Summary
Bacterial pathogens have evolved to exploit humans as a rich source of nutrients to support survival and replication. The pathways of bacterial metabolism that permit successful colonization are surprisingly varied and highlight remarkable metabolic flexibility. The constraints and immune pressures of distinct niches within the human body set the stage for understanding the mechanisms by which bacteria acquire critical nutrients. Here we discuss how different bacterial pathogens carry out carbon and energy metabolism in the host, and how they obtain or use key nutrients for replication and immune evasion.
I. The interface between two organisms is shaped by metabolic interactions
Why do organisms “eat”? This core question drives the study of biochemistry, specifically - metabolism. The short answer to this seemingly simple question is two-fold: first, eating provides cells with the physical building blocks for the generation of cellular components (i.e., growth of the physical cell – something must come from something); and second, eating is the way to extract energy to do cellular work (i.e., powering the process of growth – work is never done for free). These two processes – catabolism & anabolism – are inextricably linked. The pathways of catabolism, like glycolysis and the tricarboxylic acid (TCA) cycle, which break down molecules for energy metabolism, also branch off into anabolic pathways that generate building blocks for the cell. Bacterial metabolism is dynamic and flexible, with different bacterial species encoding different metabolic capacities within their genomes. Thus, the canonical TCA cycle may function fully in one bacterial species, while another bacterium, missing a key enzyme of the “cycle” now uses the other TCA enzymes in branched oxidative and reductive pathways. Moreover, depending on availability of carbon sources or oxygen, even if a bacterium encodes all of the enzymes for respiration, with its high-energy yield, the less energy efficient but faster process of fermentation may predominate. Thus, the flexible metabolic space of rapidly evolving bacterial genomes enables many different ways for bacteria to take advantage of nutrients in complex environments for robust replication.
The essential infrastructure of metabolism is made more complex during bacterial infection when one organism thrives by drawing nutrients from the other. From a bacterial perspective, the mammalian host is a vast ecosystem, with some regions like the intestine, heavily populated with competitors, while other niches are wide open for exploitation. To appreciate how bacterial metabolism shapes infection, it is important to consider the localized host environment where the bacteria replicate, as well as the metabolic capacity of the pathogen. The host environment is not simply a source of food for bacteria. Rather, host cells are constantly controlling their own metabolic function, using available nutrients and removing waste products. In addition, host organisms actively survey their inner spaces for invading micro-organisms. Thus, bacterial pathogens must overcome constant pressure from the predatory immune system of the host. These defining aspects of the host:pathogen interaction are drivers of disease, whether it is acute or chronic, inflammatory or silent, mild or deadly.
In this chapter, we consider three aspects of metabolism in the host:pathogen interaction: first, how bacteria within a host employ specific modules of central metabolism to generate energy, second, how bacterial pathogens exploit the host for critical nutrients required for proliferation and lastly, how these bacterial invaders use metabolic tricks to sense their environment and to evade host immunity.
II. Bacterial Energy Metabolism During Infection
This section focuses primarily on recent research that is revealing how bacterial pathogens maintain energy metabolism within the host environment – in other words, how do bacterial pathogens uniquely go about the business of eating inside a host to meet their energy demands (1)? First, a brief note on “energy”: it is worth noting that energy is not a thing, but rather, a potential, and that the energy all cells work to acquire is the energy that “lives” within the chemical bonds of what the cell eats. Ultimately, for cells to do work, chemical bond energy from some food source must be moved to a molecule that can be used directly to fuel cellular tasks – the predominant, but not exclusive, form being adenosine triphosphate (ATP). The idea that phosphate bond energy transfer is the way in which cells power most cellular work stands as a monumental paradigm shift in the natural sciences (2, 3), ushering in the full elucidation of “central metabolism”, the foundational energy yielding pathways for all cellular life. A more intimate way of viewing central metabolism is as the process that best illustrates the close kinship of all cellular life. Glycolysis shows us how we are related to E. coli. The tricarboxylic acid cycle unites us with our cousins, the fungi. And no matter what the genetic content, there is a common need for ATP.
Thus, when considering the specific role of energy-yielding metabolism during bacterial infection, it is good to remember that both the pathogen and the infected host are engaging in non-stop, regulated central metabolic activity, often competing for the same resources and usually trying to influence the behavior of the other. Here, we start by exploring how some bacterial pathogens conduct energy metabolism during infection.
What's for dinner inside the host?
Depending on where in the host an invading bacterium takes up residence, the food sources available within that niche will determine whether that microbe can successfully establish itself. Some pathogens, such as invasive streptococci, Salmonella enterica and Brucella abortus, can feed their “sweet tooth” during infection by acquiring specific carbohydrates; whereas the “low-carb” pathogen Mycobacterium tuberculosis (M. tb) prefers to fuel itself with energy-rich fatty acids during a chronic infection that can last for many decades. Recent research is revealing some unique ways in which pathogens go about feeding themselves during host colonization.
Pathogenic streptococci are a group of Gram-positive bacteria that cause a range of disease from mild (dental caries) to severe (pneumonia and sepsis). These bacteria establish themselves on extracellular surfaces of the host, such as on the tissues of the human naso-pharynx or in dental biofilms. Here, they are positioned to be fed directly by the host as the host feeds him or herself, or to acquire nutrients from the extracellular surfaces of host tissues. Streptococcus mutans, a major cause of dental caries, sets itself apart from the resident, non-pathogenic oral microbiota by having a large and flexible metabolic range with regard to nutrient acquisition. As the human diet has changed over time to become richer in carbohydrates, (especially sucrose) so has the physiological capability of S. mutans evolved (4, 5). A study that compared ancient and modern bacterial communities of dental plaque suggests that pathogenic S. mutans has become a more common resident of the human oral cavity as a result of this change in diet, especially in modern, post-industrial times (4). One of the reasons for this co-evolutionary success has been the expansion of the S. mutans genome to contain a wide range of genes involved in carbohydrate uptake, especially the EII permeases of the phosphoenolpyruvate:sugar phosphotransferase system (PTS), the multi-enzyme pathway of carbohydrate uptake present in most bacteria. Also, the Carbohydrate Catabolite Repression (CCR) system, which allows bacteria to regulate which carbohydrates to use for energy catabolism by sensing intracellular nutrient content, also appears to be unique in S. mutans in that it has more levels of CCR flexibility than other Gram-positive bacteria (5, 6).
The PTS and CCR systems also have expanded roles in the more serious and invasive Streptococcus pathogens, S. pneumoniae and S. pyogenes, by being involved in controlling levels of virulence gene expression, such as the toxin Streptolysin S (7-9). Also, S. pneumoniae is able to use a wider range of carbon sources for energy catabolism compared to other Gram-positives (10) and has the ability to obtain and use complex carbohydrates that it is able to remove from host extracellular surfaces with specialized enzymes (11-13). A comprehensive review of streptococcal carbohydrate utilization during pathogenesis can be found elsewhere; rather, these brief examples introduce and highlight the themes that; (i) the ability of pathogens to exploit the host niche and obtain preferred food sources for energy catabolism is an important aspect of bacterial virulence; (ii) pathogens often set themselves apart from non-pathogens by having an expanded and flexible capability for obtaining preferred energy sources within the site of infection; and (iii) virulence gene expression is often intimately linked to the pathways of carbon acquisition and energy metabolism.
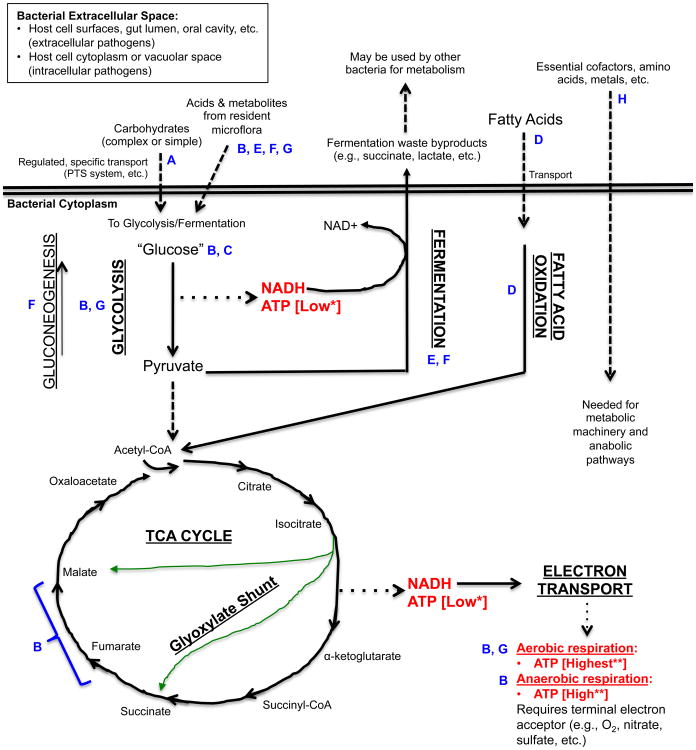
Focusing in more closely on central metabolism, it is essential to emphasize the key role played by glucose in energy-yielding catabolism. When considering energy metabolism via respiration, where glycolysis and the TCA cycle are both utilized, it is the simple sugar glucose that gives a fast and efficient energy yield in the form of ATP for host and pathogen alike. Thus, these pathways are important for many bacterial pathogens, such as Salmonella and Brucella, which have evolved to be able to feed their energy-yielding pathways during infection specifically with glucose “stolen” from the host (Figure 1).
Figure 1. An simplified view of catabolic energy-yielding pathways in bacteria and points of relevance for the indicated bacterial pathogens.
The figure shows a simplified outline of some of the different pathways for utilizing carbohydrates or fatty acids for the generation of ATP. These catabolic pathways and the anabolic pathways that they feed are under extremely complex levels of control, which have been studied mainly in non-infectious organisms (126). However, the unique metabolic strategies employed by infectious bacteria are becoming more appreciated as important aspects of bacterial pathogenesis (1). *ATP generated by substrate level phosphorylation; **ATP generated by oxidative phosphorylation; Solid lines: Metabolic Pathway; Dashed lines: Substrates that feed into catabolic pathways and products generated by bacteria as a result of metabolism or required for metabolism; Dotted lines: Main energy yielding metabolites generated from pathways.
Blue letters indicate specific points of importance for the energy-yielding metabolism of select pathogens listed below (see Table 1 for summary):
A. Pathogenic Streptococci (S. mutans, S. pyogenes, S. pneumoniae); B. Salmonella enterica serovar Typhimurium; C. Brucella abortus; D. Mycobacterium tuberculosis; E. Clostridium difficile; F. Enterohemorrhagic Escherichia coli (EHEC); G. Aggregatibacter actinomycetemcomitans; H. Listeria monocytogenes.
The pathogenic streptococci previously mentioned reside at the surface of cell tissues where glucose concentration is low (12) but where many other carbohydrates, both simple and complex, are vastly abundant. Salmonella enterica and Brucella abortus, on the other hand, are intracellular pathogens, and must go about feeding themselves from the inside of host cells where the range and concentration of available carbon sources is very limited. Therefore, rather than evolve an expanded ability to utilize a range of carbohydrates, these bacteria are able to more efficiently steal the fewer carbon sources available to them from within the host cell – primarily, glucose or glucose phosphate. Using a mouse model and a series of bacterial metabolic deletion mutants, it has been shown that Salmonella enterica serovar Typhimurium (S. Typhimurium) has a major dependence upon glucose, glycolysis and the TCA cycle both in tissue culture and during infection in mice (14, 15). The S. Typhimurium mouse model has been used to elucidate the molecular mechanism of infection for the human pathogen S. enterica serovar Typhi, which causes typhoid fever (16, 17). In their intracellular niche within host macrophages (phagocytic cells of the innate immune system), S. Typhimurium resides within a “Salmonella-containing vacuole” (SCV), where it is protected from host defenses, but where it nonetheless is able to obtain the nutrients needed for intracellular replication (18). Bacteria lacking both homologs of phosphofructokinase, a key enzyme unique to the glycolysis pathway, are unable to replicate within macrophage culture and are severely attenuated during infection of BALB/c mice (14), showing a major dependence on this pathway during intracellular infection. Also in this study, extensive use of 15 different metabolic mutants of S. Typhimurium, showed that glucose, specifically, is the preferred carbon source within macrophages and in the mouse host (14). Importantly, the key triple knockout strain from this study was lacking genes for both glucose uptake (two PTS systems) and glucose catabolism (glucokinase), resulting in a total glucose catabolic mutant. However, although this strain was highly attenuated in tissue culture and in vivo, it was still able to replicate at low levels, suggesting that although glucose is the predominant energy source in this model, there likely exists another, less preferred carbon source available within the host.
At this point, it is important to emphasize how fundamental a role the glycolysis pathway plays within the metabolic infrastructure of the cell. This one central pathway participates in a variety of activities which include: (1) providing the molecule pyruvate to the TCA cycle during aerobic or anaerobic respiration; (2) being a sole anaerobic energy yielding pathway called “fermentation”, which has a lower energy yield than respiration; and (3) providing most of the enzymes for gluconeogenesis, which is a glucose generating pathway that is basically “glycolysis in reverse”. So considering the previous finding that glycolysis and glucose are of primary importance to intracellular Salmonellae, one might ask what role the glycolysis pathway is serving for the bacterium. Again, using a systematic series of metabolic knockout strains, it has been shown that S. Typhimurium requires a fully intact TCA cycle within its host, in this case the BALB/c mouse, for full virulence (15). Just as importantly, this work revealed that certain metabolic pathways are not essential for S. Typhimurium during infection, including gluconeogenesis, fatty acid metabolism and the glyoxylate bypass (15). More recent refinement of this work has identified the specific steps in the TCA cycle that are the most important for full virulence, including the conversion of fumarate to malate, and the conversion of malate to oxaloacetate and pyruvate (19) (Figure 1). But what makes this work both complex and confusing is that different metabolic mutants show a range of virulence phenotypes, from totally avirulent, to attenuated, to fully virulent. This observation along with the wide range of metabolic flexibility and redundancy in bacterial genomes highlights the difficulty of pinpointing the precise modes of metabolism during microbe-host interactions. Regardless, this example illustrates the modularity of metabolic pathways and the presence of key vulnerable chokepoints that may be unique to each organism. In this case, the vulnerability of infectious S. Typhimurium for running a complete TCA cycle is its need for malate in order to generate pyruvate and oxaloacetate to replenish the cycle. Ablating enzymatic steps upstream of the two essential reactions for generating malate results in only slight attenuation, suggesting that the microbe is able to use molecules potentially scavenged from the host to generate the key malate precursor, succinate, outside of those two steps. The authors intriguingly hypothesize that succinate, ornithine or arginine scavenged from host cells may be used by the bacterium for malate generation; however, this idea needs further investigation.
This last point brings into focus the dynamic tension that exists between host cells and infecting microbes. To complete the discussion on the need for glucose, the following describes pathogens that rely on host cell behavior to provide adequate glucose needed for basic energy metabolism. Both S. Typhimurium and Brucella abortus infect and replicate within host cell macrophages, which are immune cells that can adopt a range of behaviors depending upon which of their many jobs they are performing. Both pathogens are able to establish a persistent, chronic infection within macrophages that are in the “M2” state, a non-inflammatory phenotype characterized by a certain repertoire of gene expression (20, 21), in particular, the regulatory proteins of the peroxisome proliferator-activated receptors (PPAR) (22). Each pathogen relies upon host expression of a different PPAR protein to promote an intracellular environment with available glucose for bacterial metabolism. For S. Typhimurium, host expression of PPAR-delta is important for persistent infection, since the bacteria are not able to establish a persistent infection in Ppar-delta -/- cells or mice (23). Host expression of PPAR-delta is not necessary for other intracellular pathogens such as Listeria monocytogenes, Francisella tularensis and M. tb, illustrating the diversity in survival strategies of different pathogens. In general, PPAR-delta controls host cell metabolism, moving the macrophage away from glucose metabolism and promoting fatty-acid oxidation, putatively freeing up intracellular glucose that S. Typhimurium can then use. Testing whether increased glucose availability in the PPAR-delta expressing, fat-metabolizing macrophage was permitting S. Typhimurium to replicate, the authors observed that a bacterial mutant deficient in glucose transport was unable to replicate within the macrophage. Additionally, the authors saw that wild type bacteria were able to acquire a fluorescent glucose analog from the macrophages, and also replicated to higher levels when wild type host cells were treated with a PPAR-delta agonist (23). Together, these data provide strong evidence that the metabolic capacity of bacterial pathogens is strongly influenced by host metabolic function.
During chronic B. abortus infection in mice, “M2” type macrophages in the spleen harbor replicating bacterial cells and express the gene for the PPAR-gamma regulator (21), another PPAR protein that tilts macrophage metabolism away from glucose usage and toward fatty acid oxidation. A PPAR-gamma agonist increased macrophage intracellular glucose concentration and also supported increased bacterial colonization, while blocking PPAR-gamma activity with a specific inhibitor decreased splenic colonization of B. abortus in mice. As in the previous study, mutant B. abortus deficient in glucose transport were greatly attenuated for growth in cell culture and in mice, strongly suggesting that glucose availability is key for this microbe to establish chronic infection. Two important observations comparing these two studies are that: (1) both studies elucidated bacterial metabolism during persistent, chronic bacterial infections, suggesting that pathogens likely implement different metabolic strategies over the lifetime of an infection; and (2) whereas S. Typhimurium may be actively inducing increased expression of PPAR-delta in the host since infected macrophages upregulate this gene in the absence of cytokine stimulation, it appears that B. abortus does not directly cause macrophages to increase PPAR-gamma. Thus, bacterial pathogens may actively or passively take advantage of host cell metabolic capacity. These studies then raise the question as to why these two pathogens benefit from host expression of different PPAR proteins, since both homologs have similar roles in the macrophage. Lastly, the dependence on glycolytic metabolism in these two bacteria and their ability to obtain host glucose demonstrate that central metabolism is a key determinant in virulence and infectious persistence.
Glucose is not the only molecule that can be oxidized by cells for energy metabolism. In fact, fatty acids harbor much more potential energy than glucose, and oxidation of fatty acids can provide a greater yield of ATP (Figure 1). However, bacteria can only establish infection if they are able to acquire usable food sources. A landmark study revealed that the major pathogen M. tb prefers to metabolize fatty acids rather than glucose during persistent infection (24). The genomic era has expanded our view of M. tb metabolism by identifying the range of metabolic genes that this organism harbors, in particular, a high degree of redundancy in genes for fatty acid beta-oxidation enzymes (25). A large-scale genetic screen has shown that several M. tb glycolysis enzymes are not needed for growth in mice (26). More recent work has refined how this pathogen makes its energetic living during infection, answering key questions as to where in the host the microbes acquire their preferred food source, and what pathogen specific pathways/proteins are key for metabolism during infection.
The M. tb bacillus establishes a particularly resilient, chronic infection in the host lung by persisting in a semi-dormant state within granulomas, conglomerations of host immune cells putatively working to wall off the spread of infection, but inadvertently serving as a stable reservoir for bacilli. Recent work has shown that oxygenated mycolic acids, molecules found on the cell surface of certain Mycobacteria, can induce lung macrophages to transform into a phenotype called the “foamy macrophage”, an important component of the granuloma characterized by a high lipid content and ablation of bactericidal activity (27). But does induction of this macrophage phenotype only help protect the bacteria from clearance, or does the foamy macrophage contribute to bacterial metabolism? Despite being in a semi-dormant state, the bacilli within the granuloma associate closely with lipid droplets inside of these lipid-rich macrophages (27), and subsequent investigation has confirmed that M. tb are able to accumulate intracellular, host-derived triacylglycerols (TAG) (28). The use of these host-derived TAG for bacterial metabolism during the dormant state is strongly supported by the observation that bacterial fatty acid oxidation genes are upregulated during residence within lipid-loaded macrophages (28). As to exactly how the bacteria are conducting metabolism in vivo, two bacterial homologs of isocitrate lyase (ICL1 and ICL2) are both needed for intracellular replication (29). These proteins are vital for the glyoxylate cycle in M. tb, a pathway needed for replenishment of TCA cycle intermediates during fatty acid metabolism when glycolysis is not being utilized (Figure 1).
Sugar and fat, and in a pinch, amino acids can be fed into central metabolism to yield energy. The questions explored in these examples regarding the preferred food sources and metabolic strategies of several bacterial pathogens are relevant for other pathogenic species as well. In the case of the human body, a wide range of tissue types and physiological environments exist where pathogens elicit disease, each one being unique in terms of carbon source content and availability. One might then speculate that pathogens that are able to infect numerous tissue types, like Staphylococcus aureus, display a range of metabolic behavior based on the variable levels of glucose, fatty acids and other carbon resources at different sites of infection. Also, do bacteria that infect immune privileged tissues, such as the brain and meninges, have the ability to acquire nutrients that are normally unavailable to other microbes? These and many other questions about energy metabolism during infection will certainly reveal more surprising capabilities of bacterial pathogens.
The host is a crowded and sometimes bountiful place
One of the most exciting developments in microbiology has been the launching of the Human Microbiome Project (HMP) (30, 31), an undertaking to identify and investigate the myriad microorganisms that reside in and on the human body. This endeavor is now possible due to the availability of new genomic tools that allow researchers to investigate metagenomes, complex microbial communities composed of multiple species. But one of the most important aspects of the HMP is that it has increased appreciation for the ways in which the non-pathogenic, resident microbiota of the body can contribute to health and disease. In other words, our bodies are complex ecosystems, and when bacterial pathogens cause disease, they are often doing it within an environment where the growth of other microbes may have an influence, for better or worse. In terms of energy metabolism, it is important to appreciate that within a species-rich niche such as the gastrointestinal tract (GI) or the oral cavity, a multitude of resident bacterial species are engaging in a wide range of metabolism, consuming different food sources and excreting different waste products, creating a niche that some bacterial pathogens have found unique ways of exploiting.
One of the most severe pathogens of the GI system is the Gram-positive endospore-forming bacterium Clostridium difficile. An obligate anaerobe, the various classical virulence factors that this microbe produces, such as toxins, have been well characterized (32). But how does C. difficile make its living, metabolically speaking, within the gut milieu? It is extremely difficult to test this question directly in hosts that harbor a vastly diverse native gut microbiota. To simplify the situation, a model system using gnotobiotic mice (mice harboring a defined and limited microbiota) has helped reveal how C. difficile benefits from the fermentation products of one microbe in order to run its own fermentative metabolism (33). The non-pathogenic, common gut microbe Bacteroides thetaiotaomicron (Bt) produces the metabolite succinate during fermentation of carbohydrates. In Bt-harboring mice, C. difficile is able to grow to higher population densities when the mice are fed a high carbohydrate diet compared to when they are fed a low-carbohydrate diet. Comparing the global transcriptional profile of C. difficile grown in Bt-harboring mice fed a standard, carbohydrate-rich diet versus a polysaccharide deficient one, the pathogen showed increased expression of multiple carbohydrate metabolism genes under the former condition, in particular, genes involved in the succinate to butyrate fermentation pathway. When mice are colonized with Bt only, high levels of succinate are detectable in the mouse gut. In mice co-colonized with Bt and C. difficile, higher levels of butyrate are generated than in mice harboring C. difficile only. Thus, the C. difficile appear to adapt their metabolic strategy to use Bt-generated succinate when it is present, generating butyrate as an end-product. But in the normal gut, succinate levels are generally kept low due to the presence of other resident bacteria that produce other fermentation end-products, such as the short chain fatty acids acetate and butyrate. However, wild type mice treated with antibiotics have higher levels of succinate in the gut, suggesting that antibiotic treatment changes the microbiota population distribution, and thus changing the metabolite content as well. In antibiotic treated mice, wild type C. difficile greatly out-competes an isogenic mutant lacking the succinate transporter (33), supporting the idea that C. difficile experiences a growth environment that supports succinate fermentation when members of the microbiota that prevent the accumulation of succinate are absent. This defined experimental approach greatly illuminates our understanding of how a pathogen like C. difficile can be present in the gut at non-pathogenic levels under normal conditions, but then can expand and cause disease after antibiotic treatment, all due to a simple switch in energy-yielding metabolism.
The benign gut resident Bacteroides thetaiotaomicron (Bt) also may assist another enteric pathogen via succinate production. Enterohemorrhagic Esherichia coli (EHEC) has a curious way of colonizing the gut by forming what are called “attaching and effacing (AE) lesions”, causing intestinal epithelial cells to remodel their cytoskeleton to form pedestal-like extensions upon which the EHEC firmly attach themselves. EHEC exhibits lower levels of virulence gene expression in conditions that promote glycolysis (i.e., carbohydrate rich), while virulence expression is increased in sugar-poor conditions where gluconeogenesis is needed by the microbe (34, 35). EHEC grown in vitro in the presence of Bt showed enhanced expression of virulence genes, and AE lesion formation was much higher in tissue culture when EHEC was co-cultured with Bt (36). Interestingly, virulence gene expression in the presence of Bt was decreased in EHEC mutants lacking the protein Cra, which is a regulatory protein that senses sugar concentrations (36). Since Cra enhances virulence gene expression in nutritional environments that suppress glycolysis but enhance gluconeogenesis, the authors hypothesized that in vivo, EHEC benefits from localized succinate production by the resident microbiota in the sugar-limited region of the gut epithelial surface, as opposed to the carbohydrate-rich gut lumen (34, 36).
A final example of the dynamic metabolic interplay between bacterial pathogens and resident microbiota shows that “cross-feeding” of preferred food sources can alter virulence behavior. The bacterium Aggregatibacter actinomycetemcomitans can colonize the human oral cavity and cause aggressive periodontitis. A facultative anaerobe, A. actinomycetemcomitans can use several carbon sources, but prefers to catabolize L-lactate over glucose, even though growth is slower using L-lactate (37). The microbe does not catabolize L-lactate anaerobically, but rather, oxidizes this food source in an oxygen dependent manner (38). When grown aerobically with glucose during in vitro co-culture with the non-pathogenic resident of the oral microbiota, Streptococcus gordonii, A. actinomycetemcomitans mutants deficient in L-lactate metabolism show diminished growth, though they are able to grow well in high-glucose mono-culture, suggesting that the ability to metabolize L-lactate is important specifically in the presence of S. gordonii. Using a mouse thigh abscess model (a common model system for examining oral bacterial pathogenesis), the authors further show that the ability to metabolize L-lactate is important only for A. actinomycetemcomitans infection during co-culture with S. gordonii, since mutants lacking the ability to catabolize L-lactate establish mono-culture infection as well as wild type, but are much less abundant in co-culture abscesses (38). These data strongly suggest that L-lactate produced by S. gordonii, and also potentially by host lactate dehydrogenase in the subgingival crevice, can affect enhance the growth of this pathogen in the human oral cavity.
These examples not only further illustrate that the ability to find and utilize preferred food sources during infection is important for bacterial pathogens, but also show that the host resident microbial landscape can have major effects on pathogen energy metabolism by providing specific nutritional environments. More importantly, these studies support the idea that changes to or perturbations in the host microbiota can create environments that can enhance or inhibit a pathogen's ability to engage in its preferred microbial metabolic pathways, thereby supporting or hindering damage to the host.
The parts that make it all go
Yet another way of looking at central metabolism for bacterial pathogens is to ask questions about the enzymatic machinery that is essential for conducting preferred modes of metabolism. Organisms vary widely in their ability to generate essential cellular components from scratch and in their ability to acquire key components from exogenous sources when de novo synthesis is not possible. The intracellular pathogen Listeria monocytogenes replicates efficiently within the cytoplasm of host cells where it is able to acquire a variety of host molecules to support growth, including glucose-6-phosphate as a carbon source (39) and the essential thiol-containing co-factor lipoate (40) that is needed for several enzymes involved in aerobic metabolism (41). The L. monocytogenes genome does not contain genes necessary for de novo lipoate biosynthesis (42). However, this bacterium does produce two functional lipoate ligase enzymes (LplA1 and LplA2) that are able to catalyze the covalent attachment of lipoate to the enzyme pyruvate dehydrogenase (PDH), a critical enzyme for bacterial respiration (40, 43). The LplA1 enzyme is specifically needed for L. monocytogenes intracellular growth and virulence in mice (41). More importantly, LplA1 but not LplA2 is needed for utilization of lipoyl-peptides, a specific form of lipoate that is available within the host cell (40). This example highlights the importance of organic accessory co-factors that bacteria need for running metabolism during infection, and also shows how pathogens may evolve to have an expanded ability for obtaining important co-factors that exist in various forms within the host.
The end of the line – using a novel electron sink to outpace the competitors
The last example illustrating the flexible and opportunistic logic of one bacterial pathogen's approach to metabolism involves S. Typhimurium, which is able to make its metabolic living inside the host cell by acquiring intracellular glucose and undergoing standard aerobic respiration (glycolysis + TCA cycle). But this bacterium is also able to replicate within the lumen of the gastrointestinal tract, outside of host cells and amongst the intestinal microbiota, where oxygen is limiting, and where competition for carbon sources is fierce. The key to understanding the significance of these findings is an appreciation of the differences between using fermentation versus respiration for ATP generation. Fermentation can be simple glycolysis run cyclically, and is characterized by a low yield of ATP per oxidized carbon source input relative to respiration (Figure 1). Unlike fermentation, respiration capitalizes on energy released during the process of electron transport to create a proton gradient that drives enzymatic ATP synthesis (oxidative phosphorylation), resulting in a much larger yield of usable energy. But the central difference is that only respiration requires a highly oxidized, terminal electron acceptor to act as a sink at the end of electron transport, whereas fermentation relies on the enzymatic re-oxidation of the electron carriers that were reduced along the pathway (NADH to NAD+), generating some waste product in the process, often an acid such as lactate. Although the electron sink that supports the highest theoretical energy yield via respiration is oxygen, many other molecules (like nitrate or sulfate) can play the role of terminal electron acceptor as well. And this brings the discussion back to S. Typhimurium. Infection of mice with S. Typhimurium results in acute intestinal inflammation that is induced by various bacterial virulence factors, including two different bacterial secretion systems (44). The gut inflammation promotes a less hospitable environment for the resident microbiota, and allows the Salmonellae to outcompete the resident microbiota (45). Part of the mechanism that supports this growth advantage for S. Typhimurium in the inflamed gut is its unique ability to use the molecule tetrathionate as a terminal electron acceptor for anaerobic respiration. Bacteria in the mouse colon, a generally anaerobic environment, produce hydrogen sulphide as a fermentation byproduct, which is converted to the less toxic molecule thiosulphate by host mucosal cells. Importantly, thiosulphate can be converted to tetrathionate by a strong oxidant. In a mouse model of colitis, C57BL/6 mice infected with S. Typhimurium show acute levels of caecal inflammation whereas infection with bacteria lacking the ability to use tetrathionate in anaerobic respiration result in inflammation with increased caecal tetrathionate concentration (46). Tetrathionate was not detected in the gut of mice infected with S. Typhimurium lacking the virulence factors that cause inflammation, strongly supporting the idea that reactive oxygen species generated by the host during inflammation are responsible for the appearance of tetrathionate that is usable by the bacteria. Bacteria able to use tetrathionate respiration showed a strong growth advantage compared to tetrathionate metabolic mutants in the mouse gut, but not in the spleen, suggesting that this metabolic advantage is site-specific in the host (46). Importantly, generation of reactive oxygen by the host NADPH oxidase during inflammation, and not nitric oxide production by the nitric oxide synthase, seems to be responsible for the generation of tetrathionate in the inflamed gut, since wild type bacteria did not outcompete tetrathionate metabolic mutants in mice lacking the gene encoding the NADPH oxidase. The ability to undergo anaerobic respiration with a terminal electron acceptor generated by the combined activity of both the host (reactive oxygen) and the resident microbiota (hydrogen sulfide) is a remarkably unique strategy to bolster a pathogenic metabolic advantage.
Tetrathionate anaerobic respiration is not the only trick used by S. Typhimurium to outcompete the microbiota in the colon. Nitrate is an even more electronegative electron acceptor than tetrathionate, and thus promotes an even stronger energy yield when used for anaerobic respiration. In a mouse model of colitis, a strain of S. Typhimurium lysogenized by a bacteriophage carrying the virulence gene sopE was able to switch to nitrate metabolism in the gut to outcompete the native microbiota (47). Like the tetrathionate example, the generation of nitrate in the gut is dependent on host inflammation. However, unlike tetrathionate, luminal nitrate concentration rises due to the host expression of the inducible nitric oxide synthase, which generates nitric oxide that then leads to nitrate. Bacteria with the sopE containing bacteriophage did not have a growth advantage in iNOS-deficient mice. Interestingly, the lysogenized bacteria suppress the genes for tetrathionate metabolism both in vitro and in vivo when nitrate is available, illustrating a remarkable level of metabolic flexibility and control (47).
The examples outlined thus far clearly show that bacterial pathogens engage in energy yielding metabolism during infection in ways that take advantage of the specific host environment at the site of infection. The process of bacterial metabolism is extremely dynamic, and pathogens exhibit a wide range of metabolic control and flexibility during infection. A key point to emphasize about energy-yielding metabolic pathways is that, although we consider them mainly in terms of their importance in ATP generation, the reality is that intermediates for both anabolism and catabolism are constantly being fed into and siphoned off these pathways for many other metabolic needs, with constant regulation based on the moment-to-moment needs of the cell. Thus, each of the stories illustrated here represent a “tip of the iceberg” situation, posing new questions as to how pathogenic catabolic behavior is connected to the anabolic needs and strategies during infection as well.
III. Ion and Nutrient Acquisition by Bacterial Pathogens
The human body is a rich reservoir of fundamental nutrients for bacteria that can exploit it, and nutrient acquisition is an essential aspect of host-pathogen interactions. The mechanisms of energy generation described in the previous section require high levels of carbon and other building blocks, and bacteria must extract these nutrients from resources found in the host, including sugars, amino acids, lipids and nitrogen-containing compounds. Moreover, transition metals like iron, zinc and manganese are essential for survival and proliferation of all living organisms. This section will focus on strategies used by bacteria to gain access to these critical nutrients from the complex host environment.
Transition metal ions – precious metals for life
Transition metals, including iron, zinc, manganese and copper among others, are nutrients required for many biological processes. These metals have unique redox potential as they can undergo change in their oxidation state, and serve as essential co-factors for many enzymes (48). In bacteria, it is estimated that 30 to 45% of enzymes required a metal co-factor for function (49). However, at high concentrations, these metals are toxic for the cells as they perturb cellular redox potential and can drive production of highly reactive hydroxyl radicals. Therefore, all organisms possess biochemical systems to sense and regulate metal levels, and mammals have evolved strategies to sequester free metal ions to limit toxicity, and also to restrict availability of these metal ions to invading microorganisms. This concept of growth restriction by limiting access to essential metals is called nutritional immunity (50, 51). As described below, pathogens have evolved numerous mechanisms for metal uptake or efflux to circumvent nutritional immunity (Table 2).
Table 2. Summary of bacterial mechanisms for metal ion acquisition.
| Pathogens | Infection site (model) | Ion | System contributing to virulence | Ref. |
|---|---|---|---|---|
| S. Typhimurium | Systemic infection (mice) | Fe | EntC, EntB, IroN (salmochelin/enterobactin) | (62) |
| Streptococcus suis | Systemic infection (mice) | Fe | FeoB, iron transporter | (56) |
| Campylobacter jejuni | Gastrointestinal tract (chick) | Fe | FeoB, iron transporter | (58) |
| Staphylococcus aureus | Systemic infection (mice) | Fe | Isd heme uptake system | (69) |
| Yersinia pestis | Systemic infection/bubonic plaque (mice) | Fe | Yef and Feo iron transporter | (127) |
| Staphylococcus aureus | Systemic infection (mice) | Mn | MntABC and MntH Mn transporters | (82) |
| Escherichia coli (UPEC) | Bladder and kidney (mice) | Zn | ZnuABC and ZupT Zn transporters | (86) |
| S. Typhimurium | Inflammed gut (mice) | Zn | ZnuABC Zn uptake system | (84) |
| Acinetobacter baumannii | Lungs (mice) | Zn | ZnuABC Zn uptake system | (83) |
| Mycobacterium tuberculosis | Differentiated human macrophages | Zn | CtpC P1-type ATPase, Zn efflux system | (85) |
| Mycobacterium tuberculosis | Lung and lymph nodes (Guinea pigs) | Cu | MctB copper transport protein B | (95) |
| Escherichia coli | Mouse macrophage-like cells | Cu | CopA ATPase, Cu+ export system | (89) |
| Escherichia coli (UPEC) | Bladder (mice) | Cu | Yersinibactin | (96) |
| Pseudomonas aeruginosa | Systemic infection (mice) | Cu | CopA1 P-type ATPase, Cu+ efflux system | (94) |
(A) Iron
Iron (Fe) is the most abundant transition metal in the human body, but free iron is almost undetectable. Bacteria need concentrations on the order of 10-6M iron to survive and proliferate, whereas circulation of free iron in human blood is approximately 10-18M. Iron is a co-factor for many enzymes involved in fundamental cellular processes, including DNA replication, transcription, metabolism and energy generation through respiration.
(1) Iron sequestration by the host
In vertebrates, most iron is stored intracellularly in complex with heme, a tetrapyrrole ring that binds a ferrous iron (Fe2+) atom. Heme is primarily found in the oxygen-transporting protein hemoglobin, contained within circulating erythrocytes. Moreover, free hemoglobin or heme is bound by the serum proteins haptoglobin and hemopexin, respectively (52). At physiological pH, free Fe2+ in the extracellular environment is rapidly oxidized to ferric iron (Fe3+) and captured by the serum protein, transferrin, or by lactoferrin, a glycoprotein found in human secretions such as saliva, tears and breast milk, rendering free ferrous iron unavailable for microorganisms. In cells, Fe3+ is captured by the storage protein ferritin. In phagocytic cells of the innate immune system, such as macrophages and neutrophils, the phagosomal membrane protein NRAMP1 (natural resistance associated macrophage protein 1), pumps Fe2+ out of the phagosomal compartment, which physically surrounds bacteria taken up by the host cell, in order to reduce metal ion availability to intracellular bacteria (53). These mechanisms of iron limitation also exist in plants and invertebrates, which suggest a conserved and universal innate immune response against invading pathogens (54, 55).
(2) Bacterial mechanisms of iron acquisition
Host mechanisms of iron sequestration discussed above efficiently limit availability of iron, but bacterial pathogens possess their own weapons to counteract host innate defenses and to acquire Fe2+ that will be used as a nutrient source. Specific strategies used by a given pathogen mainly depend on the particular sites of infection and the intracellular or extracellular life-style. Bacterial mechanisms for iron acquisition can be divided into three major categories: siderophores, heme acquisition systems and free Fe2+ transporters (50). Free Fe2+ can enter the periplasm of Gram-negative bacteria through non-specific porins and gets transported across the cytoplasmic membrane of Gram-negative and Gram-positive bacteria by the FeoB family of transporters. Members of the FeoB family of metal transporters are important for virulence of many pathogens, including Streptococcus suis and Campylobacter jejuni (56-58).
Siderophores are low-molecular mass iron chelators secreted by bacteria that bind to Fe3+ with an affinity higher than mammalian Fe-binding proteins like lactoferrin and transferrin, allowing them to steal iron from these host proteins. Iron-bound siderophores are recognized at the bacterial surface by specific bacterial receptors and transported into the cytoplasm where bound Fe3+ is reduced into Fe2+ or released by degradation of the siderophore. Epithelial cells and neutrophils can neutralize some siderophores by secreting the antimicrobial protein, lipocalin-2 (also know as neutrophil gelatinase-associated lipocalin), during acute infection (59). Lipocalin-2 expression is controlled by signaling through Toll-like receptors, pattern recognition receptors of the innate immune system responsive to microbial ligands. Lipocalin-2 can sequester the catecholate siderophore, enterobactin, produced by common pathogenic enterobacteria, such as Escherichia coli and Salmonella enterica serovar Typhimurium, causative agents of gastrointestinal diseases (59). To circumvent this immune defense, some bacteria synthesize a modified siderophore, called salmochelin, which is a glucosylated derivative of enterobactin that is not recognized by lipocalin-2 (60). Production of salmochelin is essential for efficient colonization and growth of S. Typhimurium within the inflamed gut (61, 62).
Heme represents one of the most abundant sources of iron and pathogens have evolved heme uptake systems as well as hemophore systems. To gain access to heme, which is mainly bound to hemoproteins, bacteria secrete exotoxins to degrade hemoglobin (63). Free heme is captured by specific membrane complexes (TonB-dependent heme acquisition systems in Gram-negative bacteria, and Isd systems in Gram-positive bacteria) and transported into the cytoplasm where heme-catabolizing enzymes release Fe2+ (50, 64). Heme is the preferred source of iron of Staphylococcus aureus, a member of the skin microbiome that can cause tissue abscesses, as well as more severe diseases including bacteremia and endocarditis (65-67). S. aureus can growth in vitro with erythrocytes as the sole source of iron. Hemolysins secreted by S. aureus lyse erythrocytes, releasing hemoglobin, which is targeted by the IsdB (iron regulated surface determinant B) cell-wall receptor. As alternative routes for iron acquisition, IsdH recognizes the hemogloblin-haptoglobin complex, whereas IsdA captures free heme (68). The Isd system is critical for iron acquisition during systemic S. aureus infection and for efficient colonization of spleen, kidneys and heart (69). In addition to surface heme receptors, some bacteria also secrete hemophores, which are proteins that bind heme and can be re-acquired by the bacteria. Bacillus anthracis, the causative agent of anthrax, secretes two hemophores, IsdX1 and IsdX2, which bind heme and are required for growth in iron-depleted environments (70-72).
(B) Zinc and manganese
Sequestration of zinc (Zn) and manganese (Mn) is also an important innate defense strategy used by vertebrates to fight bacterial infection. Zinc is used for its structural and catalytic roles in a large number of proteins. In bacteria, 5-6% of the proteome consists of zinc-binding proteins, justifying the need for specific acquisition systems (73). Among them, enzymes involved in central metabolic pathways, DNA repair, response to oxidative stress and antibiotic resistance require zinc as an essential co-factor (74). Manganese is also an important metal for bacteria, as many Mn-dependent enzymes encoded by pathogens are involved in central carbon metabolism and are necessary for resistance to oxidative stress (75).
(1) Control of Mn2+ and Zn2+ levels by the host
Vertebrates secrete proteins that belong to the S100 family and inhibit bacterial growth by chelating Mn2+ and Zn2+. S100A7 protein (also called psoriasin) is secreted as an homodimer by keratinocytes and inhibits E. coli growth on human skin through sequestration and chelation of Zn2+ (76). Similarly, calprotectin, which is a heterodimer formed by S100A8 and S100A9 (also known as calgranulin A and B, respectively), represent ∼40% of the cytosolic protein pool in neutrophils, and exhibit antimicrobial activity against bacterial pathogens through chelation of nutrients Mn2+ and Zn2+ (77, 78). Calprotectin is the major neutrophil-derived protein found in abscesses formed by S. aureus (77). As is true for iron, immune cells also control Mn2+ and Zn2+ availability to intracellular bacteria by multiple mechanisms. Manganese is pumped out of the phagosome by the NRAMP1 protein, whereas zinc levels are reduced by the action of members of the ZIP and ZnT zinc transporter family (79-81).
(2) Bacterial acquisition of Mn2+ and Zn2+
The fight for metals is a continuous ongoing process between bacteria and the host. As described for iron, bacterial mechanisms exist that minimize the chelation of zinc and manganese by calprotectin. To counteract this antimicrobial response, S. aureus expresses specialized manganese transporters, MntABC and MntH to compete with calprotectin for Mn2+ during systemic infection (82). Likewise, S. Typhimurium and Acinetobacter baumannii express the high-affinity zinc transporter ZnuABC to overcome the antimicrobial effect of calprotectin during infection (83, 84). S. Typhimurium, in addition to neutralizing the effect of calprotectin, also exploits the accumulation of this antimicrobial molecule during infection of the inflamed gut to advantageously compete with the resident host microbiota (84).
(3) Use of zinc in host defense
In the constant battle to limit metal access during infection, mammals also exploit the toxicity associated with elevated concentrations of transition metals as an anti-microbial strategy. Macrophages control zinc levels inside the bacteria-containing phagosome during infection by M. tuberculosis. This increase in free Zn2+ results in accumulation of the metal inside the cytoplasm of M. tuberculosis, reducing intracellular growth. Mycobacterium resolves this zinc-dependent intoxication by expressing CtpC, a metal efflux P1-type ATPase (85). Similarly, the ZnuABC and ZupT zinc exporters are required for optimal colonization of mice bladders and kidneys during urinary tract infection cause by E. coli (86). Thus, it is clear that while bacteria must acquire metal ions for replication and metabolism, levels of these key nutrients within the bacteria themselves are tightly regulated.
(C) Copper
Copper (Cu) is a redox-active metal ion that can exist in a reduced (Cu+) or an oxidized state (Cu2+) and, as a consequence, is critical for proteins involved in a wide range of cellular processes. Copper is also used by some metalloenzymes involved in electron transfer reactions, such as cytochrome oxidase (87). However, in contrast to Fe, Mn or Zn that are required for bacterial growth, copper is mainly recognized for its antimicrobial benefits and its role in innate immune defense against bacteria.
(1) Copper toxicity
In mammals, the innate immune response provoked by secretion of the pro-inflammatory cytokine, interferon-γ, induces expression of Cu pumps, resulting in an increased Cu concentration at the infection site (88). In activated macrophages, the Cu+ transport protein 1 (CTR1) takes Cu+ inside the cell, whereas the P-type ATPase ATP7A pumps Cu+ inside the phagolysosome, again emphasizing the importance of precise regulation of metal ion levels (89). Multiple mechanisms have been described to explain copper toxicity in bacteria. Macrophages use copper to increase bacterial killing by oxidative damage. In the phagosome, Cu+ can interact with hydrogen peroxide to produce the highly reactive hydroxyl anion and hydroxyl radical, which in turn causes lethal damage to lipids, proteins and nucleic acids. Unligated copper also causes disruption of iron-sulfur clusters by replacing the Fe atoms, resulting in the disruption of protein structure. Enzymes that contain a Fe-S cluster, such as dehydratases involved in branch-chain amino acid synthesis in E. coli, are targeted by Cu during infection (90). An alternative mechanism has been described during Neisseria gonorrhoeae infection where increased Cu levels result in increased sensitivity to reactive nitrogen species produced by innate immune cells (91).
(2) Copper regulation by bacteria
The recurring theme of measure:countermeasure should be now apparent, as bacteria have evolved strategies to prevent copper toxicity imposed by the host and to tightly regulate the level of cytoplasmic copper (92). In general, bacterial proteomes contain only few copper-binding proteins (∼0.3%), and these proteins are mainly localized in the periplasm or in the cyoplasmic membrane, preventing unligated copper from accessing the cytoplasm. Bacteria can also express Cu-binding proteins to sequester free Cu+, such as the Mycobacterium methallothionein, (MymT), which protects the bacterium from copper toxicity (93). Many pathogenic bacteria encode a P-type ATPase Cu+ efflux protein that pumps copper outside of the bacterial cytoplasm. For E. coli, a mutant lacking the CopA ATPase is hypersensitive to killing by macrophage-like cells (89). Similarly, a CopA1 ATPase mutant of Pseudomonas aeruginosa is attenuated in a systemic mouse model of infection (94). Mycobacterium tuberculosis also encodes a membrane copper-transporter that is required to ensure resistance to copper toxicity and intracellular survival in vivo (95). Lastly, uropathogenic E. coli (UPEC) use an unusual method to limit copper toxicity. During bladder infection, UPEC secretes the siderophore, yersiniabactin, that allows capture of tessential iron, but this compound can also bind to copper and prevent its toxicity (96). Consistently, E. coli strains that produce yersiniabactin exhibit greater resistance to copper-related toxicity.
Nutrient acquisition
In the host cell, many nutrients essential for bacterial growth are enclosed within complex molecules such as proteins and higher-order glycolipids and are not readily accessible. Moreover, depending on the particular niche colonized by the pathogen or the local inflammation state, nutrient supply might vary greatly during the course of infection. Therefore, nutritional restriction by the host is an important aspect of the innate immune defense against pathogens (97). The following section will highlight some of the mechanisms used by pathogenic bacteria to maximize nutrient acquisition in the complex environment of the host (graphically summarized in Figure 2).
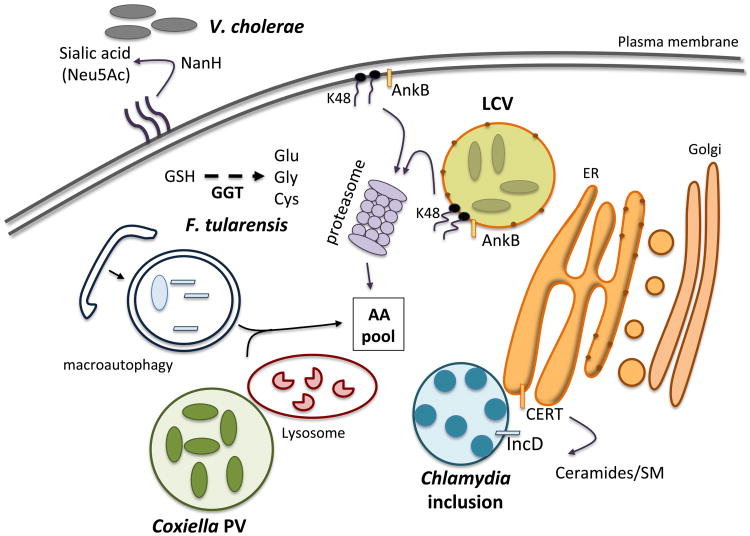
Figure 2. Graphical summary of bacterial mechanisms used to release intracellular host nutrients.
Pathogenic bacteria perturb host cell functions to provide resources for survival and replication. Abbreviations: GSH (glutathione), GGT (γ-glutamyl transpeptidase), LCV (Legionella-containing vacuole), ER (endoplasmic reticulum), AA (amino acids), CERT (ceramide transfer protein), SM (sphingomyelin).
(A) The arsenal of microbial degradative enzymes
Host macromolecules are a good source of carbon and nitrogen when bacterial enzymes, such as proteases or phospholipases, can degrade these molecules to extract essential nutrients. Vibrio cholerae, the causative agent of the severe diarrheal disease cholera, utilize host sialic acid to generate carbon, nitrogen and energy (98, 99). Sialic acids are a family of nine-carbon amino sugars found in abundance in the mammalian gut. V. cholera encodes a sialic acid catabolism gene cluster (the nan-nag cluster) located in the VPI2 pathogenicity island, which is only found in toxigenic strains that are the primary cause of disease. NanH, a neuraminidase, is required for cleavage of bound sialic acid from higher-order gangliosides found at the cell surface. The free sialic acid (mainly N-acetylneuraminic acid or Neu5Ac) is taken up by V. cholerae and converted in fructose-6-P that feeds into the glycolytic pathway. This catabolic pathway plays a significant role for V. cholera colonization of infant mice, an important animal model for studying human cholera (98). The nan cluster is also found in many other human pathogens that colonize the gut, including E. coli, S. Typhimurium, Shigella spp. and Clostridium spp., among others, and a correlation between sialic acid catabolism and bacterial fitness in the gut is now emerging (100). Of note, the use of NanH to hydrolyze host sialic acid as a carbon source is not restricted to gut pathogens, but is also required for biofilm formation and lung colonization by P. aeruginosa (101).
The γ-glutamyl transpeptidase, GGT, produced by F. tularensis is another striking example of a microbial strategy for nutrient acquisition (102). F. tularensis is a highly infectious Gram-negative bacterial pathogen that causes the deadly disease, tularemia. After entry into mammalian cells, F. tularensis replicates in the cytosol using amino acids as its major source of carbon and energy required for proliferation. F. tularensis particularly requires high levels of cysteine to support intracellular proliferation, as cysteine is metabolized into pyruvate, a key substrate of the TCA cycle. As cysteine is the most limiting amino acid in humans, F. tularensis has evolved to use host gluthathione (GSH), a tri-peptide (L-γ-L-glutamyl-L-cysteinyl-glycine) that is abundant in the cytosol. The GGT enzyme of F. tularensis cleaves GSH to release cysteine; mutants of Francisella that lack this enzyme are severely impaired for growth in mouse macrophages and attenuated for virulence in mice (102).
(B) Raiding the pantry: exploitation of host processes to provide food
Chlamydia species are Gram-negative obligate human intracellular bacteria that manipulate the host cell to enhance nutrient and lipid acquisition. Chlamydia resides in a unique Chlamydia-containing inclusion in the cytosol of host cells, and is one of the few bacterial pathogens that need host-derived lipids such as sphingomyelin (SM) for intracellular proliferation. SM is incorporated into the Chlamydia cell wall as well as into the growing inclusion membrane during bacterial replication (103). To acquired host-derived SM, Chlamydia trachomatis uses the IncD protein, an effector of the syringe-like bacterial Type III secretion system, which is inserted into the inclusion membrane. IncD mediates recruitment of the host ceramide transfer protein, CERT, at the site of contact between the bacterial inclusion and the endoplasmic reticulum (ER) membrane (104). CERT is involved in the non-vesicular transfer of ceramides, precursors of SM, from the ER to the Golgi apparatus. The recruitment of CERT to the bacterial inclusion is required for C. trachomatis intracellular replication (105). Thus, Chlamydia, with its reduced genome, illustrates how obligate intracellular pathogens are able to extensively exploit host trafficking pathways to gain essential lipids/nutrients to support replication.
Some bacteria are able to obtain nutrients by manipulating host degradation processes rather than relying on their own degradative mechanisms, including C. trachomatis and Chlamydia pneumoniae. Lysosomes are cellular degradation compartments within the host cell that contain an array of degradative enzymes that break down proteins, carbohydrates, nucleic acids and lipids. Depletion of amino acids within the lysosomal compartment results in growth inhibition of Chlamydia spp, which suggests these bacteria rely on degradation of exogenous cargo for nutrient acquisition within the inclusion (106). Coxiella burnetti, an obligate intracellular bacterium that causes human Q fever, also uses the degradative properties of the lysosome for nutrient acquisition (107). C. burnetti proliferates inside the host cell in a specialized vacuole, termed the parasitophorous vacuole (PV), which is a phagolysosome-like compartment (108). An acidic environment is required for optimal growth of these bacteria, and amino acids and others nutrients required for replication likely reach the PV via fusion of protein/peptide-loaded lysosomes to the PV.
One of the most well-studied bacterial pathogens that exploits host cell degradation capacity is Legionella pneumophila, a bacterium that replicates within alveolar macrophages causing pneumonia (Legionnaires's disease). Inside macrophages, L. pneumophila replicates within the Legionella-containing vacuole (LCV) that is remodeled by a series of secreted effectors and resembles the rough ER membrane (109). The level of amino acids, the main source of carbon and energy generation for Legionella, within the host cytosol is not sufficient to power the robust intracellular replication of this bacterium. To gain access to these key nutrients, Legionella boosts the cellular level of amino acids by subverting host proteasomal degradation processes (110). The secreted bacterial effector AnkB is anchored to the cytosolic face of the LCV membrane by a lipid modification, and triggers polyubiquitin tagging of proteins at the LCV surface. These ubiquitinated proteins are subsequently targeted to the 26S proteasome and rapidly degraded to generate a localized supply of amino acids (110). AnkB is also injected into the host cell directly after bacterial attachment and localized to the plasma membrane. This allows rapid polyubiquitination and degradation of host proteins and generates a nutrient-rich environment for the establishment of the LCV (111). Not surprisingly, the Legionella AnkB mutant has a severe growth defect for intravacuolar replication in macrophages and has a reduced ability to cause pulmonary disease in mice. L. pneumophila also encodes amino acid transporters, such as the PhtA transporter involved in import of threonine, an essential amino acid for Legionnella, into the bacteria during infection (112). The PhtA transporter is required for growth, but not survival inside macrophages.
F. tularensis, a bacterium that can replicate up to1000-fold in the host cell cytosol, also manipulates host degradation machinery to facilitate nutrient uptake. Acquisition of host amino acids is critical for this intracellular pathogen, as F. tularensis is auxotrophic for six amino acids, relying solely on the host to provide them. To promote a permissive growth environment, F. tularensis induces macroautophagy, a host recycling pathway normally stimulated by starvation that targets host proteins, organelles and invading pathogens for degradation (113). Macroautophagy inhibition results in decreased intracellular replication of Francisella, which can be rescued by adding excess amino acids, highlighting the role of this host pathway for bacterial proliferation (113). Autophagy has been studied mainly for its role in innate immune defense, but several bacterial pathogens like Francisella, have evolved mechanisms to evade killing by autophagic degradation and instead exploit this pathway to increase bacterial proliferation. Anaplasma phagocytophilum, an obligate intracellular bacterium that belongs to the Rickettsiales order and causes human granulocytic anaplasmosis, is another example of a bacterium that actively induces autophagy to stimulate the richer host nutrient environment essential for proliferation (114).
IV. Metabolic control of host:pathogen interactions
Pathogenesis is defined by the nature of the interaction between host and pathogen. As described above, bacterial pathogens are adept at exploiting the host as a chemical warehouse of sorts. However, the complex metabolic interactions of the bacteria with its host also signal regulatory processes that expand the bacterial repertoire for successful infection. In this section, we will tie in previous examples of pathogen metabolism with mechanisms used by bacteria to position themselves to take advantage of the host.
(A) Orienteering: nutrient sensors control metabolism and niche adaptations
S. Typhimurium is able to outcompete resident gut microbiota by stimulating intestinal inflammation in a mouse model of colitis, and then relying on tetrathionate respiration for robust replication, as described earlier in this chapter (46). However, this striking result does not explain how S. Typhimurium can position itself at the intestinal interface where tetrathionate is most abundant. Previous studies had determined that chemotaxis, the energy-dependent process by which bacteria move up or down chemical gradients, was important for the fitness of S. Typhimurium in models of colitis (115). Baumler and colleagues identified tetrathionate as a signal for the Aer methyl-accepting chemotaxis protein (116). Aer regulated chemotaxis towards tetrathionate, and only conferred a fitness advantage upon S. Typhimurium under conditions of tetrathionate respiration in vivo. These findings indicate that S. Typhimurium not only has the metabolic capacity to remodel the biochemical environment of the intestine to give itself a competitive edge, but also encodes the machinery to seek areas with the highest concentration of alternative electron acceptors, like tetrathionate, to enhance the opportunity for rapid replication.
In the previous case, S. Typhimurium succeeds by changing the host intestinal environment to a more inflammatory state that favors replication, however, other pathogens such as enterohaemorrhagic Escherichia coli (EHEC) take advantage of nutrient signals generated by the resident microbiota to control virulence and metabolic gene expression. EHEC encodes a two component sensing system, FusKR, that consists of a sensor kinase (FusK) and a response regulator (FusR). FusK senses fucose and is required for EHEC colonization of the mammalian intestine (35). Although fucose is present at high levels in the intestine, host cells incorporate most fucose molecules into complex carbohydrate structures, like mucin, which is abundant at the surface of the intestinal epithelium. Fucose can be liberated from mucin by the fucosidase activity of Bt, a common resident of the microbiota (117). During growth in mucin, Bt freed fucose for sensing by EHEC FusKR, regulating EHEC virulence and metabolic gene expression and thereby modulating pathogenesis (35). In this case, free fucose enabled EHEC to spatially orient in the mammalian intestine, likely limiting expression of fucose utilization and virulence genes to localized environments where such metabolic capacity provides a competitive advantage.
(B) Evading immune system predation
Metabolism can also provide powerful weapons used by bacteria to manipulate host immune responses, promoting pathogen survival. M. tb can be considered one of the most successful human pathogens, having been discovered in human remains thousands of years old. The success of M. tb likely relies in large part on the myriad ways in which the bacterium controls the immune response to its advantage. In a screen for M. tb genes that enhanced fitness in response to immune pressure, Rubin and colleagues identified genes of the tryptophan biosynthesis pathway (118). One established mechanism of nutritional immunity relies on the host enzyme, indoleamine 2,3-dioxygenase (IDO), which degrades the amino acid tryptophan, decreasing its availability to pathogens (119). IDO is induced by the pro-inflammatory cytokine, interferon-γ, and can be effective in host resistance against pathogens that are tryptophan auxotrophs, like Chlamydia or Toxoplasma spp. (120). However, M.tb can upregulate trp biosynthesis genes within the infected host to overcome the lack of tryptophan availability. A trp mutant strain was hypersusceptible to killing by IFN-γ-treated macrophages compared to wildtype M.tb, and that hypersusceptibility was reversed by inhibiting IDO, or by addition of exogenous tryptophan (118). Thus, M.tb uses a metabolic strategy to counteract a specific mechanism of host nutritional immunity.
Host immune cell metabolism is increasingly appreciated as a critical determinant of immune function (121). The nucleoside adenosine has powerful immunosuppressive effects on the immune system, promoting T cell tolerance, or anergy (122). Adenosine receptors can play an important role in protecting the host from tissue damage or organ failure in models of polymicrobial sepsis (123). The Gram-positive pathogen Staphylococcus aureus is a common colonizer of human skin that can also cause abscesses, soft tissue infections and systemic disease (124). Mutations in the S. aureus adenosine synthase adsA gene were found to predispose bacteria to killing in human blood (125). AdsA was required for staphylococcal virulence and abscess formation, indicating the importance of this protein during infection in vivo. Notably, the AdsA protein is not found in the bacterial cytosol, but rather outside the bacterial cell, anchored to the cell wall. Incubation of S. aureus in blood increased the adenosine concentration in an AdsA-dependent manner, and this enzymatic activity contributed to virulence (125). These results indicate that S. aureus is able to synthesize adenosine in human blood, promoting an immunosuppressive environment. S. aureus adsA mutants were more susceptible to neutrophil killing, suggesting that at least one mechanism of adenosine-dependent immunosuppression occurs through modulation of innate immune effector functions.
The host:pathogen interactions described above indicate that bacterial metabolism is an essential component of the disease process. Acquisition of nutrients to support energy metabolism and biosynthesis drives bacterial replication within the host. Speed and efficiency of bacterial replication likely play a determining role in the outcome of disease. However, it is also apparent that bacterial metabolism can enhance other aspects of pathogenesis, including nutrient sensing and immune evasion. A deeper appreciation for pathogen metabolic capabilities will continue to emerge as we apply more sophisticated methodologies to our understanding of pathogenesis. Defining genomes, transcriptomes and metabolomes at a global level will paint a more comprehensive picture of how the tremendous metabolic capacity of bacterial pathogens enables them to exploit the human host.
Table 1. Summary points of energy-yielding metabolism for select pathogenic bacteria.
| Label | Bacterium | Points of interest for metabolism during infection | Select references |
|---|---|---|---|
| A | Pathogenic Streptococci Streptococcus mutans S. pyogenes S. pneumoniae |
|
(1-13) |
| B |
Salmonella enterica serovar Typhimurium |
|
(14, 15, 19, 23, 44-47) |
| C | Brucella abortus |
|
(21) |
| D |
Mycobacterium tuberculosis |
|
(24, 27, 28, 29) |
| E | Clostridium difficile |
|
(33) |
| F | Enterohemorrhagic Esherichia coli (EHEC) |
|
(34-36) |
| G |
Aggregatibacter actinomycetemcomitans |
|
(38) |
| H | Listeria monocytogenes |
|
(40, 41) |
References
- 1.Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 2011;19:341–348. doi: 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipmann F. Metabolic generation and utilization of phosphate bond energy. Adv Enzymol Rel S Bi. 1941;1:99–162. [Google Scholar]
- 3.de Duve C. The other revolution in the life sciences. Science. 2013;339:1148. doi: 10.1126/science.339.6124.1148-a. [DOI] [PubMed] [Google Scholar]
- 4.Adler CJ, Dobney K, Weyrich LS, Kaidonis J, Walker AW, Haak W, Bradshaw CJ, Townsend G, Soltysiak A, Alt KW, Parkhill J, Cooper A. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet. 2013;45:450–455, 455e451. doi: 10.1038/ng.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moye ZD, Zeng L, Burne RA. Fueling the caries process: carbohydrate metabolism and gene regulation by Streptococcus mutans. Journal of oral microbiology. 2014;6 doi: 10.3402/jom.v6.24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, Burne RA. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol. 2008;190:2340–2349. doi: 10.1128/JB.01237-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kietzman CC, Caparon MG. Distinct time-resolved roles for two catabolite-sensing pathways during Streptococcus pyogenes infection. Infect Immun. 2011;79:812–821. doi: 10.1128/IAI.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One. 2011;6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One. 2012;7:e33320. doi: 10.1371/journal.pone.0033320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King SJ, Hippe KR, Weiser JN. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol. 2006;59:961–974. doi: 10.1111/j.1365-2958.2005.04984.x. [DOI] [PubMed] [Google Scholar]
- 12.Shelburne SA, Davenport MT, Keith DB, Musser JM. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 2008;16:318–325. doi: 10.1016/j.tim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnaugh AM, Frantz LJ, King SJ. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J Bacteriol. 2008;190:221–230. doi: 10.1128/JB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowden SD, Rowley G, Hinton JC, Thompson A. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar typhimurium. Infect Immun. 2009;77:3117–3126. doi: 10.1128/IAI.00093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tchawa Yimga M, Leatham MP, Allen JH, Laux DC, Conway T, Cohen PS. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect Immun. 2006;74:1130–1140. doi: 10.1128/IAI.74.2.1130-1140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dougan G, Baker S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol. 2014;68:317–336. doi: 10.1146/annurev-micro-091313-103739. [DOI] [PubMed] [Google Scholar]
- 17.Thiennimitr P, Winter SE, Baumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol. 2012;15:108–114. doi: 10.1016/j.mib.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salcedo SP, Noursadeghi M, Cohen J, Holden DW. Intracellular replication of Salmonella typhimurium strains in specific subsets of splenic macrophages in vivo. Cell Microbiol. 2001;3:587–597. doi: 10.1046/j.1462-5822.2001.00137.x. [DOI] [PubMed] [Google Scholar]
- 19.Mercado-Lubo R, Leatham MP, Conway T, Cohen PS. Salmonella enterica serovar Typhimurium mutants unable to convert malate to pyruvate and oxaloacetate are avirulent and immunogenic in BALB/c mice. Infect Immun. 2009;77:1397–1405. doi: 10.1128/IAI.01335-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime reports. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xavier MN, Winter MG, Spees AM, den Hartigh AB, Nguyen K, Roux CM, Silva TM, Atluri VL, Kerrinnes T, Keestra AM, Monack DM, Luciw PA, Eigenheer RA, Baumler AJ, Santos RL, Tsolis RM. PPARgamma-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe. 2013;14:159–170. doi: 10.1016/j.chom.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang K, Reilly SM, Karabacak V, Gangl MR, Fitzgerald K, Hatano B, Lee CH. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab. 2008;7:485–495. doi: 10.1016/j.cmet.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisele NA, Ruby T, Jacobson A, Manzanillo PS, Cox JS, Lam L, Mukundan L, Chawla A, Monack DM. Salmonella require the fatty acid regulator PPARdelta for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe. 2013;14:171–182. doi: 10.1016/j.chom.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloch H, Segal W. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J Bacteriol. 1956;72:132–141. doi: 10.1128/jb.72.2.132-141.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 26.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffe M, Emile JF, Marchou B, Cardona PJ, de Chastellier C, Altare F. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008;4:e1000204. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011;7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methe BA, Huttenhower C. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10:e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grice EA, Segre JA. The human microbiome: our second genome. Annual review of genomics and human genetics. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK. Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes. 2012;3:121–134. doi: 10.4161/gmic.19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut Microbiota-Produced Succinate Promotes C. difficile Infection after Antibiotic Treatment or Motility Disturbance. Cell Host Microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Njoroge JW, Gruber C, Sperandio V. The interacting Cra and KdpE regulators are involved in the expression of multiple virulence factors in enterohemorrhagic Escherichia coli. J Bacteriol. 2013;195:2499–2508. doi: 10.1128/JB.02252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The Gut Commensal Bacteroides thetaiotaomicron Exacerbates Enteric Infection through Modification of the Metabolic Landscape. Cell Host Microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown SA, Whiteley M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol. 2007;189:6407–6414. doi: 10.1128/JB.00554-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsey MM, Rumbaugh KP, Whiteley M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 2011;7:e1002012. doi: 10.1371/journal.ppat.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, European Listeria Genome Consortium T. Vazquez-Boland JA. Hpt, a bacterial homolog of the microsomal glucose- 6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U S A. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keeney KM, Stuckey JA, O'Riordan MX. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol Microbiol. 2007;66:758–770. doi: 10.1111/j.1365-2958.2007.05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Riordan M, Moors MA, Portnoy DA. Listeria intracellular growth and virulence require host-derived lipoic acid. Science. 2003;302:462–464. doi: 10.1126/science.1088170. [DOI] [PubMed] [Google Scholar]
- 42.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couve E, de Daruvar A, Dehoux P, Domann E, Dominguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, Portillo FG, Garrido P, Gautier L, Goebel W, Gomez-Lopez N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Perez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, Tierrez A, Vazquez-Boland JA, Voss H, Wehland J, Cossart P. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 43.Zhao X, Miller JR, Jiang Y, Marletta MA, Cronan JE. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem Biol. 2003;10:1293–1302. doi: 10.1016/j.chembiol.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 44.Santos RL, Raffatellu M, Bevins CL, Adams LG, Tukel C, Tsolis RM, Baumler AJ. Life in the inflamed intestine, Salmonella style. Trends Microbiol. 2009;17:498–506. doi: 10.1016/j.tim.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Baumler AJ. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio. 2012;3 doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 49.Klein JS, Lewinson O. Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metallomics : integrated biometal science. 2011;3:1098–1108. doi: 10.1039/c1mt00073j. [DOI] [PubMed] [Google Scholar]
- 50.Hood MI, Skaar EP. Nutritional immunity: transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg ED. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 52.Johnson EE, Wessling-Resnick M. Iron metabolism and the innate immune response to infection. Microbes Infect. 2012;14:207–216. doi: 10.1016/j.micinf.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cellier MF, Courville P, Campion C. Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 2007;9:1662–1670. doi: 10.1016/j.micinf.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 54.Franza T, Expert D. Role of iron homeostasis in the virulence of phytopathogenic bacteria: an ‘a la carte’ menu. Molecular plant pathology. 2013;14:429–438. doi: 10.1111/mpp.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geiser DL, Winzerling JJ. Insect transferrins: multifunctional proteins. Biochim Biophys Acta. 2012;1820:437–451. doi: 10.1016/j.bbagen.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 56.Aranda J, Cortes P, Garrido ME, Fittipaldi N, Llagostera M, Gottschalk M, Barbe J. Contribution of the FeoB transporter to Streptococcus suis virulence. International microbiology : the official journal of the Spanish Society for Microbiology. 2009;12:137–143. [PubMed] [Google Scholar]
- 57.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. Feo--transport of ferrous iron into bacteria. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2006;19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 58.Naikare H, Palyada K, Panciera R, Marlow D, Stintzi A. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect Immun. 2006;74:5433–5444. doi: 10.1128/IAI.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 60.Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Baumler AJ. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crouch ML, Castor M, Karlinsey JE, Kalhorn T, Fang FC. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2008;67:971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- 63.Drago-Serrano ME, Parra SG, Manjarrez-Hernandez HA. EspC, an autotransporter protein secreted by enteropathogenic Escherichia coli (EPEC), displays protease activity on human hemoglobin. FEMS Microbiol Lett. 2006;265:35–40. doi: 10.1111/j.1574-6968.2006.00463.x. [DOI] [PubMed] [Google Scholar]
- 64.Porcheron G, Garenaux A, Proulx J, Sabri M, Dozois CM. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Frontiers in cellular and infection microbiology. 2013;3:90. doi: 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coates R, Moran J, Horsburgh MJ. Staphylococci: colonizers and pathogens of human skin. Future Microbiol. 2014;9:75–91. doi: 10.2217/fmb.13.145. [DOI] [PubMed] [Google Scholar]
- 66.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 67.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 68.Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pishchany G, Dickey SE, Skaar EP. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect Immun. 2009;77:2624–2634. doi: 10.1128/IAI.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fabian M, Solomaha E, Olson JS, Maresso AW. Heme transfer to the bacterial cell envelope occurs via a secreted hemophore in the Gram-positive pathogen Bacillus anthracis. J Biol Chem. 2009;284:32138–32146. doi: 10.1074/jbc.M109.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Honsa ES, Fabian M, Cardenas AM, Olson JS, Maresso AW. The five near-iron transporter (NEAT) domain anthrax hemophore, IsdX2, scavenges heme from hemoglobin and transfers heme to the surface protein IsdC. J Biol Chem. 2011;286:33652–33660. doi: 10.1074/jbc.M111.241687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maresso AW, Garufi G, Schneewind O. Bacillus anthracis secretes proteins that mediate heme acquisition from hemoglobin. PLoS Pathog. 2008;4:e1000132. doi: 10.1371/journal.ppat.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andreini C, Banci L, Bertini I, Rosato A. Zinc through the three domains of life. J Proteome Res. 2006;5:3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- 74.Cerasi M, Ammendola S, Battistoni A. Competition for zinc binding in the host-pathogen interaction. Frontiers in cellular and infection microbiology. 2013;3:108. doi: 10.3389/fcimb.2013.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kehres DG, Maguire ME. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 76.Glaser R, Harder J, Lange H, Bartels J, Christophers E, Schroder JM. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 77.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 78.Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, Zhang Y, Betz C, Hench L, Fritz G, Skaar EP, Chazin WJ. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci U S A. 2013;110:3841–3846. doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jabado N, Jankowski A, Dougaparsad S, Picard V, Grinstein S, Gros P. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J Exp Med. 2000;192:1237–1248. doi: 10.1084/jem.192.9.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Begum NA, Kobayashi M, Moriwaki Y, Matsumoto M, Toyoshima K, Seya T. Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics. 2002;80:630–645. doi: 10.1006/geno.2002.7000. [DOI] [PubMed] [Google Scholar]
- 81.Kitamura H, Morikawa H, Kamon H, Iguchi M, Hojyo S, Fukada T, Yamashita S, Kaisho T, Akira S, Murakami M, Hirano T. Toll-like receptor-mediated regulation of zinc homeostasis influences dendritic cell function. Nat Immunol. 2006;7:971–977. doi: 10.1038/ni1373. [DOI] [PubMed] [Google Scholar]
- 82.Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, Chazin WJ, Caprioli RM, Skaar EP. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun. 2013;81:3395–3405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 2012;8:e1003068. doi: 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charriere GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe. 2011;10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sabri M, Houle S, Dozois CM. Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infect Immun. 2009;77:1155–1164. doi: 10.1128/IAI.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dupont CL, Grass G, Rensing C. Copper toxicity and the origin of bacterial resistance--new insights and applications. Metallomics : integrated biometal science. 2011;3:1109–1118. doi: 10.1039/c1mt00107h. [DOI] [PubMed] [Google Scholar]
- 88.Hodgkinson V, Petris MJ. Copper homeostasis at the host-pathogen interface. J Biol Chem. 2012;287:13549–13555. doi: 10.1074/jbc.R111.316406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Djoko KY, Franiek JA, Edwards JL, Falsetta ML, Kidd SP, Potter AJ, Chen NH, Apicella MA, Jennings MP, McEwan AG. Phenotypic characterization of a copA mutant of Neisseria gonorrhoeae identifies a link between copper and nitrosative stress. Infect Immun. 2012;80:1065–1071. doi: 10.1128/IAI.06163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Samanovic MI, Ding C, Thiele DJ, Darwin KH. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gold B, Deng H, Bryk R, Vargas D, Eliezer D, Roberts J, Jiang X, Nathan C. Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat Chem Biol. 2008;4:609–616. doi: 10.1038/nchembio.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwan WR, Warrener P, Keunz E, Stover CK, Folger KR. Mutations in the cueA gene encoding a copper homeostasis P-type ATPase reduce the pathogenicity of Pseudomonas aeruginosa in mice. Int J Med Microbiol. 2005;295:237–242. doi: 10.1016/j.ijmm.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 95.Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat Chem Biol. 2012;8:731–736. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abu Kwaik Y, Bumann D. Microbial quest for food in vivo: ‘nutritional virulence’ as an emerging paradigm. Cell Microbiol. 2013;15:882–890. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- 98.Almagro-Moreno S, Boyd EF. Sialic acid catabolism confers a competitive advantage to pathogenic vibrio cholerae in the mouse intestine. Infect Immun. 2009;77:3807–3816. doi: 10.1128/IAI.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jeong HG, Oh MH, Kim BS, Lee MY, Han HJ, Choi SH. The capability of catabolic utilization of N-acetylneuraminic acid, a sialic acid, is essential for Vibrio vulnificus pathogenesis. Infect Immun. 2009;77:3209–3217. doi: 10.1128/IAI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Almagro-Moreno S, Boyd EF. Bacterial catabolism of nonulosonic (sialic) acid and fitness in the gut. Gut Microbes. 2010;1:45–50. doi: 10.4161/gmic.1.1.10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, Singh PK, Kaneko Y, Wolfgang MC, Hsiao YS, Tong L, Prince A. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J Clin Invest. 2006;116:2297–2305. doi: 10.1172/JCI27920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alkhuder K, Meibom KL, Dubail I, Dupuis M, Charbit A. Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog. 2009;5:e1000284. doi: 10.1371/journal.ppat.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scidmore MA. Recent advances in Chlamydia subversion of host cytoskeletal and membrane trafficking pathways. Microbes Infect. 2011;13:527–535. doi: 10.1016/j.micinf.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Derre I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 2011;7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, Melancon P, Engel JN. Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development. PLoS Pathog. 2011;7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ouellette SP, Dorsey FC, Moshiach S, Cleveland JL, Carabeo RA. Chlamydia species-dependent differences in the growth requirement for lysosomes. PLoS One. 2011;6:e16783. doi: 10.1371/journal.pone.0016783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghigo E, Colombo MI, Heinzen RA. The Coxiella burnetii parasitophorous vacuole. Adv Exp Med Biol. 2012;984:141–169. doi: 10.1007/978-94-007-4315-1_8. [DOI] [PubMed] [Google Scholar]
- 108.Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Host Proteasomal Degradation Generates Amino Acids Essential for Intracellular Bacterial Growth. Science. 2011 doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 111.Bruckert WM, Price CT, Abu Kwaik Y. Rapid nutritional remodeling of the host cell upon attachment of Legionella pneumophila. Infect Immun. 2014;82:72–82. doi: 10.1128/IAI.01079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sauer JD, Bachman MA, Swanson MS. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc Natl Acad Sci U S A. 2005;102:9924–9929. doi: 10.1073/pnas.0502767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Steele S, Brunton J, Ziehr B, Taft-Benz S, Moorman N, Kawula T. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 2013;9:e1003562. doi: 10.1371/journal.ppat.1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci U S A. 2012;109:20800–20807. doi: 10.1073/pnas.1218674109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun. 2004;72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rivera-Chavez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio SP, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, Bevins CL, Tsolis RM, Harshey R, Adams LG, Baumler AJ. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS Pathog. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 118.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell. 2013;155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schmidt SV, Schultze JL. New Insights into IDO Biology in Bacterial and Viral Infections. Frontiers in immunology. 2014;5:384. doi: 10.3389/fimmu.2014.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.MacKenzie CR, Gonzalez RG, Kniep E, Roch S, Daubener W. Cytokine mediated regulation of interferon-gamma-induced IDO activation. Adv Exp Med Biol. 1999;467:533–539. doi: 10.1007/978-1-4615-4709-9_66. [DOI] [PubMed] [Google Scholar]
- 121.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zarek PE, Powell JD. Adenosine and anergy. Autoimmunity. 2007;40:425–432. doi: 10.1080/08916930701464939. [DOI] [PubMed] [Google Scholar]
- 123.Thiel M, Caldwell CC, Sitkovsky MV. The critical role of adenosine A2A receptors in downregulation of inflammation and immunity in the pathogenesis of infectious diseases. Microbes Infect. 2003;5:515–526. doi: 10.1016/s1286-4579(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 124.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46 Suppl 5:S344–349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 125.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J Exp Med. 2009;206:2417–2427. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chubukov V, Gerosa L, Kochanowski K, Sauer U. Coordination of microbial metabolism. Nat Rev Microbiol. 2014;12:327–340. doi: 10.1038/nrmicro3238. [DOI] [PubMed] [Google Scholar]
- 127.Fetherston JD, Mier I, Jr, Truszczynska H, Perry RD. The Yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect Immun. 2012;80:3880–3891. doi: 10.1128/IAI.00086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]