Abstract
In flowering plants, self-incompatibility is an effective genetic mechanism that prevents self-fertilization. Most Prunus tree species exhibit a homomorphic gametophytic self-incompatibility (GSI) system, in which the pollen phenotype is encoded by its own haploid genome. To date, no identification of S-alleles had been done in Prunus africana, the only member of the genus in Africa. To identify S-RNase alleles and hence determine S-genotypes in African cherry (Prunus africana) from Mabira Forest Reserve, Uganda, primers flanking the first and second intron were designed and these amplified two bands in most individuals. PCR bands on agarose indicated 26 and 8 different S-alleles for second and first intron respectively. Partial or full sequences were obtained for all these fragments. Comparison with published S-RNase data indicated that the amplified products were S-RNase alleles with very high interspecies homology despite the high intraspecific variation. Against expectations for a locus under balancing selection, frequency and spatial distribution of the alleles in a study plot was not random. Implications of the results to breeding efforts in the species are discussed, and mating experiments are strongly suggested to finally prove the functionality of SI in P. africana.
Introduction
Prunus africana (Hook. f) Kalkman is a medicinal tree indigenous to the montane regions of West, Central, East and Southern Africa, including Madagascar. It is the only member of the genus Prunus which comprises of more than 200 species that are indigenous to Africa [1] where are found in fragmented populations. The species is economically important in Uganda, Cameroon, Madagascar and Kenya for its bark extract that is used to treat benign prostatic hypertrophy. However, unsustainable harvesting has threatened the survival of the tree and it is currently classified as an endangered species by the Convention for International Trade in Endangered Species (CITES) [2]
Taking into account the economic importance of P. africana, a number of in-situ and ex-situ initiatives aimed at sustainably conserving and managing the species have been initiated. However, long term sustainability of ex-situ initiatives may be constrained if the species is self-incompatible as are many other Prunus species. Most of these species operate a strictly homomorphic gametophytic self-incompatibility (GSI) system, in which specificity of self/non self-recognition is controlled by products encoded within the S -locus. Self incompatibility may be a hindrance to breeding for pure lines and the distribution and frequency of S-alleles may affect mating success, and consequently gene dispersal patterns, spatial genetic structure and genetic diversity [3]. Compatible crosses require distinct alleles so that the population-level rare alleles are favored. On the other hand, SI is of evolutionary importance in flowering plants due to its effectiveness in avoiding inbreeding and encouraging outcrossing which helps to promote heterozygosity and fitness [4].
S-RNase genes have been identified and characterized in several Prunus species including Prunus dulcis, P. avium, P.cerasus and P. mume [5–8]. Characterization of S-RNases in Rosaceae shows that there can be in excess of thirty different S-alleles. The aim of this work was to identify and characterize the S-RNase gene associated with gametophytic SI in P. africana. The results will be of use in breeding and seed production of P. africana during selection of individuals for cross-fertilization.
Materials and Methods
Study area and Population description
Leaf samples were collected from Mabira forest reserve, located in South Central Uganda between 0o 22’–0o 35’N and 32o56’–33o 02’E. It is a mid-elevation forest located between 1070 and 1340 m a.s.l, with a total area of 300 km², occupying gently undulating plains with numerous flat-topped hills and wide shallow valleys. About 95% of the area is covered by medium altitude moist semi-deciduous forest [9]. The remaining portion is occupied by medium altitude moist evergreen forest. The three recognized sub-climaxes are: colonizing forest, mature mixed forest and Celtis mixed forest. Although some parts of the forest are degraded, there is still an undisturbed nature reserve.
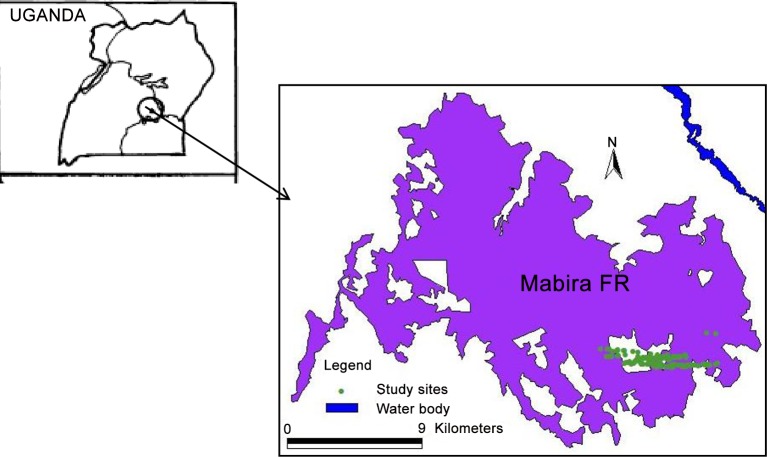
The initial reconnaissance survey of P. africana in Mabira forest showed that it occurs in demes separated by a minimum of 30 m (Fig 1). The isolated demes have a minimum number of 50 mature trees. Samples for this study were collected from the intensively studied plot in the more intact nature reserve. A total of 150 trees were sampled.
Fig 1. Map showing the location of Mabira forest and the individual trees of P. africana sampled for S-allele analysis.
Sample Collection and DNA extraction
From the intensively studied plot, young and disease free leaf samples of 150 trees were collected. The leaves were then cleaned with absolute ethanol and then sliced into small pieces before they were put into zip lock bags filled with self indicating blue silica gel. The individual tree locations were geo-referenced with a Global Positioning System (GPS) for easy monitoring and for spatial genetic structure analysis (Table 1). The leaf samples were shipped to the Federal Research and Training Centre for Forests, Natural Hazards and Landscape (BFW) genetic laboratory for DNA extraction and analysis. Total genomic DNA was extracted from about 10g of all the leaf samples following documented protocols [10]. The quality and purity of genomic DNA was evaluated by a ND-1000 spectrophotometer (NanoDrop, USA)) and by gel electrophoresis. For gel electrophoresis, 1 μl of each DNA extract was analysed in a 1.5% agarose gel containing 0.5% ethidium bromide and was visualized by U.V. illumination.
Table 1. GPS coordinates of all the Prunus africana trees that were sampled for analysis of S-alleles.
| TREE NO | LONGITUDE | LATITUDE |
|---|---|---|
| 1 | 50.165 | 0.42677 |
| 2 | 50.1633 | 0.42678 |
| 3 | 50.1643 | 0.42685 |
| 4 | 50.1641 | 0.42664 |
| 5 | 50.1627 | 0.42664 |
| 6 | 50.1584 | 0.42627 |
| 7 | 50.1528 | 0.42599 |
| 8 | 50.1472 | 0.4246 |
| 9 | 50.1519 | 0.42578 |
| 10 | 50.1438 | 0.42507 |
| 11 | 50.1551 | 0.42471 |
| 13 | 50.1511 | 0.42388 |
| 15 | 50.1571 | 0.42392 |
| 16 | 50.1593 | 0.42412 |
| 17 | 50.159 | 0.42395 |
| 19 | 50.1668 | 0.42331 |
| 20 | 50.1658 | 0.42313 |
| 21 | 50.1673 | 0.42317 |
| 22 | 50.1704 | 0.42323 |
| 23 | 50.1747 | 0.42313 |
| 24 | 50.1754 | 0.42308 |
| 25 | 50.1785 | 0.42305 |
| 26 | 50.1796 | 0.4231 |
| 27 | 50.1788 | 0.42322 |
| 28 | 50.177 | 0.42343 |
| 29 | 50.1764 | 0.42334 |
| 30 | 50.176 | 0.42332 |
| 31 | 50.1716 | 0.42352 |
| 32 | 50.1712 | 0.42342 |
| 33 | 50.1695 | 0.42343 |
| 34 | 50.1679 | 0.42343 |
| 35 | 50.168 | 0.42342 |
| 36 | 50.1673 | 0.42345 |
| 37 | 50.1667 | 0.42368 |
| 38 | 50.1637 | 0.42644 |
| 41 | 50.1608 | 0.42298 |
| 42 | 50.1815 | 0.4227 |
| 43 | 50.1552 | 0.42278 |
| 44 | 50.1596 | 0.42209 |
| 45 | 50.1569 | 0.42155 |
| 46 | 50.1657 | 0.42173 |
| 47 | 50.1619 | 0.42217 |
| 48 | 50.1636 | 0.42224 |
| 49 | 50.1669 | 0.42241 |
| 50 | 50.1736 | 0.4227 |
| 52 | 50.1771 | 0.42264 |
| 53 | 50.1746 | 0.42244 |
| 54 | 50.1737 | 0.42236 |
| 55 | 50.1734 | 0.42253 |
| 56 | 50.1721 | 0.4225 |
| 59 | 50.1858 | 0.42324 |
| 60 | 50.1875 | 0.42313 |
| 61 | 50.1869 | 0.42308 |
| 62 | 50.1888 | 0.42318 |
| 65 | 50.1857 | 0.42329 |
| 66 | 50.1862 | 0.42329 |
| 69 | 50.1875 | 0.42366 |
| 70 | 50.1862 | 0.42376 |
| 71 | 50.1863 | 0.44238 |
| 72 | 50.1923 | 0.42398 |
| 73 | 50.1921 | 0.44215 |
| 74 | 50.1931 | 0.42403 |
| 78 | 50.1847 | 0.42368 |
| 83 | 50.1469 | 0.4249 |
| 84 | 50.1405 | 0.4252 |
| 85 | 50.1403 | 0.4246 |
| 86 | 50.1405 | 0.42482 |
| 87 | 50.1435 | 0.42507 |
| 88 | 50.1401 | 0.42458 |
| 89 | 50.1385 | 0.42456 |
| 90 | 50.1374 | 0.42433 |
| 91 | 50.143 | 0.42405 |
| 92 | 50.1416 | 0.4242 |
| 94 | 50.1413 | 0.42364 |
| 95 | 50.1475 | 0.42368 |
| 96 | 50.1509 | 0.42362 |
| 97 | 50.1523 | 0.42395 |
| 98 | 50.1487 | 0.42385 |
| 99 | 50.138 | 0.42917 |
| 100 | 50.1678 | 0.42781 |
| 101 | 50.1682 | 0.42763 |
| 2A | 50.1676 | 0.42783 |
| 3A | 50.1681 | 0.42812 |
| 4A | 50.1695 | 0.42801 |
| 5A | 50.168 | 0.42788 |
| 6A | 50.1705 | 0.42787 |
| 7A | 50.1701 | 0.42782 |
| 8A | 50.1745 | 0.42799 |
| 10A | 50.1699 | 0.42818 |
| 11A | 50.1685 | 0.42817 |
| 12A | 50.1714 | 0.42819 |
| 14A | 50.1687 | 0.42804 |
| 15A | 50.1667 | 0.42811 |
| 17A | 50.1634 | 0.42944 |
| 23A | 50.1427 | 0.43112 |
| 25A | 50.1455 | 0.43104 |
| 27A | 50.1532 | 0.42984 |
| 28A | 50.1573 | 0.42938 |
| 29A | 50.1732 | 0.42873 |
| 31A | 50.174 | 0.42922 |
| 34A | 50.1633 | 0.42802 |
| 35A | 50.1608 | 0.42793 |
| 36A | 50.1608 | 0.4279 |
| 37A | 50.1592 | 0.42767 |
| 38A | 50.1593 | 0.42755 |
| 39A | 50.1604 | 0.42763 |
| 41A | 50.1598 | 0.42749 |
| 42A | 50.1624 | 0.42765 |
| 43A | 50.16 | 0.42762 |
| 44A | 50.1601 | 0.42743 |
| 46A | 50.1522 | 0.42735 |
| 47A | 50.1511 | 0.42736 |
| 48A | 50.1506 | 0.4272 |
| 49A | 50.1515 | 0.42726 |
| 50A | 50.1453 | 0.42809 |
| 51A | 50.1481 | 0.42675 |
| 52A | 50.1472 | 0.4266 |
| 53A | 50.1443 | 0.42646 |
| 54A | 50.1462 | 0.4264 |
| 55A | 50.1493 | 0.42751 |
| 56A | 50.1506 | 0.42764 |
| 57A | 50.1511 | 0.42766 |
| 58A | 50.1504 | 0.42795 |
| 59A | 50.1508 | 0.42779 |
| 61A | 50.153 | 0.42767 |
| 62A | 50.153 | 0.42761 |
| 63A | 50.1548 | 0.42754 |
| 64A | 50.1563 | 0.42767 |
| 66A | 50.1583 | 0.42811 |
| 67A | 50.1453 | 0.42792 |
| 68A | 50.1483 | 0.42774 |
| 69A | 50.1232 | 0.43271 |
| 71A | 50.166 | 0.42758 |
| 72A | 50.1607 | 0.42844 |
| 73A | 50.1608 | 0.42843 |
| 74A | 50.1273 | 0.433 |
| 75A | 50.1283 | 0.43295 |
| 76A | 50.1374 | 0.43195 |
| 77A | 50.1368 | 0.43206 |
| 78A | 50.1328 | 0.43163 |
| 79A | 50.1328 | 0.4317 |
| 80A | 50.1305 | 0.43159 |
| 81A | 50.1332 | 0.43167 |
| 82A | 50.1332 | 0.43249 |
| 100A | 50.1337 | 0.42864 |
| 101A | 50.1334 | 0.42867 |
| 102A | 50.1268 | 0.42822 |
| 103A | 50.1278 | 0.42825 |
| 104A | 50.1288 | 0.4282 |
| 105A | 50.1297 | 0.42819 |
PCR amplification for initial screening
Various consensus polymerase chain reaction (PCR) primers that have been used for identifying S-alleles in other Prunus species [6][11][12] were tested on 20 samples. The amplicons were purified, sequenced and the results used to design specific primers that were used in this study. Two specific primer pairs (Pacons2F/PAI2-2R and PaI2-2F/PaI2-2R,) flanked the second intron of the S-RNase and three pairs (PaCons1F/PaCons1R, PaSI1f/PaSI1R and Paintron1F/Paintron1R) [12] were used to amplify the first intron. To amplify the first intron, reactions were carried out in a reaction volume of 25 μL containing PCR buffer, 0.4 mM dNTPs, 0.3 μM of each primer and 1.25 U of TaqDNA polymerase (Peqlab). The thermal cycling conditions were similar to those used in documented protocols [12] except that the annealing temperature was raised to 59.8°C. The PCR products were separated on 1.5% agarose gel with the help of Ethidium Bromide (EtBr) staining and the gel image was captured using gel documentation system (Bio-rad).
The second intron was initially amplified using a pair of documented primers [11]. Amplification was carried out in a 25 μL volume containing PCR buffer, 2.5 mM MgCl2, 0.3 mM dNTPs, 1.25 U Taq polymerase and 0.3 μM of each primer. Apart from the annealing temperature which was changed from 58.5°C to 49.3°C the other thermal cycling conditions remained similar to the documented protocols [11]. The extension time was also increased by 12 seconds per cycle. The PCR products were separated on 1.5% agarose gel at 100 V for 30 minutes after ethidium bromide staining. The gel image was taken using Bio-Rad gel documentation system.
Cloning, sequencing and primer design
For amplifications containing two alleles (most of the cases), bands were cut from the gel, purified and subsequently cloned via a pGEM vector (Fermentas) into Escherichia coli competent cells using a heat shock protocol [13] After visually selecting out the cells that were successfully transfected, 7 plasmids per allele were sequenced using universal M13 primers. PCR products of the expected size were purified using the QIAprep Spin Miniprep Kit (Qiagen, Hilden, Germany). In some instances, depending on the length of the insert, internal primers designed from the partial sequences were also used. Specific primers for P. africana were designed based on the sequences obtained (Table 2).
Table 2. Specific primers developed for amplification of S-alleles in P. africana.
Pattern indicates primer combinations.
| Primer Pairs | Primer name | Sequence (5’-3’) | Amplified region |
|---|---|---|---|
| Pair 1 | Pacons2F | ggccaagtaattattcaaacc | 2nd Intron |
| PAI2-2R | ttcgccttcccaaaayyttg | 2nd Intron | |
| Pair 2 | PaI2-2F | ggccaagtaaytattcaaaccca | 2nd Intron |
| PaI2-2R | ttcgccttcccaaaayyttg | 2nd Intron | |
| Pair 3 | PaCons1F | mcttgttcttgstttygctttcttc | 1st Intron |
| PaCons1R | catgratggtgaartwttgtaatgg | 1st Intron | |
| Pair 4 | PaSI1f | gttcttggtttygctttcttc | 1st Intron |
| PaSI1R | catgratggtgaagtwttgtaatgg | 1st Intron | |
| Pair 5 | Paintron1F | ttatgagcastggtgggttg | 1st Intron |
| Paintron1R | actgggcatckttgggtttg | 1st Intron |
Data Analysis
Different size fragments as estimated on agarose were manually scored and distinct allele combinations determined the different genotypes. The fit of the expected and observed distribution was tested using χ2 –test. Mate availability was estimated as percentage of compatible alleles from the observed genotypes [14].
“Sequences” of 7 other Prunus species were obtained from GenBank (http://www.ncbi.nlm.nih.gov). Homologous DNA sequences were aligned using ClustalX [15] and manually adjusted when necessary. Regions that aligned poorly were excluded from the data set. To get a perfect comparison of P. africana S-RNases with other Rosaceous S-RNases, a phylogenetic analysis that grouped S-RNases [16] was used. The minimum evolution method for inferring phylogeny using distance matrices with pair wise deletion obtained following the number of differences, as implemented in MEGA software [17] was used. A close-neighbour-interchange heuristic search (level 2) was performed. For the heuristic search, the initial tree was obtained using the distance based Neighbour-Joining method [17]-[18]. The reliability of clustering patterns of the phylogenetic trees was tested by bootstrapping with 1000 pseudo-replicates [19]. Spatial Genetic Structure (SGS) in the population was analyzed with the program SPAGeDi [20]. Closely related pairs of alleles that occupied paired terminal branches on the species-level phylogeny were chosen for analysis of synonymous and non-synonymous substitution rates. Numbers of synonymous substitutions per synonymous sites (Ks) and non-synonymous substitutions per non-synonymous sites (Ka) were calculated using DnaSP [21].
Results
Statement of Ethics
Authorization for research within the forest was granted by National Forestry Authority, the managers of Uganda’s Central Forest Reserves. Phytosanitary clearance for the leaf samples was given by the crop protection division of the Ministry of Agriculture, Animal Industries and Fisheries of the government of Uganda.
S-RNase intron polymorphism and genotype frequencies
A total of 142 out of 150 samples successfully amplified using specifically designed primers. Primers developed [6][11],[22]for other Prunus species did not effectively amplify the S-alleles in most samples indicating that there are pronounced differences in the loci under investigation between P. africana from other Prunus species. As expected of individuals with functional gametophytic SI systems, the alleles were highly polymorphic (Table 3). Of the 142 individuals that were assessed, 98% were heterozygous at the S-locus. However, the dual bands of first intron could not be distinctively separated on agarose gel, implying that the two fragments had very small nucleotide differences.
Table 3. List of the 26 alleles detected in the second intron of the S-RNase of P. africana.
| Allele No. | Product Size | Count |
|---|---|---|
| S1 | 230 | 1 |
| S2 | 250 | 2 |
| S3 | 270 | 22 |
| S4 | 300 | 20 |
| S5 | 400 | 4 |
| S6 | 480 | 7 |
| S7 | 500 | 23 |
| S8 | 510 | 35 |
| S9 | 550 | 4 |
| S10 | 600 | 15 |
| S11 | 650 | 2 |
| S12 | 700 | 13 |
| S13 | 800 | 8 |
| S14 | 900 | 3 |
| S15 | 950 | 2 |
| S16 | 980 | 18 |
| S17 | 1000 | 20 |
| S18 | 1200 | 6 |
| S19 | 1350 | 18 |
| S20 | 1500 | 2 |
| S21 | 1700 | 13 |
| S22 | 1800 | 11 |
| S23 | 2000 | 1 |
| S24 | 2500 | 15 |
| S25 | 2800 | 4 |
| S26 | 3000 | 15 |
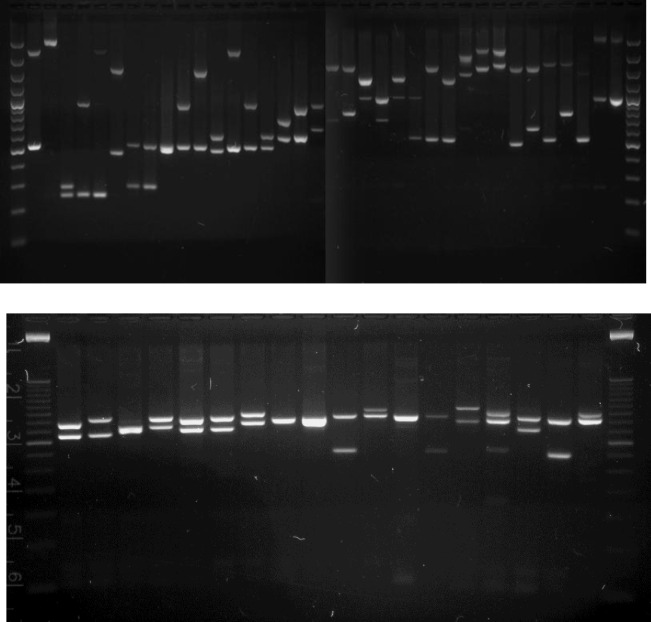
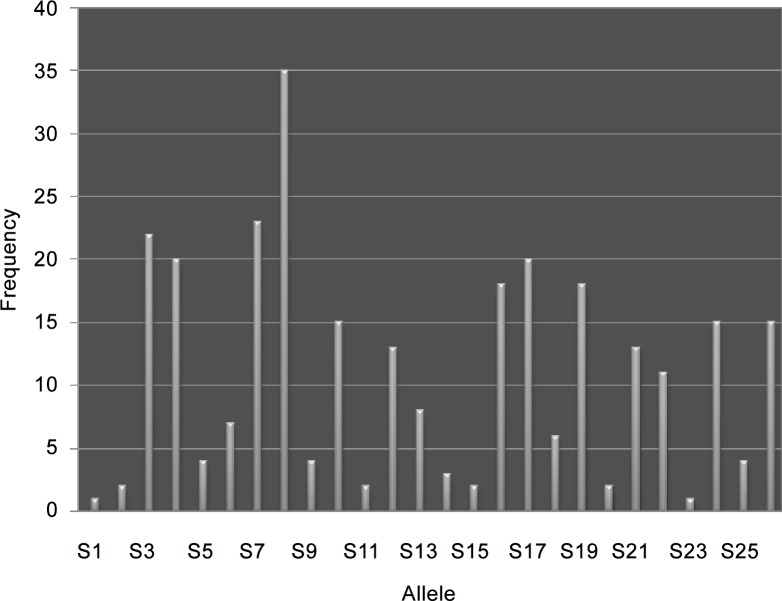
PCR fragments ranged from 210 bp to 3000 bp. Fragments from the first intron ranged from 210 to 550 bp (Fig 2B) whilst second intron lengths ranged from 245 to 3000 bp (Fig 2A). While 26 different alleles were scored for the second intron, only 8 alleles were identified on the agarose gels for the first intron. The overall S-allele frequencies from the second intron are given in Fig 3 and their approximate sizes as scored both on agarose gels and as sequenced product in Table 2 above. The most common allele was S8 (12.3%), while S1 and S23 were least represented (0.3% each). Although, the alleles were expected to be represented in equal frequency under frequency-dependent selection, this was not the case for alleles from both introns as indicated by the Mantel statistic (χ2 = 11.5; P < 0.05). In total, the allele patterns resulted in 54 different genotypes (Fig 2). In this Prunus africana population, mating availability ranged from 85% to 100% given the large number of alleles.
Fig 2.
(a) Agarose gel showing amplification products of second intron sequences. First and last lane is the 100 bp size standard. (b) Agarose gel showing amplification products of first intron sequences. First and last lane is the 50 bp size standard
Fig 3. Graph indicating the frequencies of different S-alleles in P. africana.
Sequence analysis
Verified using BLAST searches (http://blast.ncbi.nlm.nih.gov), all sequences were from the signal peptide or C1–C5 regions. Several P. africana S-RNase sequences were very similar to other Prunus sequences, both on the intra- and interspecific level. 35 sequences of P. africana were similar to published sequences of P. Speciosa, P. dulcis, P. avium, and P. Salicina. Others were similar to alleles found in P. pseudocerasus, P. webbii, P. mume, P. argentea, P. ceracifera and P. cerasus. Although sequences especially those from the second intron could not be reasonably aligned due to their large variation in length, comparison of these sequences with ClustalX [15] showed that there were 40 different sequences. All of these S-RNase alleles had a considerably high homology within the coding region and displayed an intron/exon structure characteristic of Prunus ribonucleases that is, they contained the five conserved regions, C1, C2, C3, RC4 and C5, as well as the hyper-variable region, RHV, associated with the deduced amino acid sequence of rosaceous S-RNases [5].
The regions of the gene that were compared have highly diversified sequences that contained different amino acids ranging from 1 to 10. Pair wise similarity comparison of exons at the nucleotide sequence level showed minimum identity of 50% to maximum identity of 87%, whilst identity at amino acid level was between 20% and 50%. Most identical sequences were generated from more than two representatives of each haplotype. Microsatellites (tandem repeats) mainly AT, GAA, TA were found in several sequences from both introns. Tajiman’s test of neutrality was estimated at 6.2. Distinct sequences were deposited in the GenBank (http://www.ncbi.nlm.nih.gov/) with accession numbers from KT985615 to KT985636.
Identification of S-RNase regions exposed to selection
Closely related pairs of alleles were chosen for analysis of synonymous and non-synonymous substitution rates. Analysis focused on sister alleles that occupied paired terminal branches on the species-level phylogeny. The overall mean values for Ka, Ks and Ka/Ks were 0.1129, 0.096 and 1.18 respectively. An excess of non-synonymous substitutions over synonymous substitutions occurred. The matrix of amino acid differences indicates that the degree of sequence divergence within species is on the same order as the sequence divergence between species. In fact, some alleles appear to be more similar to alleles in other species than to other alleles in the same species.
Phylogenetic tree analysis
Within population S-RNase sequences for P. africana did not show any distinct clusters (Fig 4). However, to place the new S-RNase alleles in context with previously identified Prunus S-RNase alleles, a phylogenetic tree was constructed comparing S-RNases from a representative range of Prunus species (Fig 5). Some sequences were excluded after failure to find a reliable root position because of the large genetic distances between alleles from some Prunus species. Generally, S-RNases of P. africana often displayed closer relationships to those of other Prunus species. This is consistent with trans-specific S-allele evolution within Prunus as has previously been pointed out [5]. Trans-specific polymorphism involving the new alleles was evidenced by high bootstrap support for grouping of for example S-5 of P.africana and S-10 of P. salicina. No specific subgroups within Prunus were detected although some pairs of alleles showed high percentages of pair wise amino acid similarity, e.g. Prunus pseudocerasus and S-5 of P.africana (92%). There are also a few monospecific sister pairs while some alleles showed monophyletic clades. Several close pairs of alleles even seem to represent divergent copies of the same specificity yet quite a few alleles did not cluster with any other known Prunus alleles.
Fig 4. Phylogenetic (minimum evolution) tree of P. africana S-RNase intron 2 sequences, showing the relationship among alleles.
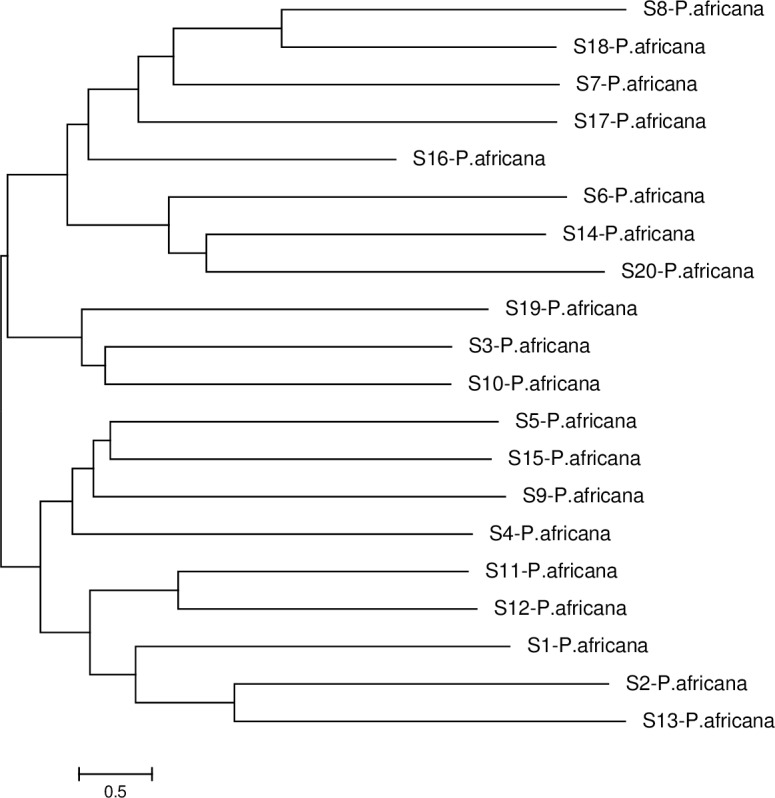
Fig 5. Phylogenetic tree (minimum evolution) of P. africana and other Prunus second intron S-RNase sequences depicting relationships with other congeneric species.

Allele sequences are highly diverged and partially are similar to S-allele sequences obtained in other Prunus species, a pattern commonly observed as alleles are older than the species radiation.
Spatial Genetic Structure (SGS)
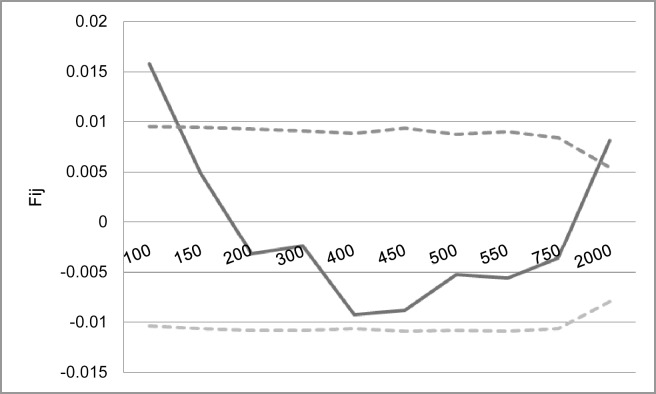
Based on the second intron, the results of fine scale spatial genetic structure (SGS) analysis calculated as correlation between Loiselle‘s kinship coefficient [23] and spatial distance between individuals with 95% confidence limits derived from bootstraps are depicted in Fig 6. Significant SGS was detected only in the first distance class (up to 100 m). No significant SGS was observed in longer distances.
Fig 6. Results of spatial genetic structure analysis in the second intron S-alleles of P. africana.
Significant autocorrelation was detected in the first distance class (up to 100 m) between individuals. X-axis is spatial distance between individuals in metres, y-axis is Loiselle’s kinship coefficient for the second intron S-alleles
Discussion
S-RNase polymorphism and genotype frequencies
The identified S-alleles in P. africana were highly divergent both in length and sequence variation. The number of alleles did not deviate from those observed in other wild and domesticated species of the Rosaceae family which ranges from 12 to 45 [24]. Hoebee [25] for example identified a total of 26 different S-alleles from 138 individuals of Pyrus pyraster. A total of 22 alleles were identified in Prunus lannesiana var. speciosa [26].Twenty alleles were retrieved from 20 Sorbus aucuparia individuals, whereas 16, 17 and 22 alleles were found in Prunus avium [27], Crataegus monogyna [28] and Malus domestica [29] respectively. All these above Rosaceae species being diploid, a similar number of specificities was also found in the polyploidy Prunus spinosa [16]. The total number of alleles observed in these species represents a higher than expected S-allele diversity based on theoretical expectations for gametophytic self-incompatibility system as predicted by the balanced model [30]. For GSI systems, the model stated that “under a moderate mutation/immigration rate of 10−3, a population of 50 individuals should harbour about eight different S-alleles, whereas a population of 200 individuals would have about 15 S-alleles” [30]. However, it is a common occurrence that the number of S-alleles found within populations is usually greater than that predicted by theory [24]. A high S-allele diversity is often indicative of high immigration rate of new S-alleles, which would subsequently experience positive frequency dependent selection [31].
Correspondingly, the mate availability was also expectedly as high as in other Rosaceae species e.g. in Pyrus pyraster [32]. This is in agreement with theoretical expectations on mate availability in small populations of plants with a gametophytic self-incompatibility system. Theoretically, a functional gametophytic self-incompatibility (SI) system in plants should exhibit high polymorphism at the SI controlling S-locus because individuals with rare alleles have a higher probability to successfully pollinate other plants than individuals with more frequent alleles [3] [32]. Pollen carrying a rare allele will not be rejected by incompatibility reactions of recipient plants and therefore will have higher reproductive success and consequently increased frequency of rare S-alleles in subsequent generations. In other words, opportunities for mating of each S-allele are inversely related to the alleles' frequencies [30]. Based on these theories, Only populations harboring less than 5 different S-alleles should show reduced mate availabilities below 90% [33]. This negative-frequency-dependent selection is explicitly expected to maintain many S-alleles in effectively large populations. The very long evolutionary time detected in phylogenetic analyses of P. africana [34] has also allowed for inclusion of many mutations and new migratory alleles to increase the observed diversity.
Unequal specificity frequencies were observed in P. africana, although most species with gametophytic SI support the equal allelic frequencies (isoplethy) hypothesis [27], [31] e.g. isoplethy was observed in Prunus avium [27]. However, earlier results indicated no departure from the identical allelic frequencies hypothesis in 16 out of 19 sampled populations from 12 species with gametophytic SI [24]. The deviation from isoplethy as of Prunus africana may have been caused by the fragmentation of populations as a result of forest degradation, death from overharvesting of the species for bark and the species distribution irregularities. P. africana is thus found as isolated demes which as a consequence may have suffered different founder events resulting in differentiation at the S-locus. Limited dispersal and pollen flow as well as plant size variation that often follow degradation can cause deviation from the expected frequencies [35].
However, it is also urged that it is almost impossible to find equal frequencies when a large sample size is used in analysis [36]. Most studies that have proved the isoplethy theory have used small sample sizes far much less than what was used in this study [36]. As such unequal frequencies have been found in many species that have used large sample sizes for example in Prunus lannesiana var. speciosa [26], Prunus spinosa [16] and Prunus avium [36]. Due to lack of isoplethy, it was therefore not possible to estimate the total number of alleles.
Sequence analysis
The lengths of most of the sequences from both introns were similar to those from previously observed Prunus S-RNases [25] [37]. Sequences of the first intron of P. africana ranged from 210 to 550 bp whilst those of second intron lengths ranged from 245 to 3000 bp. However, P. dulcis sequences from the first intron are longer (up to 1061 bp) than observed for P. africana and a size not recorded in any other Prunus species [37]. For the second intron, P. cerasifera has alleles up to 3400 bp [38] relative to the 3000 bp alleles observed in P. africana. These sequence lengths are also comparable to those detected in Pyrus pyraster, P. pyrifolia and P. communis with average lengths between 334 and 2000 bp, 352 and 1347 bp and 641 and 2217 bp [39] respectively. Nevertheless, interpretation of allele number as obtained solely by molecular techniques is the question of whether different sequences necessarily correspond to different specificities.
Synonymous and non-synonymous substitutions
Finding genomic regions under selection is a critical step in elucidating adaptability and adaptiveness of traits and history of populations. The overall Ka/Ks ratio for 14 P. africana S-RNases was 1.18 which is substantially higher than has been found in both the cultivated P. avium and P. dulcis as well as other wild Prunus species like P. cerasifera, P. lannesiana (0.793) [26], and P. tenella (0.99) [40]. Although these wild species have consistently had a higher Ka/Ks, this has always been <1, indicating negative selection, which is typical of S-alleles that are always under negative frequency dependent selection. Such selection allows rare alleles in a population to increase relative to more common ones by allowing rare alleles to get more compatible mate advantage and also favouring immigrant alleles. Observations made on single populations indicated that the structure of S-alleles is less dependent on mutation rate [30]. In contrast, P. africana Ka/Ks > 1 implying a positive selection for advantageous new mutations in the population. Ecological factors that trigger a great number of adaptive amino acid changes on the S-locus have not yet been explored although drastic reduction in effective population size that P. africana has undergone through logging and habitat degradation can modify the intensity of genetic drift and thus the efficiency of selection against deleterious mutations. This certainly results in a relative excess of non-synonymous changes.
Phylogenetic analysis
Population size should be revealed not only by number of alleles, but also by divergence time. The S-RNases in Prunus africana vis a vis other Prunus species have shown higher intrasubfamilial similarities. These results corroborate the findings of previous studies [5], that suggest that the S-locus of Prunus species diverged more recently or evolved at a slower rate, and thus retains a relatively conserved structure. A comparison of the S-locus region of the Sc and Sd-haplotypes in almond (Prunus dulcis) by Southern blot analysis found that outside of the divergent genomic region around the Sc-RNase gene (≈ 70 kb), the genomic sequence was highly conserved between the two S-haplotypes, suggesting that the ≈ 70 kb region contains the entire complement of S-determinant genes. Comparatively, P. africana appears to have older S-alleles than the S-alleles of other Prunus species with some alleles not being able to form any clade with other alleles. Probably this may support the “out of Africa hypothesis” for P. africana just like many other plants and animals.
P. africana S-RNase spatial genetic structure
The expected patterns of spatial genetic structure (SGS) at target loci will strongly depend on the type of selection involved. Self-incompatibility (SI) system is a frequency-dependent mechanism that is expected to shape the number, frequency distribution, and spatial distribution of self-incompatibility alleles in natural populations. On the contrary though, it is occasionally indicated that SI in a species tends to eliminate existing spatial genetic structures through its obstruction to self-fertilization and cross-fertilization with close individuals that share the same incompatibility alleles [41]. In P. africana, a positive spatial structure was observed. A significant spatial genetic structure for the S-locus was also observed in P. avium [3]
In P. africana, a positive spatial structure over scales of approximately 100 m implies a number of interplaying ecological, demographic and other genetic factors, rather than solely SI. In small populations, such as that of P. africana in this study, interactions between balancing selection and population structure shape the distribution of S-alleles, sometimes in nontrivial ways. Although generally in such populations genetically controlled SI systems may impose significant demographic constraints leading to loss of S-allele richness that further suppresses mate availability [42] [43]. Drift has, for example, played an important part in the population history of P. africana [34]. It has also been suggested that in small populations, there is a common tendency for ecological and demographic factors to override genetic influences [3]. Prunus africana populations in Mabira forest were observed to have very limited seed dispersal and survival of seedlings.
All P. africana trees that were monitored in this study showed a prolific fruit and seed set but seedlings exhibited limited survival and were commonly found under the mother trees. This indicated that either limited seed dispersal or the microenvironment after dispersal doesn’t favour recruitment, with strong implication on the SGS. Reduced reproductive success has also been associated with local genetic effects in Prunus virginiana [44]. The population of P. africana has also recently dramatically declined due to overharvesting of the bark for commercial use in treatment of prostate cancer. When populations decline, genetically controlled SI systems may impose significant demographic constraints leading to loss of S-allele richness; further suppressing mate availability [42] [43] with implications to SGS and this can also be verified by the positive values of the Tajiman’s test. Drift has, for example, played an important part in the population history of P. africana [34].
Besides constrained dispersal and regeneration, the observed SGS for P. africana may also be attributed to the inefficiency of the SI system at preventing selfing or crossing between relatives. Observations indicate that P. africana may exhibit mixed mating system especially when clumped [45] as it was observed in the Mabira forest population. Results of the ratio of synonymous and non-synonymous substitutions, Ka/Ks > 1, imply a positive selection for advantageous new mutations in the population and allowing novel S-alleles into the system may disrupt functionality of the SI system of the plant [46].
Conclusions
This study has provided important insights into the SI system of P. africana. The observed similarity of P. africana S-alleles to other Prunus species strengthens the taxonomic placement of the species within this genus. Although empirical evidence is missing that the observed alleles operate a functional SI system, the high number of alleles detected largely indicate an active SI in P. africana. Crossing experiments involving individuals with known S-genotype are strongly encouraged to finally solve this functionality issue.
The results also have important implications for future breeding purposes. To ensure reproductive success and hence sustainability of the species, genebank establishment and clonal development has to maximally diversify the S- alleles. As indicated above, further empirical studies are needed to finally prove SI in P. africana.
Supporting Information
(XLS)
Acknowledgments
Appreciation goes to all the reviewers, for the constructive criticisms. Thanks go to Dr. Samson Gwali of the National Forestry Resources Research Institute (NaFORRI), Caroline Kadu, Thomas Thalmayr and all the colleagues in BFW lab for their valuable contributions. This research was funded by the Austrian Development Agency. It is the result of a collaboration between Bioversity International and the Federal Research and Training Centre for Forests, Natural Hazards and Landscape (BFW) in Austria, and of a partnership with the School of Forestry, Environmental and Geographical Sciences of Makerere University within the framework of the project 'Development of strategies for the conservation and sustainable use of Prunus africana to improve the livelihood of small-scale farmers.’ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Data are available in GenBank KT985615-KT985634.
Funding Statement
This research was funded by the Austrian Development Agency. It is the result of a collaboration between Bioversity International and the Federal Research and Training Centre for Forests, Natural Hazards and Landscape (BFW) in Austria, and of a partnership with the School of Forestry, Environmental and Geographical Sciences of Makerere University within the framework of the project 'Development of strategies for the conservation and sustainable use of Prunus africana to improve the livelihood of small-scale farmers.' The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Simons AJ, Dawson IK, Duguma B, Tchoundjeu Z. Passing problems: prostate and Prunus. Herbalgram 1998; 43: 49–53 [Google Scholar]
- 2.Cunningham M, Cunningham AB, Schippmann U. Trade in Prunus africana and the implemation of CITES. German Federal Agency for Nature Conservation, Bonn. 1997; 52 pp [Google Scholar]
- 3.Schueler S, Tusch A, Scholz F. Comparative analysis of the within-population genetic structure in wild cherry (Prunus avium L.) at the self-incompatibility locus and nuclear microsatellites. Molecular Ecology 2006; 15: 3231–3243 [DOI] [PubMed] [Google Scholar]
- 4.Shuri K, Saika K, Junko K, Michiharu K, Nagamitsu T, Iwata H, Mukai Y. Impact of negative frequency-dependent selection on mating pattern and genetic structure: a comparative analysis of the S-locus and nuclear SSR loci in Prunus lannesiana var. speciosa. Heredity 2012, 109(3), 188–198 10.1038/hdy.2012.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ushijima K, Sassa H, Hirano H. Characterization of the flanking regions of the S-RNase genes of Japanese pear (Pyrus serotina) and apple (Malus x domestica). Genetics 1998; 211:159–167 [DOI] [PubMed] [Google Scholar]
- 6.Tao R, Yamane H, Sugiura A, Murayama H, Sassa H, Mori H. Molecular typing of S-alleles through identification characterization and cDNA cloning for S-RNases in sweet cherry. Journal of the American Society for Horticultural Sciences 1999; 124: 224–233 [Google Scholar]
- 7.Yaegaki H, Shimada T, Moriguchi T, Hayama H, Haji T, Yamaguchi M. Molecular characterization of S-RNase genes and S-genotypes in the Japanese apricot (Prunus mume Sieb. et Zucc.). Sexual Plant Reproduction 2001; 13: 251–257 [Google Scholar]
- 8.Yamane H, Tao R, Murayama H, Sugiura A. Determining the S-genotypes of several sweet cherry cultivars based on PCR-RFLP analysis. Journal of Horticultural Science and Biotechnology 2000; 75: 562–567 [Google Scholar]
- 9.Langdale-Brown I, Osmaston HA Wilson JG. The vegetation of Uganda and its bearing on land use 1964; Government of Uganda, Entebbe, Uganda [Google Scholar]
- 10.Dumolin S, Demesure B, Petit RJ. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoretical and Applied Genetics 1995; 91: 1253–1256 10.1007/BF00220937 [DOI] [PubMed] [Google Scholar]
- 11.Sutherland BG, Robbins TP, Tobutt KR. Primers amplifying a range of Prunus S-alleles. Plant Breeding 2004; 123: 582–584 [Google Scholar]
- 12.Sonneveld T, Tobutt KR, Robbins TP. Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theoretical and Applied Genetics 2003; 107: 1059–1070 [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J, Russell DW. Molecular cloning: a laboratory manual 3rd edition 2001; Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA [Google Scholar]
- 14.Glemin S, Petit C, Maurice S, Mignot A. Consequences of low mate availability in the rare self-incompatible species Brassica insularis. Conservation Biology 2008; 22:216–221 10.1111/j.1523-1739.2007.00864.x [DOI] [PubMed] [Google Scholar]
- 15.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vieira CP. An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. Journal of Molecular Evolution 2008; 67: 179–190 10.1007/s00239-008-9137-x [DOI] [PubMed] [Google Scholar]
- 17.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinformatics 2004; 5:150–163 [DOI] [PubMed] [Google Scholar]
- 18.Saitou N, Nei M. The neighbor joining method: a new method of reconstructing phylogenetic trees. Mol Biol Evol 1987; 4(4) 406–25 [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985; 39:783–791 [DOI] [PubMed] [Google Scholar]
- 20.Hardy OJ, Vekemans X. SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes 2: 2002; 618–620 [Google Scholar]
- 21.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009; 25: 1451–1452 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 22.Wiersma P, Wu Z, Zhou L, Hampson C, Kappel F. Identification of new self-incompatibility alleles in sweet cherry (Prunus avium L.) and clarification of incompatibility groups by PCR and sequencing analysis. Theoretical and Applied Genetics 2001; 102: 700–708 [Google Scholar]
- 23.Loiselle BA, Sork VL, Nason J, Graham C. Spatial genetic structure of a tropical understorey shrub Psychotria officinalis (Rubiaceae). American Journal of Botany 1995; 82: 1420–1425 [Google Scholar]
- 24.Lawrence MJ. Population genetics of the homomorphic self-incompatibility polymorphisms in flowering plants. Annals of Botany 2000, 85: 221–226 [Google Scholar]
- 25.Hoebee SE, Angelone A, Csencsics D, Maattanen K, Holderegger R. Diversity of S-alleles and mate availability in 3 populations of self incompatible wild pear (Pyrus pyraster). Journal of Heredity 2011; 10.1093/jhered/esr126 [DOI] [PubMed] [Google Scholar]
- 26.Kato S, Mukai Y. Allelic diversity of S-RNase at the self-incompatibility locus in natural flowering cherry populations (Prunus lannesiana var. speciosa). Heredity 2004; 92:249–256 [DOI] [PubMed] [Google Scholar]
- 27.Ganopoulos I, Aravanopoulos F, Argiriou A, Tsaftaris A. Genome and population dynamics under selection and neutrality: an example of S-allele diversity in wild cherry (Prunus avium L.). Tree Genetics & Genomes 2012; 8:1181–1190 [Google Scholar]
- 28.Raspe O, Kohn JR. S-allele diversity in Sorbus aucuparia and Crataegus monogyna (Rosaceae: Maloideae). Heredity 2002; 88(6):458–65 [DOI] [PubMed] [Google Scholar]
- 29.Larsen B, Ørgaard M, Toldam-Andersen TB, Pedersen C. A high-throughput method for genotyping S-RNase alleles in apple. Molecular Breeding 2016; 36, 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright S. The distribution of self-sterility alleles in populations. Genetics 24 1939; 538–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castric V, Vekemans X. Plant self-incompatibility in natural populations: a critical assessment of recent theoretical and empirical advances. Molecular Ecology 2004; 13: 2873–2889 [DOI] [PubMed] [Google Scholar]
- 32.Holderegger R, Häner R, Csencsics D, Angelone S, Hoebee SE. S‐allele diversity suggests no mate limitation in small populations of a self‐incompatible plant. Evolution, 2008; 62(11), 2922–2928 10.1111/j.1558-5646.2008.00498.x [DOI] [PubMed] [Google Scholar]
- 33.Vekemans X, Schierup MH, Christiansen FB. Mate availability and fecundity selection in multi-allelic self-incompatibility systems in plants. Evolution, 1998; 52:19–29. [DOI] [PubMed] [Google Scholar]
- 34.Kadu CAC, Schueler S, Konrad H. Phylogeography of the Afromontane Prunus africana reveals a former migration corridor between East and West African highlands Molecular Ecology 2011; 20:165–178 10.1111/j.1365-294X.2010.04931.x [DOI] [PubMed] [Google Scholar]
- 35.Brooks RJ, Tobias AM, Lawrence MJ. The population genetics of the self-incompatibility polymorphism in Papaver rhoeas. XI. The effects of limited pollen and seed dispersal, overlapping generations and variation in plant size on the variance of S-allele frequencies in populations at equilibrium. Heredity 1996; 76: 367–376 [Google Scholar]
- 36.Stoeckel S, Castric V, Mariette S, Vekemans X. Unequal allelic frequencies at the self incompatibility locus within local populations of Prunus avium L. an effect of population structure? J Evol Biol 2008; 21: 889–899 10.1111/j.1420-9101.2008.01504.x [DOI] [PubMed] [Google Scholar]
- 37.Ortega E, Boskovic RI, Sargent DJ, Tobutt KT. Analysis of S-RNase alleles of almond (Prunus dulcis): characterization of new sequences, resolution of synonyms and evidence of intragenic recombination. Molecular Genetics and Genomics 2006; 276: 413–426 [DOI] [PubMed] [Google Scholar]
- 38.Sutherland BG, Tobutt KR, Robbins TP. Trans-specific S-RNase and SFB alleles in Prunus self-incompatibility haplotypes. Molecular Genetics 2008; 279: 95–106 [DOI] [PubMed] [Google Scholar]
- 39.Sanzol J. Genomic characterization of self-incompatibility ribonucleases (S-RNases) in European pear cultivars and development of PCR detection for 20 alleles. Tree Genet Genomes 2009; 5:393–405 [Google Scholar]
- 40.Surbanovski N, Tobutt KR, Konstantinovic´ M, Maksimovic´ V, Sargent DJ, Stevanovic´ V, et al. Self-incompatibility of Prunus tenella and evidence that reproductively isolated species of Prunus have different SFB alleles coupled with an identical S-RNase allele. Plant Journal 2007; 50:723–734 [DOI] [PubMed] [Google Scholar]
- 41.Doligez AC, Baril H, Joly I. Fine-scale spatial genetic structure with nonuniform distribution of individuals. Genetics 1998; 148: 905–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young AG, Broadhurst LM, Thrall PH. Non-additive effects of pollen limitation and self-incompatibility reduce plant reproductive success and population viability. Annals of Botany 2012; 109(3), 643–653 10.1093/aob/mcr290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagenius S, Lonsdorf E, Neuhauser C. Patch aging and the S-Allee effect: breeding system effects on the demographic response of plants to habitat fragmentation. American Naturalist 2007; 169:383–397 10.1086/511313 [DOI] [PubMed] [Google Scholar]
- 44.Suarez-Gonzalez A, Good SV. Pollen limitation and reduced reproductive success are associated with local genetic effects in Prunus virginiana, a widely distributed self-incompatible shrub. Annals of Botany 2014; 113 (4): 595–605 10.1093/aob/mct289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berens DG, Braun C, González-Martínez SC, Griebeler EM, Nathan R, Böhning-Gaese K. Fine-scale spatial genetic dynamics over the life cycle of the tropical tree Prunus africana. Heredity 2014; 10.1038/hdy.2014.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charlesworth D, Vekemans X, Castric V, Glemin S. Plant self-incompatibility systems: a molecular evolutionary perspective. New Phytologist 2005; 168: 61–69 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
Data Availability Statement
Data are available in GenBank KT985615-KT985634.