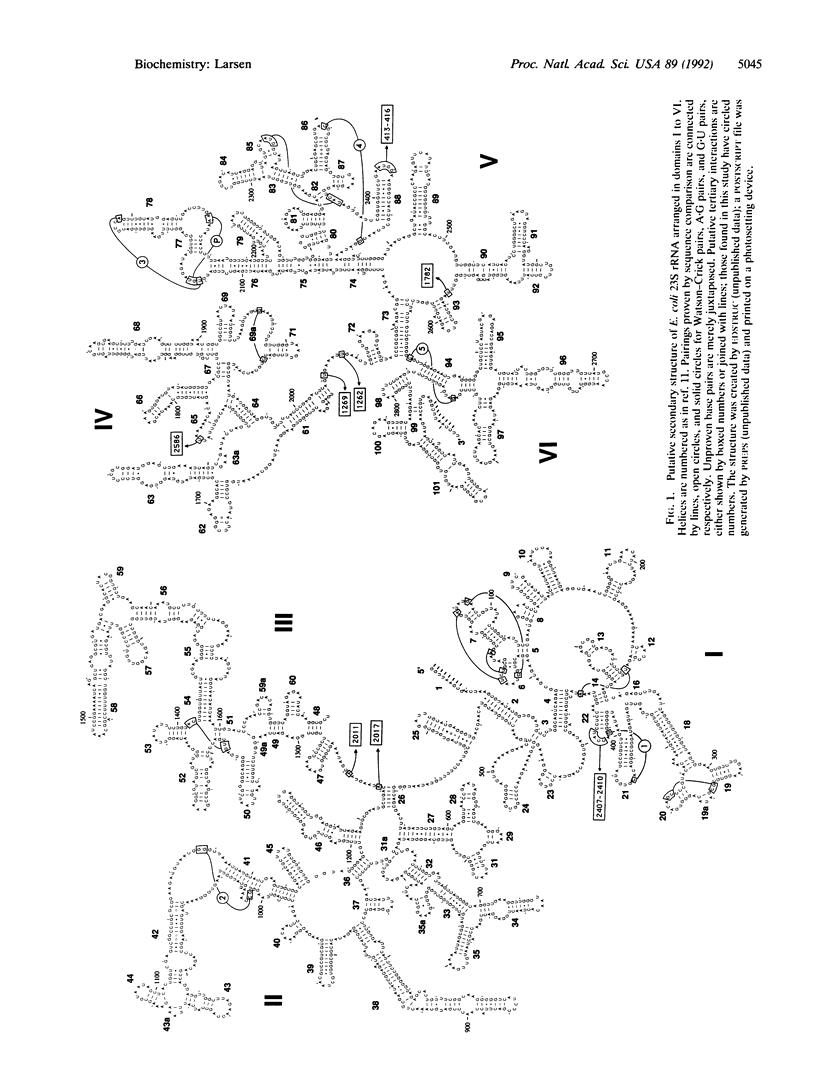
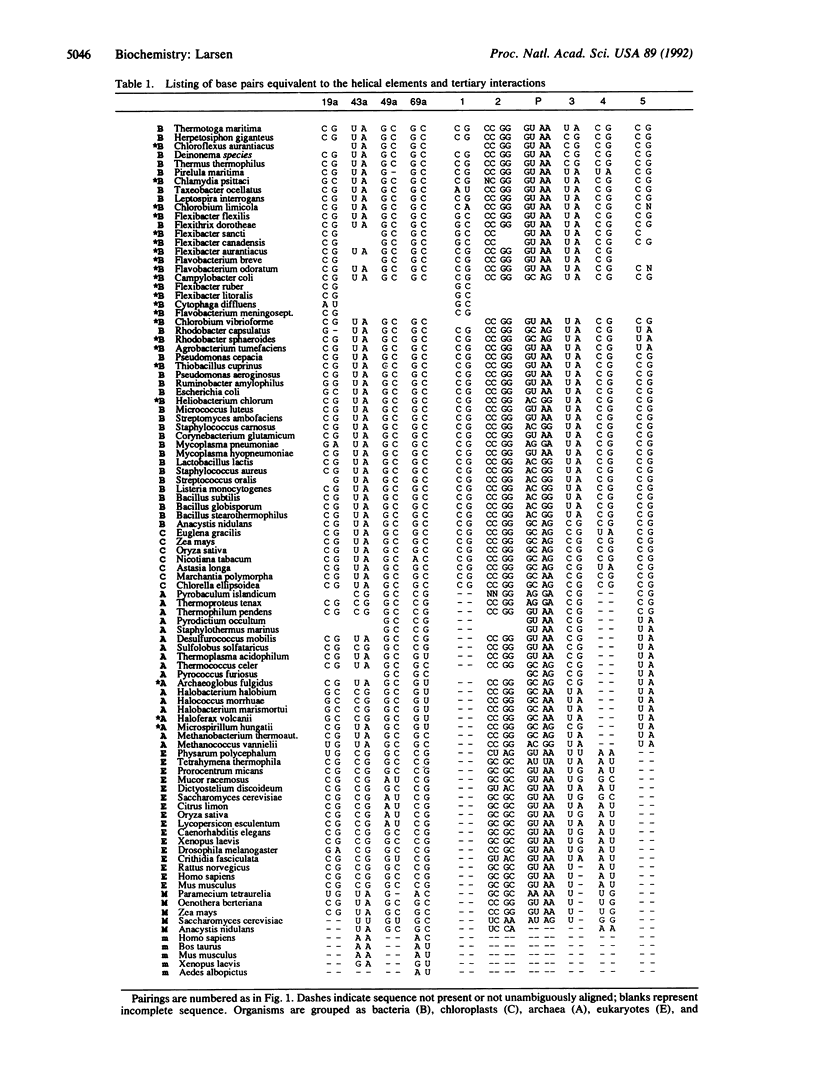
An alignment of 75 phylogenetically diverse large subunit ribosomal RNA sequences was created and searched for secondary and tertiary structure elements by computer. The search revealed four unknown secondary structural pairings, two internal loop closings, and five short-range tertiary interactions--two of which were pairings of an unusual type. One brings a loop together with two other loops previously known to be paired, and one involves a nucleotide within a presumed tetraloop. The latter interaction constrains the RNA structure near the ribosomal E-site, where two base pairs previously suggested to be in parallel orientation are now proven. No clear phylogenetic evidence for direct base pairing between the large and small subunit rRNA was found.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branlant C., Krol A., Sriwdada J., Ebel J. P., Sloof P., Garrett R. A. The binding site of protein L1 ON 23-S ribosomal RNA of Escherichia coli. 2. Identification of the rna region contained in the L1 ribonucleoproteins and determination of the order of the RNA subfragments within this region. Eur J Biochem. 1976 Nov 15;70(2):457–469. doi: 10.1111/j.1432-1033.1976.tb11037.x. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Egebjerg J., Douthwaite S. R., Liljas A., Garrett R. A. Characterization of the binding sites of protein L11 and the L10.(L12)4 pentameric complex in the GTPase domain of 23 S ribosomal RNA from Escherichia coli. J Mol Biol. 1990 May 20;213(2):275–288. doi: 10.1016/S0022-2836(05)80190-1. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Fox G. E. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res. 1988;16 (Suppl):r175–r269. doi: 10.1093/nar/16.suppl.r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Woese C. R. Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc Natl Acad Sci U S A. 1990 Jan;87(2):663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers H., Kjems J., Ostergaard L., Larsen N., Garrett R. A. Evolutionary relationships amongst archaebacteria. A comparative study of 23 S ribosomal RNAs of a sulphur-dependent extreme thermophile, an extreme halophile and a thermophilic methanogen. J Mol Biol. 1987 May 5;195(1):43–61. doi: 10.1016/0022-2836(87)90326-3. [DOI] [PubMed] [Google Scholar]
- Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969 Nov 22;224(5221):759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- Mitchell P., Osswald M., Schueler D., Brimacombe R. Selective isolation and detailed analysis of intra-RNA cross-links induced in the large ribosomal subunit of E. coli: a model for the tertiary structure of the tRNA binding domain in 23S RNA. Nucleic Acids Res. 1990 Aug 11;18(15):4325–4333. doi: 10.1093/nar/18.15.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Conformation of an RNA pseudoknot. J Mol Biol. 1990 Jul 20;214(2):437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirup S., Larsen N. E. ALMA, an editor for large sequence alignments. Proteins. 1990;7(3):291–295. doi: 10.1002/prot.340070310. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]



