Abstract
Bactofilins constitute a recently discovered class of bacterial proteins that form cytoskeletal filaments. They share a highly conserved domain (DUF583) of which the structure remains unknown, in part due to the large size and noncrystalline nature of the filaments. Here, we describe the atomic structure of a bactofilin domain from Caulobacter crescentus. To determine the structure, we developed an approach that combines a biophysical model for proteins with recently obtained solid-state NMR spectroscopy data and amino acid contacts predicted from a detailed analysis of the evolutionary history of bactofilins. Our structure reveals a triangular β-helical (solenoid) conformation with conserved residues forming the tightly packed core and polar residues lining the surface. The repetitive structure explains the presence of internal repeats as well as strongly conserved positions, and is reminiscent of other fibrillar proteins. Our work provides a structural basis for future studies of bactofilin biology and for designing molecules that target them, as well as a starting point for determining the organization of the entire bactofilin filament. Finally, our approach presents new avenues for determining structures that are difficult to obtain by traditional means.
Introduction
The cellular cytoskeleton may perform a wide range of functions, including maintaining cell shape and polarity, intracellular transport, and segregation of chromosomes during cell division. It was long thought that cytoskeletons were only found in eukaryotic cells, where proteins such as actin, tubulin, and intermediate filaments have been thoroughly studied both biochemically and structurally (1, 2, 3). Subsequently, however, prokaryotic cells were also shown to have a cytoskeleton. For example, based on sequence and structural similarities, the bacterial proteins FtsZ and MreB were found to be tubulin and actin homologs, respectively (4). In addition to these protein families, which are found in both eukaryotes and prokaryotes, recent discoveries have revealed the existence of cytoskeletal proteins specific to bacteria. Examples of such bacterium-specific proteins are the deviant Walker A-motif ATPases (5) and bactofilins (6), underlining the importance of specific and diverse cytoskeletal proteins in bacteria.
Bactofilins are a recently discovered class of proteins that are widely found across many different bacterial species (6). They are characterized by a conserved domain of unknown function (DUF583), ∼100 amino acids long and flanked by variable terminal regions. Although much remains to be discovered about this novel class of proteins, bactofilins have been shown to have distinct but varying functions across different species. For example, a bactofilin called CcmA in Helicobacter pylori is required for the cell to maintain its characteristic helical shape, and a deletion of CcmA affects the bacterium’s ability to infect the stomachs of mice (7). Deletion of the bactofilin BacM from Myxococcus xanthus causes the bacterium to change into a crooked morphology and be more sensitive to cell-wall-targeting antibiotics (8). In Caulobacter crescentus, two bactofilin paralogs, BacA and BacB, form filamentous structures focused at one of the cell poles carrying the stalk, possibly serving as molecular scaffolds for multiple cellular pathways (6).
Bactofilins have been shown to form filaments spontaneously (6, 9), which in addition to highlighting the evolutionary selection toward filament formation, at the same time makes them challenging targets for classical structure-determination approaches using x-ray crystallography or liquid-state NMR spectroscopy. Studies using template-based modeling have suggested that the DUF583 domain has a β-helical structure with an extensive network of hydrophobic interactions, explaining the high stability of the filaments (9, 10). The modeled structures were presented as left-handed helices, although their orientation appears to have been dictated by the templates used to model the structures rather than by direct information, since both left- and right-handed β-helices are found in nature (11, 12). Vasa et al. (9) also studied the structural properties of bactofilins more directly using solid-state NMR (ssNMR) spectroscopy of isotope-labeled filaments of the BacA protein from C. crescentus. In that work, the authors were able to obtain a complete assignment of the backbone chemical shifts in the 103-residues-long DUF583 domain, and also measured a small number (n = 20) of ambiguous long-range contacts. Their analysis of the chemical shifts using TALOS+ (13) suggested that the structure mostly contains β-sheet structure, although the exact number and location of the strands remained unclear.
We recently described an approach to determine structures of naturally occurring fibrillar proteins using amino acid contacts derived from an analysis of multiple sequence alignments (MSAs) (14). In particular, we leveraged recent improvements in methods for predicting amino acid contacts from covariation of amino acid pairs during evolution (15, 16, 17, 18) and combined those contacts with an efficient molecular force field called Profasi (19). We applied this approach to the functional amyloid protein, CsgA, which is the main constituent of bacterial curli fibrils that play an important role in biofilm formation. We found that CsgA forms a flat β-helical structure with two strands per turn, and that each turn corresponds to previously found sequence subrepeats.
Here, we extend that approach by including experimentally obtained NMR chemical shifts to determine the structure of the DUF583 domain from C. crescentus. Previous studies demonstrated that it is possible to determine the structures of small, soluble proteins using NMR chemical shifts alone (20, 21, 22, 23, 24), but for larger and more complex proteins, chemical shifts need to be supplemented by distance information, e.g., from nuclear Overhauser effect (NOE) experiments (25, 26, 27). In particular, we combined a recently described NMR chemical-shift-driven molecular sampling method (24) with contacts obtained from an analysis of an MSA and the Profasi force field. In agreement with previous models, we found that the DUF583 domain took on a β-helical structure, although in contrast to those models, it assumed a right-handed topology. A search of structural databases did not reveal any clear structural homologs within eukaryotic cytoskeletal proteins, further strengthening the hypothesis that bactofilins are specific to bacteria. Our structure provides an experimentally derived model of this novel class of proteins, and should be useful for understanding their diverse biochemical functions and developing ways to interfere with those functions using small molecules.
Materials and Methods
Simulation details
We simulated the 103-amino-acids-long DUF583 domain in BacA from C. crescentus using the Phaistos software (28). Experimental chemical shifts and predicted contacts were integrated with the Profasi force field via a hybrid energy function, (details below), in MC simulations starting from an extended structure. To set the scale between the different terms, we used the previously determined relative weight between and (14), as well as the previously determined weight between and (24). Profasi is an efficient implicit-solvent, all-atom energy function that has been used successfully in simulations of protein folding and aggregation (19).
We used amino acid contacts obtained from a covariance analysis performed using Gremlin (29, 30) applied to an MSA generated using HHblits (31). HHblits is a fast, sensitive, and accurate sequence-search tool, which we used (with an E-value cutoff of ) to perform four sequence search iterations, obtaining 847 sequences of bactofilin homologs. Gremlin, in turn, provides a method to derive a statistical model of the sequences in an MSA. In particular, Gremlin allows us to extract pairs of amino acids that covary during bactofilin evolution and thus are likely to be in close spatial proximity. In analogy to our work on CsgA (14), we used the 50 top-scoring contacts based on the covariance analysis and constructed a pseudoenergy function that favors the contact formation of the amino acid pairs.
Previously measured backbone NMR chemical shifts (9) (N, CO, Cα, and Cβ) were used as input to CS-TORUS (24), which is a continuous and probabilistic molecular fragment model and a natural choice to avoid the discretization of angles associated with other fragment models. We used CS-TORUS as a generative model to directly sample backbone dihedral angles that were in agreement with the chemical shifts in a way that effectively corresponded to sampling from a biased force field, albeit significantly more efficiently (24). Here, we denote the contribution of the model as .
We performed 32 independent simulations using in an enhanced sampling method (32). We further refined the lowest-energy structure obtained by performing all-atom molecular-dynamics (MD) simulations and incorporating the predicted contacts and experimental chemical shifts as restraints. Specifically, we solvated the lowest-energy structure in a dodecahedral box, with a minimum of 6 Å from the solute to the box boundary in each dimension. We added Na+ and Cl− ions to neutralize the overall electric charge and model an ion concentration of 150 mM. We used the CHARMM22∗ force field (33) and smoothly shifted the van der Waals interactions to zero at distances between 0.8 and 1.0 nm. To calculate long-range electrostatic interactions, we used the particle mesh Ewald algorithm with a grid spacing of 0.16 nm and an interpolation order of 4. Bonds involving hydrogen atoms were constrained using the LINCS algorithm to allow for a 2 fs time step. After a steepest-descent energy minimization, we equilibrated the system for 1 ns in the NPT ensemble controlled by a Parrinello-Rahman barostat and V-rescale thermostat (34). All simulations were run in GROMACS 5.0.5 (35), and PLUMED 2.1.2 (36) was used to include chemical-shift and distance restraints. NMR chemical-shift restraints were applied in the MD simulations as previously described (37). Contact restraints between Cα atoms were applied using the NOE potential implemented in PLUMED2, with an upper limit of 8.0 Å. To improve sampling, we used a periodic simulated annealing strategy with temperatures between 440 K and 10 K. Four annealing cycles were performed, with each cycle lasting 10 ns, and the final structure was used for analyses. Overall, the MD-refined structure is very similar to that obtained from the MC simulations, but has somewhat improved packing of side chains.
Comparison with ambiguous distance restraints from ssNMR
We define a measure of discrepancy (mean deviation) to quantify the agreement between a structure and 20 previously measured semiquantitative distance measurements by ssNMR:
where is the distance of contact i from a simulated structure and is for the HN-HA contacts and for the C-C contacts. As most of the contacts were ambiguous, we calculated the distances between the various possible assignments and used the shortest distance for comparison with experiments.
Results and Discussion
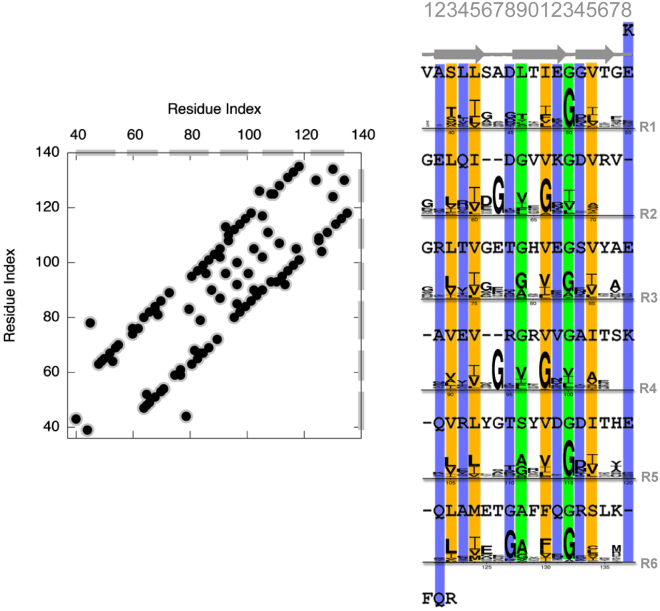
We combined predicted contacts and experimental NMR chemical shifts with an efficient force field to determine the structure of the DUF583 domain of a bactofilin. Sequence-based predictions of contacts are particularly useful when homology modeling is not feasible. This can be quantified by calculating the HHΔ-value (30), with values close to unity indicating there are not any proteins with known structure that have a high sequence similarity to the target. For the bactofilin domain, we calculated HHΔ-value to be 0.87, suggesting that sequence-based contact prediction contains information that can add to that available from template-based modeling. We therefore analyzed an MSA to predict evolutionarily conserved contacts (Fig. 1, left). The result reveals a distinct diagonal pattern with the occasional offset of one or two amino acids, suggestive of an overall β-helical structure with a periodicity of ∼16–18 amino acids. As the predicted contacts report on the structure within the evolutionarily selected for filamentous state, they might conceivably correspond to both inter- and intramolecular contacts. Earlier work on a fibrillar protein with a similar contact pattern (CsgA), however, demonstrated that the contacts most likely occur within a single protein subunit (14). Further, it was previously proposed that bactofilin subunits stack laterally with relatively large horizontal distances between subunits (9), making it more unlikely to find intermolecular contacts.
Figure 1.
Contact prediction and amino acid sequence of the bactofilin DUF583 domain. Left: the 50 top-scoring amino acid contacts obtained from a covariation analysis of an MSA using Gremlin. The gray bars on the axes represent the six internal repeats. Right: the amino acid sequence of the DUF583 domain has six internal repeats (R1–R6) that we aligned using the structure we determined. Gaps are added where neighboring repeats bulge to maintain the phase. Between the repeats, we show sequence logos that represent the conservation pattern in the MSA of the bactofilin domains. The individual columns are color-coded depending on the kind of amino acids that are dominant, with orange (columns 3, 5, 11, and 15) representing mostly hydrophobic amino acids and blue (columns 2, 4, 8, 12, and 14) representing hydrophilic and charged amino acids. Two columns that contain many glycine residues are shown in green (columns 9 and 13) and are located in the loops that connect the different strands. To see this figure in color, go online.
The sequence of BacA (Fig. 1, right) reveals six internal repeats (R1–R6), suggesting that each repeat corresponds to one turn of a helical structure. A previous analysis of the chemical shifts (9) using TALOS+ (13) suggested mostly β-sheet structure, but it was difficult to determine the exact location of loops and the number of strands. The location of highly conserved glycine residues within each repeat suggests the location for turn residues, but not all loops have clear sequence signatures. Therefore, we used a recently developed generative model of backbone dihedral angles that allows us to sample local structures that are compatible with the chemical shifts. Combined with the long-range information inferred from the contacts and the structural information contained in the molecular energy function, we performed Monte Carlo (MC) simulations using a hybrid energy function, , that integrates the three sources of information.
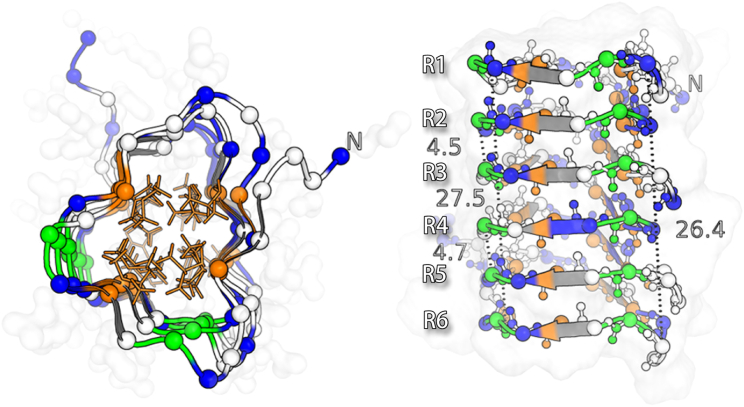
We performed 32 independent simulations that consistently resulted in right-handed β-helical structures, with a few (higher-energy) structures having some fraying near the ends. Our finding of a β-helical structure is consistent with previously described template-based models (9, 10), although in contrast to those models, which are all left-handed, we suggest that the BacA DUF583 domain takes on a right-handed orientation. We further refined the lowest-energy structure using restrained MD simulations. The final structure reveals a highly regular fold containing six repeats of slightly varying lengths (Fig. 2). When viewed from the top, the structure shows a triangular shape of the backbone, with hydrophobic amino acids pointing toward the core, and hydrophilic and charged amino acids covering the surface of the protein. This repetitive network of hydrophobic interactions in the core and abundant backbone hydrogen bonding between strands suggests a very stable protein that is often associated with filament-forming proteins. Together with glycines that align structurally throughout all six repeats, these structurally conserved hydrophilic and hydrophobic residues dictate the lateral alignment (phase) of the repeats, allowing for varying repeat lengths that realign in the loop regions. One of the turns between the strands is very tight (corresponding to position 13 in the alignment in Fig. 1) and has highly conserved glycine residues. We find that these residues adopt a conformation corresponding to the lower right-hand corner of the Ramachandran map, a region that is essentially allowed only for glycine (38), explaining the conservation at this position. All residues are found in generally allowed regions of the Ramachandran map.
Figure 2.
Structure of the DUF583 domain from the BacA bactofilin from C. crescentus. The structure was obtained by MC simulations using chemical shifts and contact restraints and was further refined by all atom MD simulations. The amino acid residues are colored to match the scheme in Fig. 1, right. To see this figure in color, go online.
The repetitive nature of the bactofilin sequence and structure is illustrated further in a structure-based sequence alignment of the six repeats (Fig. 1, right). Four positions within each repeat (3, 5, 11, and 15) show especially high hydrophobic conservation, whereas position 9 contains mostly glycines or other small amino acids, aiding to the flexibility needed in positions 6–8 (loop region) where the repeats often realign. Furthermore, we find that the evolutionary conservation agrees strongly with structural conservation in terms of hydrophobicity. As an example, we look at positions 3, 5, 11, and 15 which have almost exclusively hydrophobic residues structurally aligned as well as hydrophobic residues dominating the associated sequence logo plots, indicating that our structure indeed aligns the residues correctly. Similarly, a number of positions with conserved polar and charged amino acids correspond to repeating positions on the outside of the protein.
To examine how the different sources of information contributed to the structure determination and in particular the right-handed orientation, we performed a series of control simulations. If we exclude the chemical shifts, we obtain both left- and right-handed β-helices with comparable Profasi energies (and with roughly equal probability), indicating that the Profasi energy function on its own does not lead to a specific handedness. This observation is in agreement with our previous findings on CsgA (14). Using only the chemical shifts and Profasi, but excluding the predicted contacts, resulted in a flat, sheet-like structure (Fig. S1 in the Supporting Material). Finally, we performed simulations combining the chemical shifts and predicted contacts with a simple energy function that only represents the excluded volume. Given the drastic change in the force field, here we used an ∼2-fold greater force constant for the contact potential compared with the simulations described above. In these simulations, we consistently obtained right-handed β-helical structures, suggesting again that the chemical shifts contain information directing the structures to the right-handed orientation. The resulting structures are, not surprisingly, less well defined than those obtained using the full force field, and do not have as well defined hydrophobic cores. When we increased the weight of the contact energies in these simulations by 5, thus decreasing the contribution of the chemical shifts, we began to observe structures with both orientations.
Altogether, the simulations described above suggest that the chemical shifts enabled us to find the right-handed structure of bactofilin. To examine this issue further, we analyzed the lowest-energy left- and right-handed β-helix obtained using the Profasi energy function and amino acid contacts. As these structures were generated without the use of chemical shifts, we first calculated to evaluate which one was in best agreement with the NMR data. As expected, we found the right-handed β-helix to be in better agreement (1406 vs. 1432 log-likelihood units (24)). We found a similar difference (6.5 log-likelihood units) after relaxing both the right- and left-handed structures in the presence of the chemical-shift restraints. We note, however, that the left-handed structures, but not the right-handed ones, deformed substantially when they were relaxed to be more compatible with the chemical-shift data.
As an alternative and independent way to compare these left- and right-handed structures, we turned to the more accurate Rosetta energy function. In particular, we minimized the two structures using the RosettaRelax (39) algorithm, and found that the right-handed helix had a significantly lower energy (−206 Rosetta energy units) than the left-handed structure (−185 units). Not surprisingly, we also found that the lowest-energy structure we obtained when we included the chemical shifts in the structure determination had an even lower Rosetta energy (−253 units), providing further evidence that the chemical shifts improved the quality of our structure. We also repeated these calculations using a different energy minimization algorithm (RosettaBackrub (40)) and obtained comparable results (47, 109, and 29 energy units, respectively; note that these two Rosetta energy functions differ slightly, explaining the different magnitude).
We also examined which of the chemical-shift data helped us determine the handedness of the helix. We performed two sets of simulations in which we excluded the chemical shifts in either the β-strands or the intervening loops. Surprisingly, we found that both sets of simulations resulted in structures with both left- and right-handed helices. Thus, it appears that the chemical shifts have a more global effect and that the handedness is not easily pinned down to the effect at a few residues. We note, however, that the chemical-shift model we used is a dynamic Bayesian network in which the chemical shift of a residue may exert its effect over six residues on average (24). Additional studies are needed to examine in more detail which of the different backbone chemical shifts provide the most information to guide the handedness of the structure and therefore would be the most informative to measure in other systems.
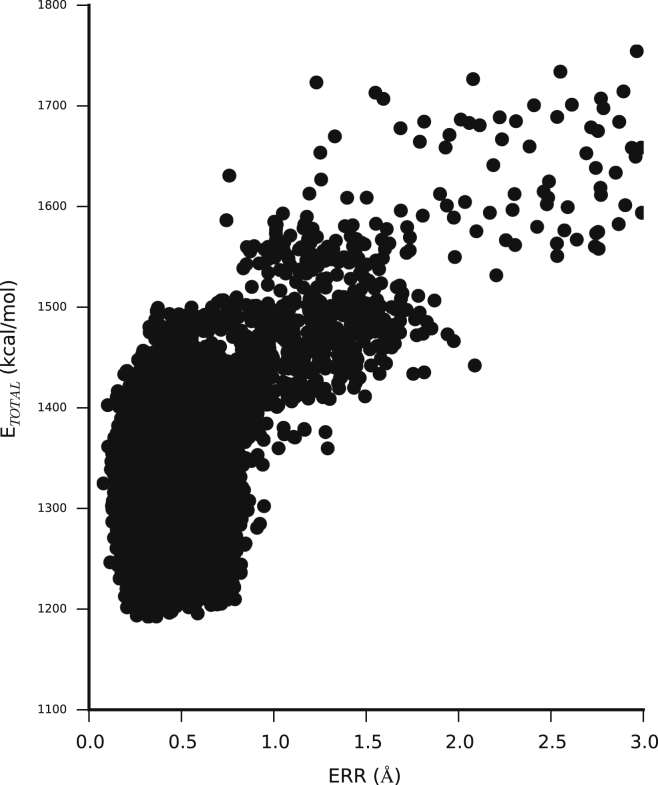
In addition to the chemical-shift assignment, the recent ssNMR experiments provided a set of 20 interatomic contacts, 15 of which were ambiguous (9). Although the experiments were only semiquantitative, employing long mixing times that prohibited accurate distance estimations, they provide a means to gauge the accuracy of our structures. In particular, we compared the structures obtained during our simulations with the previous distance measurements, using values of 5.5 Å and 7.5 Å for the measured HN-HA and C-C contacts, respectively. A calculation of the mean error for the structures obtained during the MC simulations reveals that the refinement procedure that uses only the force field, chemical shifts, and predicted contacts simultaneously results in structures that are in good agreement with the semiquantitative distance measurements (Fig. 3). The lowest-energy MC structures have mean deviations on the order of 0.5 Å, as does the final MD-refined structure. A more detailed analysis is hampered by the lack of quantitative measurements, including the possibility of spin diffusion. Nevertheless, we note that of the 20 experimental contacts, only two (contacts 8 and 13) are substantially violated (Table S1). Contact 13 (I60CD1-S43C) is positioned in the first repeat, suggesting that the violation might be the result of only simulating the monomer (even if the contact is intermolecular). By not modeling interactions with a neighboring chain (for which we have no information), we might not capture minor rotameric reorientations of, for example, I60, which could resolve the violation. Until such information is available, we assume it to be a violation. The amino acids that form contact 8 (R93HN-G109HA) are located in two loop regions and are laterally aligned in the structure, which could suggest that the loop regions are too flexible to model accurately, and that a contact formation might be in agreement with our overall structure.
Figure 3.
Validation using independently determined distance constraints from ssNMR. We calculated the mean deviation (ERR; see Materials and Methods) between distances in the structures obtained during our MC simulations and a set of 20 semiquantitative contacts obtained by ssNMR. These values are plotted against the total energy, , which includes the Profasi energy function, chemical shifts, and predicted contacts.
In addition to obtaining ssNMR distance information, Vasa et al. (9) used scanning transmission emission microscopy experiments to determine the mass-per-length of the BacA subunits within the filament, resulting in size estimates of 27–30 Å, values that are also in good agreement with our structure (Fig. 2, right). Finally, we validated our structure by mapping onto it residues that Vasa et al. (9) mutated and studied functionally (Supporting Materials and Methods; Table S2). In their study, they created 27 mutations of bactofilin and introduced them into C. crescentus to examine whether these mutations supported filament formation. In general, we find a very good agreement between the location of those mutations in the structure, the nature of the mutation (e.g., changing charge or polarity), and the ability of the mutant bactofilin to assemble into filaments (Table S2), again supporting the structure we determined.
Earlier work on bacterial cytoskeletal proteins revealed a structural similarity between FtsZ and eukaryotic tubulin, and between MreB and actin (4). To examine whether the structure of bactofilin we determined provides clues as to potential eukaryotic homologs, we used both DALI (41) and deconSTRUCT (42) to search for structural homologs. Although both searches returned many proteins with β-helical structures (including the left-handed p27 domain in dynactin, a tubulin folding cofactor and filamentous hemagglutinin), we did not find any obvious proteins with such structures forming fibrillar structures in the cytoskeleton. Thus, our structure supports the hypothesis that bactofilins are indeed specific to bacteria.
Finally, we note that during the revision of our manuscript, an independent experimental ssNMR structure of the same bactofilin domain was published (43). In that study, the final structure was determined using almost 2000 NMR-derived atomic distance restraints, as well as torsion angle restraints, in a simulated annealing simulation. Similarly to our work, the authors used chemical shifts to infer torsion angle restraints, but whereas we used easily obtainable amino acid contacts and a more detailed force field, they used atomic distances derived from advanced and demanding ssNMR experiments. We find the two structures to be very similar, with an all-heavy-atom (backbone and side chains) root mean-square deviation of 2.2 Å between the two structures (Fig. S2). Further, we compared our structure with the NMR-derived distance restraints that were used to determine the structure, and found a root mean-square violation of 0.2 Å. This value can be compared to the 0.05 Å that we calculated from the structure that was determined using these data (43), also suggesting that our structure provides a highly accurate model of bactofilin.
Conclusions
We determined the structure of the DUF583 domain from a bactofilin by combining a molecular force field with experimental NMR backbone chemical shifts and amino acid contacts obtained by sequence variation. As opposed to previously described template-based models of the domain, which describe left-handed β-helices, we let the chemical shifts determine the orientation, which, interestingly, suggested that the domain takes on a right-handed orientation. Our structure is in good agreement with independent ssNMR data as well as mass-per-length measurements. A comparison of the sequence conservation and structural similarity between each of the repeats in the bactofilin shows nearly perfect agreement, indicating that the repeats are coordinated correctly in our structure and that the repeat structure is important for stability and function. From a methodological point of view, we have presented a new, to our knowledge, method for combining NMR chemical shifts with predicted contacts and a molecular energy function. The idea is similar to a very recently described approach that also relies on NMR-derived distance information (44). Our results reveal that all three sources of information (chemical shifts, contacts, and energy function) act constructively to allow us to determine the structure of bactofilin. We expect that the improved accuracy afforded by better methods for determining molecular energy functions (45) and predicting contacts (18), as well as developments in techniques such as ssNMR (46), will expand the scope of our approach to other proteins for which classical structure determination has proven difficult. Finally, we expect that the availability of the structure of the bactofilin domain will aid in elucidating the many important roles of this new class of proteins, and possibly in the development of molecules that perturb their functions.
Author Contributions
M.M.K. performed the MC simulations, Y.W. performed MD simulations, W.B. provided new methods and assisted in MC simulations, and K.L.-L. conceived and supervised the project. All authors were involved in data analysis and contributed to writing the manuscript.
Acknowledgments
We thank Adam Lange for helpful discussions regarding the NMR experiments on bactofilin.
This research was supported by grants from the Lundbeck Foundation and the Villum Foundation, and a Hallas-Møller stipend from the Novo Nordisk Foundation (to K.L.-L.).
Editor: Rohit Pappu.
Footnotes
Supporting Materials and Methods, two figures, two tables, and one data file are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30242-9.
Supporting Material
References
- 1.Popp D., Robinson R.C. Many ways to build an actin filament. Mol. Microbiol. 2011;80:300–308. doi: 10.1111/j.1365-2958.2011.07599.x. [DOI] [PubMed] [Google Scholar]
- 2.Teixidó-Travesa N., Roig J., Lüders J. The where, when and how of microtubule nucleation - one ring to rule them all. J. Cell Sci. 2012;125:4445–4456. doi: 10.1242/jcs.106971. [DOI] [PubMed] [Google Scholar]
- 3.Goldman R.D., Cleland M.M., Kuczmarski E.R. Inroads into the structure and function of intermediate filament networks. J. Struct. Biol. 2012;177:14–23. doi: 10.1016/j.jsb.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingerson-Mahar M., Gitai Z. A growing family: the expanding universe of the bacterial cytoskeleton. FEMS Microbiol. Rev. 2012;36:256–266. doi: 10.1111/j.1574-6976.2011.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koonin E.V. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 6.Kühn J., Briegel A., Thanbichler M. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sycuro L.K., Pincus Z., Salama N.R. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–833. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koch M.K., McHugh C.A., Hoiczyk E. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol. Microbiol. 2011;80:1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasa S., Lin L., Lange A. β-Helical architecture of cytoskeletal bactofilin filaments revealed by solid-state NMR. Proc. Natl. Acad. Sci. USA. 2015;112:E127–E136. doi: 10.1073/pnas.1418450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuckerman D.M., Boucher L.E., Hoiczyk E. The bactofilin cytoskeleton protein BacM of Myxococcus xanthus forms an extended β-sheet structure likely mediated by hydrophobic interactions. PLoS One. 2015;10:e0121074. doi: 10.1371/journal.pone.0121074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasmer C., Lange A., Meier B.H. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 12.Creze C., Castang S., Gouet P. The crystal structure of pectate lyase peli from soft rot pathogen Erwinia chrysanthemi in complex with its substrate. J. Biol. Chem. 2008;283:18260–18268. doi: 10.1074/jbc.M709931200. [DOI] [PubMed] [Google Scholar]
- 13.Shen Y., Delaglio F., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian P., Boomsma W., Lindorff-Larsen K. Structure of a functional amyloid protein subunit computed using sequence variation. J. Am. Chem. Soc. 2015;137:22–25. doi: 10.1021/ja5093634. [DOI] [PubMed] [Google Scholar]
- 15.Morcos F., Pagnani A., Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. USA. 2011;108:E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marks D.S., Colwell L.J., Sander C. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6:e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones D.T., Buchan D.W.A., Pontil M. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28:184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- 18.Taylor W.R., Hamilton R.S., Sadowski M.I. Prediction of contacts from correlated sequence substitutions. Curr. Opin. Struct. Biol. 2013;23:473–479. doi: 10.1016/j.sbi.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Irbäck A., Mitternacht S., Mohanty S. An effective all-atom potential for proteins. PMC Biophys. 2009;2:2. doi: 10.1186/1757-5036-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalli A., Salvatella X., Vendruscolo M. Protein structure determination from NMR chemical shifts. Proc. Natl. Acad. Sci. USA. 2007;104:9615–9620. doi: 10.1073/pnas.0610313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y., Lange O., Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. USA. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart D.S., Arndt D., Lin G. CS23D: a web server for rapid protein structure generation using NMR chemical shifts and sequence data. Nucleic Acids Res. 2008;36:W496–W502. doi: 10.1093/nar/gkn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen Y., Vernon R., Bax A. De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boomsma W., Tian P., Vendruscolo M. Equilibrium simulations of proteins using molecular fragment replacement and NMR chemical shifts. Proc. Natl. Acad. Sci. USA. 2014;111:13852–13857. doi: 10.1073/pnas.1404948111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raman S., Lange O.F., Baker D. NMR structure determination for larger proteins using backbone-only data. Science. 2010;327:1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange O.F., Rossi P., Baker D. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc. Natl. Acad. Sci. USA. 2012;109:10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavalli A., Vendruscolo M. Analysis of the performance of the CHESHIRE and YAPP methods at CASD-NMR round 3. J. Biomol. NMR. 2015;62:503–509. doi: 10.1007/s10858-015-9940-9. [DOI] [PubMed] [Google Scholar]
- 28.Boomsma W., Frellsen J., Hamelryck T. PHAISTOS: a framework for Markov chain Monte Carlo simulation and inference of protein structure. J. Comput. Chem. 2013;34:1697–1705. doi: 10.1002/jcc.23292. [DOI] [PubMed] [Google Scholar]
- 29.Balakrishnan S., Kamisetty H., Langmead C.J. Learning generative models for protein fold families. Proteins. 2011;79:1061–1078. doi: 10.1002/prot.22934. [DOI] [PubMed] [Google Scholar]
- 30.Kamisetty H., Ovchinnikov S., Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc. Natl. Acad. Sci. USA. 2013;110:15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remmert M., Biegert A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 32.Ferkinghoff-Borg J. Optimized Monte Carlo analysis for generalized ensembles. Eur. Phys. J. B. 2002;29:481–484. [Google Scholar]
- 33.Piana S., Lindorff-Larsen K., Shaw D.E. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 35.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 36.Tribello G.A., Bonomi M., Bussi G. PLUMED 2: new feathers for an old bird. Comput. Phys. Commun. 2014;185:604–613. [Google Scholar]
- 37.Robustelli P., Kohlhoff K., Vendruscolo M. Using NMR chemical shifts as structural restraints in molecular dynamics simulations of proteins. Structure. 2010;18:923–933. doi: 10.1016/j.str.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Hovmöller S., Zhou T., Ohlson T. Conformations of amino acids in proteins. Acta Crystallogr. D Biol. Crystallogr. 2002;58:768–776. doi: 10.1107/s0907444902003359. [DOI] [PubMed] [Google Scholar]
- 39.Tyka M.D., Keedy D.A., Baker D. Alternate states of proteins revealed by detailed energy landscape mapping. J. Mol. Biol. 2011;405:607–618. doi: 10.1016/j.jmb.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauck F., Smith C.A., Kortemme T. RosettaBackrub—a web server for flexible backbone protein structure modeling and design. Nucleic Acids Res. 2010;38:W569–W575. doi: 10.1093/nar/gkq369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z.H., Bharatham K., Mihalek I. deconSTRUCT: general purpose protein database search on the substructure level. Nucleic Acids Res. 2010;38:W590–W594. doi: 10.1093/nar/gkq489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi C., Fricke P., Lange A. Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR. Sci. Adv. 2015;1:e1501087. doi: 10.1126/sciadv.1501087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Y., Huang Y.J., Montelione G.T. Protein structure determination by combining sparse NMR data with evolutionary couplings. Nat. Methods. 2015;12:751–754. doi: 10.1038/nmeth.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindorff-Larsen K., Maragakis P., Shaw D.E. Systematic validation of protein force fields against experimental data. PLoS One. 2012;7:e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Habenstein B., Loquet A. Solid-state NMR: an emerging technique in structural biology of self-assemblies. Biophys. Chem. 2016;210:14–26. doi: 10.1016/j.bpc.2015.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.