Abstract
Acetate accumulation during the fermentation process of Escherichia coli FB-04, an L-tryptophan production strain, is detrimental to L-tryptophan production. In an initial attempt to reduce acetate formation, the phosphate acetyltransferase gene (pta) from E. coli FB-04 was deleted, forming strain FB-04(Δpta). Unfortunately, FB-04(Δpta) exhibited a growth defect. Therefore, pta was replaced with a pta variant (pta1) from E. coli CCTCC M 2016009, forming strain FB-04(pta1). Pta1 exhibits lower catalytic capacity and substrate affinity than Pta because of a single amino acid substitution (Pro69Leu). FB-04(pta1) lacked the growth defect of FB-04(Δpta) and showed improved fermentation performance. Strain FB-04(pta1) showed a 91% increase in L-tryptophan yield in flask fermentation experiments, while acetate production decreased by 35%, compared with its parent FB-04. Throughout the fed-batch fermentation process, acetate accumulation by FB-04(pta1) was slower than that by FB-04. The final L-tryptophan titer of FB-04(pta1) reached 44.0 g/L, representing a 15% increase over that of FB-04. Metabolomics analysis showed that the pta1 genomic substitution slightly decreased carbon flux through glycolysis and significantly increased carbon fluxes through the pentose phosphate and common aromatic pathways. These results indicate that this strategy enhances L-tryptophan production and decreases acetate accumulation during the L-tryptophan fermentation process.
Introduction
L-tryptophan, an essential amino acid that is also a precursor of other important biomolecules, such as the neurotransmitter serotonin, is widely used in medicine, food, and animal feed [1, 2]. The L-tryptophan biosynthetic pathway in microorganisms, which involves central metabolism, the common aromatic pathway and the L-tryptophan branch pathway, is long and its regulation is complicated [3–5]. The high industrial relevance of Escherichia coli has stimulated efforts to improve L-tryptophan yield by analyzing the underlying metabolic regulatory networks of L-tryptophan biosynthesis and altering them through targeted modifications [6–8]. Despite substantial effort, by-product accumulation continues to limit L-tryptophan production.
The accumulation of acetate, a main by-product of fermentation process, can slow cell growth [9, 10]. In E. coli, two metabolic pathways are involved in acetate formation: the PoxB oxidase pathway and the Pta-AckA pathway [11]. In the PoxB oxidase pathway, pyruvate oxidase (PoxB; EC 1.2.5.1), encoded by the poxB gene, catalyzes the formation of acetate [11]. In the Pta-AckA pathway, which is the primary acetate formation pathway, phosphate acetyltransferase (Pta; EC 2.3.1.8) and acetate kinase (AckA; EC 2.7.2.1), encoded by the pta and ackA genes, respectively, catalyze the formation of acetate from acetyl-CoA in two successive steps [12, 13]. Pta, the key enzyme in this pathway, catalyzes the conversion of acetyl-CoA to acetyl phosphate, which plays an important cellular role as a phosphorus donor [14]. Knockout of pta is the most common genetic manipulation used to diminish acetate accumulation. Although a pta deletion mutant has shown increased biomass and a higher capacity for producing L-tryptophan, compared with its parental strain [15], most previous reports indicate that Pta has a significant physiological function in E. coli, and that deletion of pta adversely impacts cell growth [14, 16–18]. Remarkably, no study examining the effect of pta mutation, rather than deletion, on acetate formation, has been reported, even though this strategy may decrease acetate formation without the deleterious effects of a pta knockout.
Recombinant E. coli FB-04 was constructed in our laboratory for the production of L-tryptophan [5]. However, problems with acetate accumulation during the fermentation process hinder its usefulness. In the initial phase of this study, the native pta in the FB-04 genome was knocked out, forming strain FB-04(Δpta). This knockout led to a growth defect. Then, the pta in the FB-04 genome was replaced with the mutant pta1 identified from E. coli CCTCC M 2016009, which accumulates less acetate during fermentation process, to form the strain FB-04(pta1). This substitution not only reversed the growth defect caused by pta deletion, it also diminished acetate accumulation and improved L-tryptophan production in FB-04(pta1). For the sake of completeness, the performance of FB-04(pta1) was also compared with that of FB-04(ΔackA), an AckA deletion mutant. Metabolomics analysis, a powerful method that has proven to be an important tool for the analysis of changes in intracellular metabolite levels [19, 20], was used to investigate the metabolic distinctions among the L-tryptophan production strains.
Materials and Methods
Strains, plasmids, and genetic methods
The background, genotypes, and sources of all strains used or produced in this study, along with the primers and plasmids used in their construction, are listed in Table 1. E. coli strains JM109 and BL21(DE3), which were used for plasmid construction and protein expression, respectively, were obtained from Novagen (Madison, USA). E. coli CCTCC M 2016009 was isolated from the soil at Kanas Lake, Xinjiang Uygur Autonomous Region, China (48°43’N 87°01’E) and stored in our laboratory. Bacillus subtilis ATCC 6051a, obtained from the American Type Culture Collection (ATCC), supplied the genomic DNA used during the construction of the plasmid pMD-SK. Recombinant E. coli FB-04 was previously constructed for L-tryptophan production [5]. E. coli strains FB-04(Δpta), FB-04(ΔackA) and FB-04(pta1) are derivatives of FB-04 (Table 1). The tool plasmids pKD13 and pKD46 used for gene knockout procedures were purchased from the E. coli Genetic Stock Center (Yale University, New Haven, USA). The pMD18-T simple vector and pET24a vector were obtained from Takara (Dalian, China).
Table 1. Strains, plasmids and primers used in this study.
| Strains/plasmids/primer | Description/Genotype/Sequence | source |
|---|---|---|
| Strains | ||
| FB-04 | E. coli W3110 k12, ΔtrpR::FRTΔtnaA::FRTΔpheA::FRT | [5] |
| ΔtyrA::FRT, harboring pSTV-03 plasmid | ||
| B. subtilis ATCC 6051a | Providing template for sacB | ATCC |
| E.coli CCTCC M 2016009 | Isolated from the soil | CCTCC |
| BL(DE3) | E. coli host for protein expression | Novagen |
| BL21(DE3)/pET24a-pta | BL21(DE3) harboring pET24a-pta | This study |
| BL21(DE3)/pET24a-pta1 | BL21(DE3) harboring pET24a-pta1 | This study |
| FB-04(Δpta) | Δpta::kan-sacB; pta deletion mutant derived from strain FB-04 | This study |
| FB-04(ΔackA) | ΔackA::kan-sacB; ackA deletion mutant derived from strain FB-04 | This study |
| FB-04(pta1) | pta1 genomic substitution in FB-04 | This study |
| Plasmids | ||
| pKD13 | Amp and kan markers | [21] |
| pKD46 | Amp markers, helper plasmid | [21] |
| pSTV-03 | Based on plasmids of pACYC177 and pND707; p15A replicon, kan | [5] |
| marker, PR and PL promoters, carrying aroFfbr and trpEfbrD | ||
| pET24a | Protein expression vector in E. coli BL(DE3) | Novagen |
| pET24a-pta | pET24a harboring pta | This study |
| pET24a-pta1 | pET24a harboring pta1 | This study |
| pMD18-T | Cloning vector | Novagen |
| pMD-SK | pMD18-T harboring kan and sacB | This study |
| Primersa | ||
| C-pta | 5’-aaacatatgtcccgtattattatgctgatcc-3’ | This study |
| 5’-cccaagcttttactgctgctgtgcagactg-3’ | ||
| P-pta | 5’-gtgtcccgtattattatgctgatccctaccggaaccagcgtcggtctgac | This study |
| attccggggatccgtcgacc-3’ | ||
| 5-ttactgctgctgtgcagactgaatcgcagtcagcgcgatggtgtagacga | ||
| tgccaataggatatcggcat-3’ | ||
| P-ackA | 5’-aggtacttccatgtcgagtaagttagtactggttctgaactgcggtagtt | This study |
| attccggggatccgtcgacc-3’ | ||
| 5-accgccagctgagctggcggtgtgaaatcaggcagtcaggcggctcgcgt | ||
| tgccaataggatatcggcat-3’ | ||
| V-ackA | 5’-atgtcgagtaagttagtact-3’ | This study |
| 5’-tcaggcagtcaggcggctcgcgt-3’ | ||
| V-pta | 5’-aagcggctttaggtgcaggc-3’ | This study |
| 5’-tttactgctgctgtgcagactgaat-3’ | ||
| V-poxB | 5’-atgaaacaaacggttgcagc-3-3’ | This study |
| 5’-ttaccttagccagtttgtt-3’ | ||
| SK-kan-sacB | F:5’-aaacatatgattccggggatccgtcgacc-3’ | This study |
| R:5’-cccaagctttgccaataggatatcggcat-3’ | ||
| Overlap-F:5’-cgaagcagctccagcctacagcaactttatgcccatgca-3’ | ||
| Overlap-R:5’-tgcatgggcataaagttgctctaggctggagctgcttcg-3’ |
The sequences of primers V-pta, V-ackA, and V-poxB, which were designed for PCR amplification of the genes pta (GenBank accession number gi 946778), ackA (GenBank accession number gi 946775), and poxB (GenBank accession number gi 946132), respectively, are shown in Table 1. These primers were used to amplify the genes pta, ackA and poxB using genomic DNA from E. coli FB-04 as the template. They were also used to amplify the genes pta1, ackA1, and poxB1 using genomic DNA from E. coli CCTCC M 2016009 as the template. To express Pta and its mutant Pta1, the PCR products pta and pta1, obtained from E. coli FB-04 and CCTCC M 2016009, respectively, using the primer set C-pta (Table 1), were digested with NdeI and HindIII, and then ligated into plasmid vector pET24a to form plasmids pET24a-pta and pET24a-pta1. These two plasmids were used to transform the expression strain E. coli BL21(DE3), resulting in strains BL21(DE3)/pET24a-pta and BL21(DE3)/pET24a-pta1.
Genetic manipulations were performed in mutant alleles by using a two-step scarless gene replacement technique employing λ Red recombination [21, 22]. For scarless gene replacement, plasmid pMD-SK was created by cloning two separate PCR products, kan and sacB, into the plasmid vector pMD18-T. First, kan and sacB were PCR amplified using plasmid pKD13 and the genomic DNA of B. subtilis ATCC 6051a as templates, respectively, and the primer pair SK-kan-sacB (Table 1). The two PCR products were ligated by overlap extension PCR, then the resulting kan-sacB fragment was cloned into pMD18-T, forming pMD-SK. Next, the kan-sacB fragment plus homologous sequences was PCR amplified from plasmid pMD-SK using P-pta or P-ackA primers (Table 1). The underlined letters represent homologous sequences. The single-gene pta knockout mutant FB-04(Δpta) and single-gene ackA knockout mutant FB-04(ΔAckA) were constructed by transducing the corresponding kan-sacB fragment into cells harboring the helper plasmid pKD46, using kanamycin resistance as the positive selection marker. For the scarless insertion of pta1 in the genome of E. coli FB-04, forming mutant FB-04(pta1), the pta1 fragment, which was amplified from the E. coli CCTCC M 2016009 genome using the primer pair V-pta (Table 1), was transduced into the pta single-gene knockout mutant harboring plasmid pKD46, using 15% sucrose as the negative selection marker. PCR and DNA sequencing were also performed to verify the identities of all alleles and plasmids.
Media and culture conditions
For enzyme expression, E. coli strains BL21(DE3)/pET24a-pta and BL21(DE3)/pET24a-pta1 were grown at 37°C in LB medium. Portions of these seed cultures (10% (v/v)) were used to inoculate 100 mL portions of TB medium supplemented with 30 μg/mL kanamycin in 500 mL flasks at 37°C. After 2 h, isopropyl β-d-thiogalactopyranoside was added to a final concentration of 0.4 mM to induce protein expression. Enzyme expression was performed at 25°C for 24 h in a reciprocal shaker (200 rpm).
For flask cultivation, seed cultures of the L-tryptophan-producing strain FB-04 and its derivatives were grown in LB medium for 10 h at 37°C in a rotary shaker. Aliquots (200 μL) of these seed cultures were used to inoculate 100 mL portions of flask fermentation medium containing (per liter): 24 g K2HPO4, 9.6 g KH2PO4, 1g MgSO4·7H2O, 5 g (NH4)2SO4, 10 g glucose, 15 g yeast, 2 g citric acid, and 3 ml trace element solution. The ingredients of trace element solution have been described elsewhere [5]. These portions of fermentation medium, which were supplemented with 30 μg/mL kanamycin, were incubated in 500 mL flasks at 37°C for 48 h. During the fermentation, NH4OH and 10 g/L glucose were added to the medium every 8 h to maintain the pH at 6.5–7.2 and supply a source of carbon, respectively. For metabolomic analysis, cells were collected at 26 h.
For fermentor cultivations, 100 mL seed cultures of FB-04 and its derivatives were first prepared in 500 mL flasks in flask fermentation medium for 10 h at 37°C in a rotary shaker, then transferred to a 3 L BioFlo 110 fermentor (New Brunswick Scientific, USA) containing 900 mL fed-batch fermentation medium containing (per liter) (pH 6.5): 15 g K2HPO4, 2g MgSO4·7H2O, 1.6 g (NH4)2SO4, 2 g yeast, 7.5 g glucose, 2 g citric acid, 0.0129 g CaCl2, 0.075 g FeSO4·7H2O. The fed-batch was performed for 54 h, and samples were taken every 3 h. The pH was maintained at 6.5 by automatically adding NH4OH. The temperature was kept at 37°C. The dissolved oxygen level was controlled at 20% by adjusting agitation speed. When glucose in the medium was exhausted, glucose (800 g/L) was introduced using an exponential feeding program [23], with cell growth controlled at a specific growth rate of 0.15 h-1. All fermentation processes were repeated three times. Fermentation parameters are reported as the mean ± standard deviation of these three replicates.
Purification of wild-type Pta and its mutant Pta1
Extraction and purification of phosphate acetyltransferase were performed as previously described [24], with slight modification. All operations were performed at 4°C. E. coli cells harvested from expression cultures were suspended in 20 mM KHCO3, then disrupted by sonication for 15 min on ice. Cellular debris was removed by centrifugation at 12,000 × g for 30 min. At this point, saturated (NH4)2SO4 was added slowly to the supernatant to attain a final concentration of 50% saturation. The precipitates were stirred overnight at 4°C, collected by centrifugation, then suspended in 50 mM Tris-HCl (pH 8). This solution was dialyzed overnight against 0.2 mM ammonium sulfate in 10 mM Tris-HCl (pH 7.6). Protein purification was performed using a fast protein liquid chromatography system (AKTA FPLC system; GE Healthcare) equipped with a DEAE-Sepharose anion-exchange column (160 mm by 10 mm; Pharmacia Biotech). The dialyzed enzyme solution was diluted 4 times with 10 mM Tris-HCl (pH 8.5), and then loaded onto the DEAE-Sepharose column, which had been pre-equilibrated with 0.05 M ammonium sulfate in 10 mM Tris-HCl (pH 7.8). A linear gradient of 0.05 to 0.2 M ammonium sulfate in 10 mM Tris-HCl (pH 7.8) was used to elute the adsorbed proteins. Fractions containing enzyme were pooled and stored at -20°C.
Analytical methods
The purified phosphate acetyltransferase was analyzed by SDS-PAGE, and the Bradford method was used to determine protein concentration [25]. Phosphate acetyltransferase activity was measured as previously described [26]. The kinetic parameters (Km, Vmax, and kcat values) of the purified phosphate acetyltransferase were determined by using acetyl-CoA as the substrate (at concentrations of 20, 40, 80, 125, 150, 200, 250, 300, 360, and 400 μmol/L) at 25°C in 100 mM phosphate buffer (pH 7.4). Km and Vmax values were estimated by fitting the initial rate data to the Michaelis-Menten equation using nonlinear regression with GraphPad Prism software (GraphPad Software Inc., San Diego, CA). Enzyme assays were repeated three times. Kinetic data are reported as the mean ± standard deviation of these three replicates.
The optical density at 600 nm (OD600) was measured after appropriate dilution. For detection of extracellular metabolites, the supernatant obtained after centrifugation of the fermentation broth was stored at -20°C until analyzed using high performance liquid chromatography (HPLC). The concentrations of glucose and acetate were determined using HPLC with an Aminex HPX-87H column (300 mm × 7.8 mm; Bio-Rid, Hercules, CA) at 50°C, 50 mM H2SO4 as the mobile phase and a flow rate of 0.5 mL/min. The concentration of L-tryptophan was determined using the method described previously [5], with an Agilent Eclipse XDB-C18 column (15 mm × 4.6 mm).
Metabolome analysis by GC-MS
Gas chromatography-mass spectrometry (GC-MS) was used to characterize the intracellular metabolite profiles of FB-04, FB-04(Δpta) and FB-04(pta1). Five biomass samples of each strain were taken manually from the shaker, quickly filtered through a nitrocellulose filter (pore size 45 μm, Pall Corporation), and then immediately quenched in liquid nitrogen. Cells (100 mg wet weight) were suspended in 1.1 ml 90% methanol (-40°C, v/v) and mixed quickly. The cell suspension was frozen and thawed three times, sonicated for 15 min at 4°C, and then held at -20°C for 1 h. After centrifugation at 14,000 × g and 4°C for 15 min, isotopically enriched internal standard (L-phenylalanine-13C9-15N) was added into the supernatant to a final concentration of 10 μg/mL, then this mixture was dried using a moderate stream of nitrogen. Methoxylamine hydrochloride in pyridine (20 mg/mL; 30 μL) was added to the powder, and then 30 μL of N,O-Bis(trimethylsilyl)trifluoroacetamide containing 1% trimethylchlorosilane was added to the mixture. Derivatization was allowed to proceed at 70°C for 60 min.
The GC/MS analysis was performed as previously described [27], with slight modifications. The oven temperature was initially held at 70°C for 2 min, then ramped to 160°C at a rate of 6°C/min, to 240°C at a rate of 10°C/min, to 300°C at a rate of 20°C/min, and finally held at 300°C for 8 min. AMDIS software from NIST (National Institute of Standards and Technology) was employed to deconvolute the mass spectra obtained from the raw GC/MS data. The deconvoluted mass spectra were automatically matched with a standard library as previously described that included retention time and mass spectra [27]. Unmatched peaks were analyzed using NIST MS 2.0 software, which automatically searches for compound information from the NIST 11 library and the Golm Metabolome Database. The final data were subjected to principal component analysis using commercial software Simca-P 11.0 (Umetrics AB, Umeå, Sweden) to holistically observe the general clustering and trends among all samples.
Results
Sequence comparison of genes involved in the formation of acetate in E. coli FB-04 and E. coli CCTCC M 2016009
When they are grown under the same fermentation conditions, less acetate is secreted by E. coli CCTCC M 2016009 than by E. coli FB-04 (data not shown). To address this issue, genes involved in acetate formation were analyzed. The pta, ackA, and poxB genes from E. coli FB-04 and the pta1, ackA1, and poxB1 genes from E. coli CCTCC M 2016009 were amplified using PCR and then sequenced. Comparison of the three pairs of genes (pta/pta1, ackA/ackA1, poxB/poxB1) revealed a single base difference (c206t) between pta and pta1; the sequences of ackA and ackA1 were identical, as were those of poxB and poxB1. This difference leads to a single amino acid substitution (Pro69 to Leu) in Pta1, compared with Pta.
Enzymatic analysis of Pta and Pta1
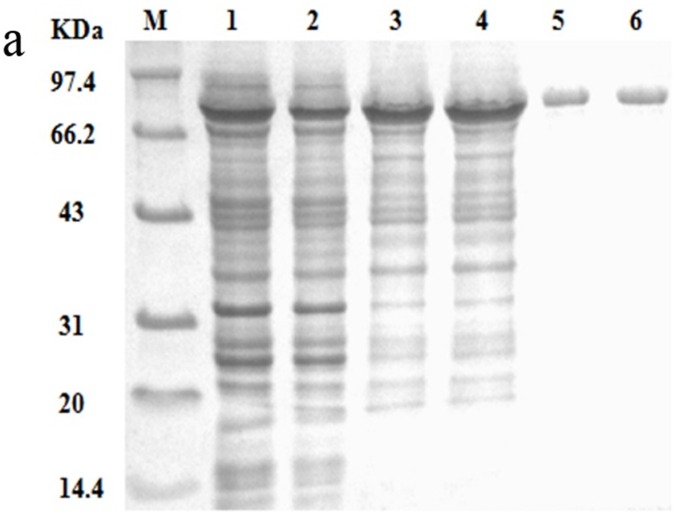
To evaluate the activities of Pta and Pta1, the two enzymes were overexpressed in E. coli BL21(DE3). SDS-PAGE analysis of cell lysates revealed protein bands at approximately 77 kDa, which correspond to those observed with purified recombinant Pta and Pta1(Fig 1). The positions of these bands are consistent with their predicted molecular masses. The protein concentrations were almost the same, indicating that the difference between the expression levels of Pta and Pta1 was not significant. Purified Pta and Pta1 were subjected to kinetic analysis. The catalytic constant (kcat) of Pta1 was 38% lower than that determined for Pta, while its Km was 190% higher than that of Pta. Thus, the kcat /Km value of Pta1 was only about 21% that of Pta (Table 2).
Fig 1. SDS-PAGE analysis of enzymes.
(a) SDS-PAGE analysis of Pta and Pta1 followed by the protein concentration. Lanes contain: cell lysates from expression cultures of E. coli BL21(DE3)/pET24a-pta (1.2 mg/mL) (lane 1) and E. coli BL21(DE3)/pET24a-pta1 (1.1 mg/mL) (lane 2); 50% saturated (NH4)2SO4 precipitations of Pta (0.7 mg/mL) (lane 3) and Pta1(0.8 mg/mL) (lane 4); Purified enzymes: Pta (0.2 mg/mL) (lane 5) and (0.2 mg/mL)Pta1 (lane 6); and protein molecular weight markers (lane M).
Table 2. Kinetic parameters for Pta and Pta1.
| Enzyme | kcat (s-1) | Km (μM) | kcat /Km (μM-1 s-1) |
|---|---|---|---|
| Pta | 2,380 ±120 | 89 ± 6.3 | 27 ± 3.6 |
| Pta1 | 1,480 ± 74 | 260 ±13 | 5.7 ± 0.5 |
All data are the average (± standard deviation) of three independent experiments.
Effect of pta1 genomic substitution on acetate formation and L-tryptophan production
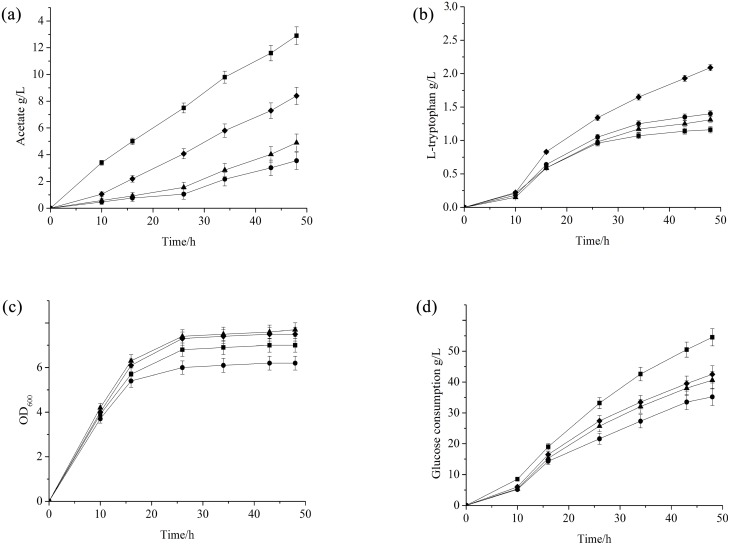
To investigate the effect of Pta-AckA pathway alteration on acetate formation, E. coli strain FB-04(Δpta), in which pta has been deleted, FB-04(ΔackA), in which ackA has been deleted, and FB-04(pta1), in which pta has been replaced by pta1, were constructed from E. coli FB-04 using λ Red recombination. Then, the fermentation performances of FB-04(Δpta), FB-04(ΔackA) and FB-04(pta1) were compared with that of parent strain FB-04 in a series of shake-flask cultures. Throughout the entire fermentation processes, the acetate levels in FB-04(Δpta), FB-04(ΔackA) and FB-04(pta1) cultures were lower than that in cultures of their parent strain FB-04 (Fig 2a). Fermentation parameters for different strains are summarized in Table 3. Strains FB-04(Δpta), FB-04(ΔackA) and FB-04(pta1) showed remarkable decreases in acetate formation, corresponding to 72%, 62%, and 35% that of the parent strain FB-04, respectively.
Fig 2. Flask cultivation of different strains.
(a) Acetate levels; (b) L-tryptophan levels; (c) biomass levels; (d) glucose consumption levels. FB-04 (square); FB-04(Δpta) (circle); FB-04(ΔackA) (triangle); FB-04(pta1) (diamond).
Table 3. Comparison of fermentation parameters for different strains in flask cultivation and fed-batch fermentation.
| Strain | Glucose | Maximum | Maximum | L-tryptophan | L-tryptophan | Maximum |
|---|---|---|---|---|---|---|
| consumption | biomass | L-tryptophan | productivity | yield | acetate | |
| (g/L) | (OD600) | (g/L) | (g/L/h) | per glucose (g/g) | (g/L) | |
| Flask fermentation | ||||||
| FB-04 | 54.5±5.0 | 7.0±0.4 | 1.1±0.1 | 0.020±0.002 | 0.020±0.003 | 12.9±0.7 |
| FB-04(Δpta) | 35.2±2.5 | 6.2±0.3 | 1.4±0.1 | 0.029±0.002 | 0.040±0.005 | 3.6±0.2 |
| FB-04(ΔackA) | 40.6±4.1 | 7.7±0.6 | 1.3±0.1 | 0.027±0.001 | 0.032±0.003 | 4.9±0.3 |
| FB-04(pta1) | 42.5±4.3 | 7.5±0.6 | 2.1±0.2 | 0.044±0.005 | 0.049±0.003 | 8.4±0.5 |
| Fed-batch fermentation | ||||||
| FB-04 | 357±22 | 73.6±0.6 | 38.1±2.3 | 0.70±0.04 | 0.11±0.01 | 4.3±0.3 |
| FB-04(Δpta) | 292±19 | 52.5±3.8 | 21.8±1.5 | 0.40±0.03 | 0.08±0.01 | 0.5±0.3 |
| FB-04(ΔackA) | 338±20 | 76.8±5.8 | 40.2±2.0 | 0.74±0.04 | 0.12±0.01 | 1.5±0.2 |
| FB-04(pta1) | 346±27 | 82.8±6.5 | 44.0±2.8 | 0.82±0.06 | 0.13±0.01 | 2.1±0.3 |
All data are the average (with standard deviation) of three independent experiments.
L-tryptophan production levels were also determined (Fig 2b). The L-tryptophan titer of strain FB-04(pta1) (2.1 g/L) was significantly higher than that of FB-04 (1.1 g/L), FB-04(Δpta) (1.4 g/L) and FB-04(ΔackA) (1.3 g/L), corresponding to increases of 91%, 50%, and 62%, respectively (p < 0.01, Student’s t-test). As shown in Fig 2c, the growth of FB-04(Δpta) was restricted, while FB-04(ΔackA), FB-04(pta1) and FB-04 displayed similar growth curves. The final biomass level (OD600) of FB-04(Δpta) was 11% less than that of FB-04, while the final biomass levels of FB-04(ΔackA) and FB-04(pta1) were slightly higher (Table 3). During cell growth, glucose consumption increased with time for all strains (Fig 2d). The glucose consumption rate of FB-04(pta1) was lower than that of FB-04, while the glucose consumption rate of FB-04(Δpta) was lowest. In addition, FB-04(pta1) displayed L-tryptophan productivity and L-tryptophan yield per glucose that were significantly greater than those of FB-04, FB-04(ΔackA), and FB-04(Δpta) (p<0.01, Student’s t-test) (Table 3).
Fed-batch fermentation of FB-04(pta1)
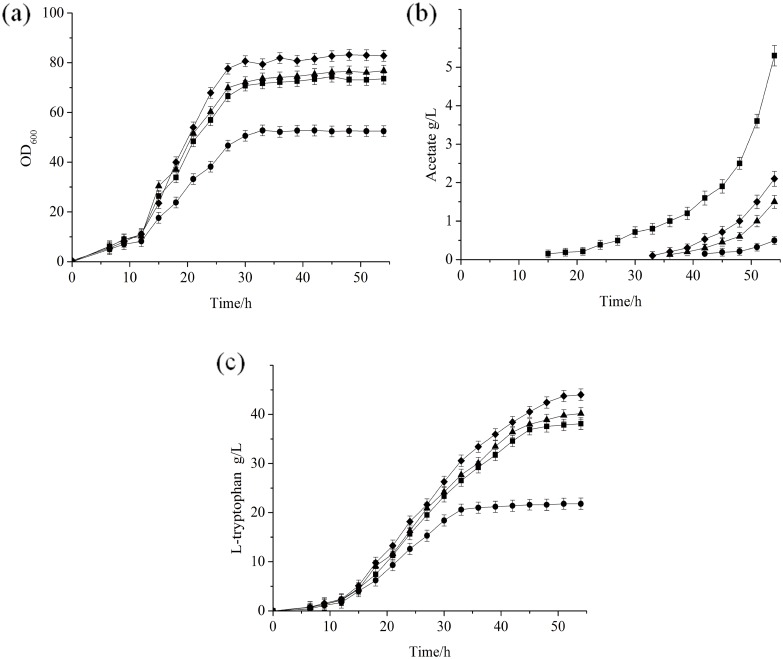
Since FB-04(pta1) produced more L-tryptophan, it was further evaluated in fed-batch fermentation, comparing its performance with those of FB-04, FB-04(ΔackA) and FB-04(Δpta) (Fig 3). The growth of FB-04(Δpta) was impaired, while the growth curves for FB-04, FB-04(ΔackA) and FB-04(pta1) were similar, although FB-04(pta1) achieved the highest total biomass (Fig 3a). Levels of acetate produced by all strains are shown in Fig 3b. The acetate content of FB-04 was undetectable during the first 14 h, and then increased with time, reaching about 2.0 g/L at 27 h and 4.3 g/L at the end of fermentation. The acetate levels of FB-04(Δpta) and FB-04(ΔackA) were extremely low throughout the fermentation processes. In contrast, the acetate content of FB-04(pta1) began to increase noticeably at 32 h and reached a final concentration of 2.1 g/L at the end of fermentation.
Fig 3. Fed-batch fermentation of different strains.
(a) Biomass levels; (b) Acetate levels; (c) L-tryptophan levels. FB-04 (square); FB-04(Δpta) (circle); FB-04(ΔackA) (triangle); FB-04(pta1) (diamond).
L-tryptophan production by all strains increased over the first 30 h of fermentation processes (Fig 3c). The L-tryptophan yield of FB-04 continued to increase during the subsequent 14 h, but remained almost unchanged after the acetate content reached about 2.0 g/L. The L-tryptophan yield of FB-04(Δpta) was lower than those of FB-04, FB-04(ΔackA) and FB-04(pta1) throughout the entire fermentation processes. It reached its maximum at 30 h despite the fact that the acetate concentration was still extremely low. L-tryptophan yields of FB-04(ΔackA) and FB-04(pta1) also increased with time, while FB-04(pta1) achieved a more significant increase in L-tryptophan production. The final L-tryptophan titer of FB-04(pta1) (44 g/L) was significantly greater than those of FB-04, FB-04(ΔackA) and FB-04(Δpta), corresponding to increases of 15%, 9.5% and 100%, respectively (p < 0.01, Student’s t-test) (Table 3). The L-tryptophan yield per glucose of FB-04(pta1) was also significantly greater than those of FB-04, FB-04(ΔackA) and FB-04(Δpta) (p < 0.01, Student’s t-test) (Table 3).
The principal component differences caused by genetic modification
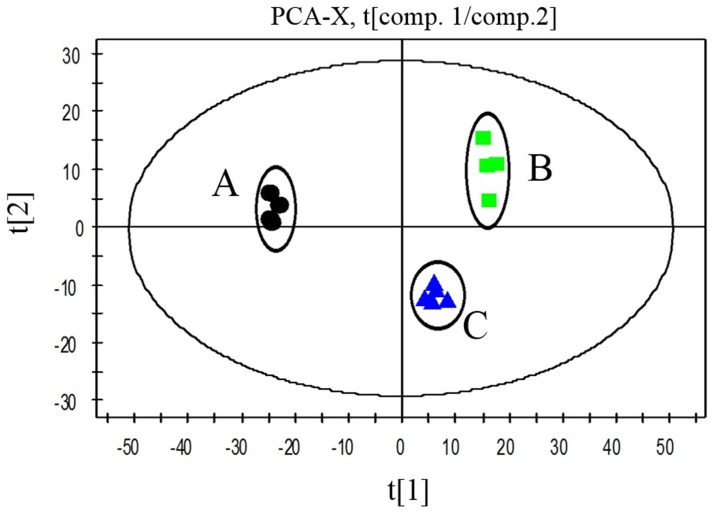
Principal component analysis (PCA) was carried out to gain insight into the multivariate data and evaluate biological alteration. Clustering of biological samples was based on their similarities and differences in the metabolite dataset. In the PCA score plot, each data point reflects a linear combination of the total metabolites from each sample. The distances between the groups give a measure of the overall variation among the metabolic profiles of the different strains. As shown in Fig 4, three groups of FB-04, FB-04(Δpta) and FB-04(pta1) were distributed in different areas of the PCA score plot.
Fig 4. PCA of FB-04, FB-04(Δpta), and FB-04(pta1).
The x axis represents the first principal component, PC1 (t[1]), which explains the major differences; the y axis represents the second principal component, PC2 (t[2]), which explains the minor differences. A, FB-04; B, FB-04(Δpta); C, FB-04(pta1)
Changes of carbon flux in strain carrying a pta deletion or a pta1 genomic substitution
In the present study, more than 80 intracellular metabolites that showed different levels in FB-04, FB-04(Δpta) and FB-04(pta1) were identified using GC-MS. Of these 80 metabolites, 23 of them were involved in L-tryptophan biosynthesis. To identify the potential reasons for the significant diversity in metabolic profiles caused by genetic modification, the relative levels of the 19 intracellular metabolites were determined (see S1 Table).
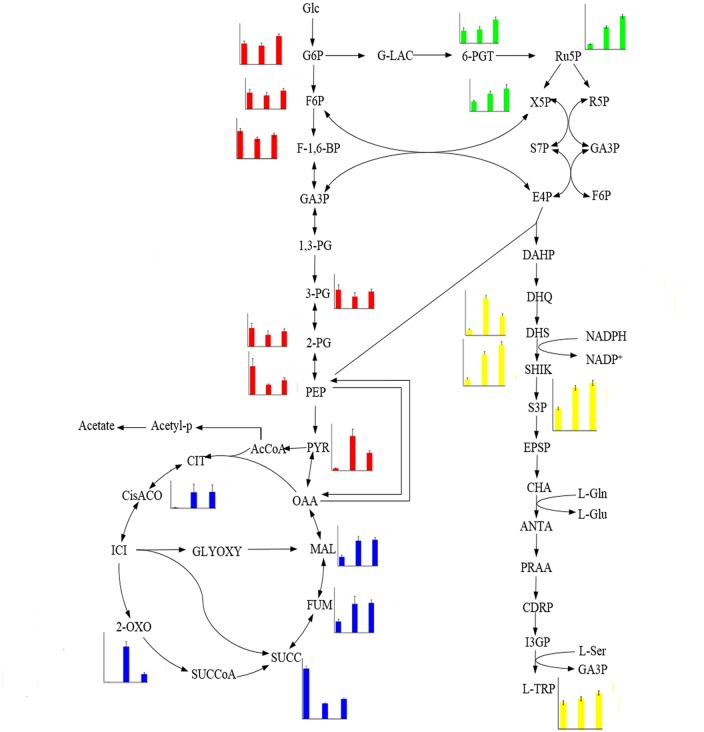
The levels of intracellular intermediates in the three strains FB-04, FB-04(Δpta) and FB-04(pta1) are shown in Fig 5. Interestingly, metabolic intermediate levels in these three strains changed remarkably. Comparing FB-04(Δpta) with the parent strain FB-04, the overall levels of glycolysis intermediates were reduced; however, pyruvate accumulated significantly. Moreover, the overall levels of TCA cycle intermediates and pentose phosphate pathway intermediates were increased. Notably, levels of common aromatic pathway intermediates, such as 3-dehydroshikimate, shikimate and shikimate-3-phosphate, were increased in FB-04(Δpta).
Fig 5. Levels of intermediates involved in L-tryptophan biosynthesis detected in FB-04, FB-04(Δpta), and FB-04(pta1).
The strains along the x-axes are FB-04, FB-04(Δpta), and FB-04(pta1), successively. The y-axes reflect the relative abundance of each intermediate, which was calculated by normalization of the peak area of each metabolite against total peak area within sample. Glc, glucose; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; 1,6-BP, fructose-1,6-bisphosphate; GA3P, glyceraldehyde-3-phosphate; 3-PG, 3-phosphoglycerate; 2-PG, 2-phosphoglycerate; PEP, phosphoenolpyruvate; PYR, pyruvate; AcCoA, acetyl coenzyme A; CIT, citrate; ICI, isocitrate; 2-OXO, 2-oxoglutarate; SUCCoA, succinyl coenzyme A; SUCC, succinate; FUM, fumarate; MAL, malate; OAA, oxaloacetate; G-LAC, 6-phosphoglucono-1,5-lactone; 6-PGT, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; X5P, xylulose-5-phosphate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; E4P, erythrose- 4-phosphate; DAHP, 3-deoxy-d-arabinoheptulosonate-7-phosphate; DHQ, 3-dehydroquinate; Quin, quinone; DHS, 3-dehydroshikimate; SHIK, shikimate; S3P, shikimate-3-phosphate; EPSP, 5-enolpyruvoylshikimate; CHA; chorismate; ANTA, anthranilate; PRAA, phosphoribosyl anthranilate; CDRP, 1-(o-carboxyphenylamino)-1-deoxyribulose-5-phosphate; I3GP, indole 3-glycerolphosphate; L-Trp, L-tryptophan; Acetyl-p, acetyl phosphate; GLYOXY, glyoxylate pathway.
Compared with FB-04, FB-04(pta1) displayed a metabolic performance similar to that of FB-04(Δpta). The overall levels of glycolysis intermediates were slightly decreased, while the overall levels of pentose phosphate pathway intermediates, TCA cycle intermediates, and common aromatic pathway intermediates were significantly increased. Moreover, compared with FB-04(Δpta), the overall levels of intermediates in glycolysis and the pentose phosphate pathway were increased in FB-04(pta1), while levels of TCA cycle intermediates were reduced. In addition, the levels of shikimate and shikimate-3-phosphate, were higher in FB-04(pta1), although that of 3-dehydroshikimate was reduced.
Discussion
L-tryptophan is mainly produced by microbial fermentation using Escherichia coli or Corynebacterium glutamicum. A randomly mutagenized E.coli strain was shown to produce up to 54.6 g/L L-tryptophan when fed L-tryptophan precursors [6]. With the recent advances in molecular technology, several studies have been conducted in an effort to construct L-tryptophan-producing strains with defined genetic modifications [15, 28–30]. For example, genetic modification of a classically derived L-tryptophan-producing Corynebacterium glutamicum strain increased L-tryptophan production to 58 g/L [28]. E.coli strain Dpta/mtr-Y, developed by Wang et al. [15], achieved an L-tryptophan yield of 48.68 g/L.
In this study, mutant strains FB-04(Δpta) and FB-04(ΔackA) were constructed to decrease acetate accumulation. Deletion of pta or ackA led to substantially lowered acetate formation (Table 3). Pta plays a more important role in the Pta-AckA pathway, in view of the fermentation performance of FB-04(Δpta) and FB-04(ΔackA) (Figs 2a and 3b). Reduced acetate levels were conducive to L-tryptophan biosynthesis, as improved L-tryptophan titers were observed in FB-04(Δpta) and FB-04(ΔackA), compared with FB-04 (Fig 2b). Notably, deletion of pta achieved a more significant increase in L-tryptophan production than deletion of ackA in shake-flask fermentations (Fig 2b). However, FB-04(Δpta) exhibited seriously restricted growth, which was consistent with previous findings [14, 17].
To avoid the physiological defects caused by pta deletion, we identified a mutant Pta (Pta1) from E. coli CCTCC M 2016009. Kinetic analysis showed that the Km of Pta1 was 190% higher than that of Pta, and the kcat /Km value of Pta1 was only about 21% that of Pta (Table 2). These data indicate that Pta1 possesses lower catalytic activity and substrate binding affinity. In this study, we constructed FB-04(pta1), in which pta was replaced with pta1. This substitution not only resulted in a noticeably lower ability to secrete acetate, it also reversed the growth defect caused by pta deletion (Fig 3a and 3b). It has been reported that the growth defect caused by pta deletion results from disturbing acetyl-CoA flux, and that any method that relieves the oversupply of acetyl-CoA would compensate for this defect [17]. The pta1 genomic substitution might alleviate the accumulation of acetyl-CoA to a certain degree by weakening the Pta-AckA pathway. In addition to its lowered acetate accumulation and normal growth characteristics, strain FB-04(pta1) showed a substantial improvement in L-tryptophan yield over those of FB-04 and FB-04(Δpta) (Fig 3c).
To investigate the effect of Pta alteration on cells, changes in metabolic flow were explored. Because the deletion of pta reduces the flux through glycolysis and increases the flux through the TCA cycle caused by the accumulation of acetyl-coA and pyruvate [17], FB-04(Δpta) displayed decreased levels of glycolytic intermediates and increased levels of TCA cycle intermediates (Fig 5). The activity of glucose-6 phosphate dehydrogenase, the first enzyme in the pentose phosphate pathway, is known to be upregulated by pta deletion [31]. This contributes to the increased flux through the pentose phosphate pathway, which is consistent with the increased levels of pentose phosphate pathway intermediates seen in this study (Fig 5). The metabolic performance of FB-04(pta1) differed from that of FB-04 in ways similar to the metabolic differences between FB-04 and FB-04(Δpta). FB-04(pta1) displayed decreased levels of glycolytic intermediates and increased levels of TCA cycle and pentose phosphate pathway intermediates (Fig 5). In addition, the levels of pentose phosphate pathway intermediates in FB-04(pta1) were greater than those of FB-04(Δpta)). This difference may be ascribed to the more reasonable distribution of metabolic flow among the central metabolic pathways caused by the pta1 genomic substitution. Increased flux through the pentose phosphate pathway not only supplies additional erythrose 4-phosphate, an important L-tryptophan precursor, it also allows the full utilization of another precursor, phosphoenolpyruvate, which boosts carbon flux through the common aromatic pathway [32]. This is consistent with the increased levels of common aromatic pathway intermediates in FB-04(pta1) (Fig 5). The level of 3-dehydroshikimate, an intermediate in the common aromatic pathway, was lower in FB-04(pta1) than in FB-04(Δpta) (Fig 5). The improved metabolic flow in the pentose phosphate pathway caused by the pta1 genomic substitution could increase the formation of NADPH, an essential cofactor in the common aromatic pathway [32]. This would drive carbon flux from 3-dehydroshikimate to shikimate, decreasing the level of 3-dehydroshikimate.
In view of the length of the L-tryptophan biosynthetic pathway and its complicated regulation mechanism, relying only on decreased acetate production and changes in metabolism caused by the pta1 genomic substitution would not achieve a remarkable improvement in L-tryptophan production. The metabolic alterations caused by the pta1 genomic substitution also increased levels of L-tryptophan precursors, which further contributed to L-tryptophan biosynthesis. In future studies, overexpression of the genes ppsA and tktA, which are involved in the biosynthesis of phosphoenolpyruvate and erythrose 4-phosphate, respectively [32], may significantly improve L-tryptophan production.
Conclusion
A pta gene knockout was constructed in E. coli FB-04 using λRed recombination to reduce the formation of acetate, but the resulting strain (E. coli FB-04(Δpta)) exhibited a growth defect. Then, a mutant (Pta1) that exhibits lower catalytic capacity and substrate affinity than Pta because of a single substitution (Pro69Leu) was identified in E. coli CCTCC M 2016009. This variant (pta1), was used to replace the pta gene of E. coli FB-04, forming strain FB-04(pta1). FB-04(pta1) not only lacked the growth defect of FB-04(Δpta) and showed improved fermentation performance, it also displayed a 91% increase in L-tryptophan yield during flask fermentation, compared with FB-04, while acetate production decreased by 35%. Moreover, acetate secretion by FB-04(pta1) was slower than that by FB-04 throughout the fed-batch fermentation processes, and finally the L-tryptophan yield of FB-04(pta1) represented a 15% increase over that of FB-04. Metabolomics analysis showed that the pta1 genomic substitution slightly decreased carbon flux through glycolysis and significantly increased carbon flux through the pentose phosphate and common aromatic pathways, contributing to the biosynthesis of L-tryptophan.
Supporting Information
To ensure the accuracy of the data, five biomass samples of each strain were subjected to GC-MS. Most of the samples were within 95% confidence interval, except samples A1 and B1 (date not show). A1 and B1 were judged to be abnormal samples and were omitted from the analysis to ensure the reliability of the results. The rt_mz values express the mass-to-charge ratio of chromatographic retention time.
(DOC)
Acknowledgments
This work was supported by grants from the National Science Fund for Distinguished Young Scholars (31425020), the National Natural Science Foundation of China (31271813 and 31401636), the project of outstanding scientific and technological innovation group of Jiangsu Province (Jing Wu), the Natural Science Foundation of Jiangsu Province (BK20140142), the 111 Project (No. 111-2-06), and the Research and Innovation Project for College Graduates of Jiangsu Province (no. KYLX15-1143). We would also like to thank Shanghai ProfLeader Biotech Co, Ltd for assistance with the GC-MS experiments and data analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Science Fund for Distinguished Young Scholars (31425020) to JW, The National Natural Science Foundation of China (31271813 and 31401636) to JW, The project of outstanding scientific and technological innovation group of Jiangsu Province (Jing Wu) to JW, The Natural Science Foundation of Jiangsu Province (BK20140142) to JW, The 111 Project (No. 111-2-06) to JW, The Research and Innovation Project for College Graduates of Jiangsu Province (no. KYLX15-1143) to LL. The authors would also like to thank Shanghai ProfLeader Biotech Co, Ltd for assistance with the GC-MS experiments and data analysis. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ikeda M: Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Applied Microbiology and Biotechnology 2006, 69:615–626. [DOI] [PubMed] [Google Scholar]
- 2.Leuchtenberger W, Huthmacher K, Drauz K: Biotechnological production of amino acids and derivatives: current status and prospects. Applied Microbiology and Biotechnology 2005, 69:1–8. [DOI] [PubMed] [Google Scholar]
- 3.Bongaerts J, Krämer M., Müller U, Raeven L, Wubbolts M: Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metabolic Engineering 2001, 3:289–300. [DOI] [PubMed] [Google Scholar]
- 4.Flores N, Xiao J, Berry A, Bolivar F, Valle F: Pathway engineering for the production of aromatic compounds in Escherichia coli. Nature Biotechnology 1996, 14:620–623. [DOI] [PubMed] [Google Scholar]
- 5.Zhao ZJ, Zou C, Zhu YX, Dai J, Chen S, Wu D, et al. : Development of L-tryptophan production strains by defined genetic modification in Escherichia coli. Journal of Industrial Microbiology and Biotechnology 2011, 38:1921–1929. 10.1007/s10295-011-0978-8 [DOI] [PubMed] [Google Scholar]
- 6.Azuma S, Tsunekawa H, Okabe M, Okamoto R, Aiba S: Hyper-production of L-trytophan via fermentation with crystallization. Applied Microbiology and Biotechnology 1993, 39:471–476. [Google Scholar]
- 7.Chan EC, Tsai HL, Chen SL, Mou Due-Gang: Amplification of the Tryptophan Operon Gene in Escherichia. coli chromosome to Increase L-Tryptophan Biosynthesis. Applied Microbiology and Biotechnology 1993, 40:301–305. [Google Scholar]
- 8.Tribe DE, Pittard J: Hyperproduction of tryptophan by Escherichia coli: genetic manipulation of the pathways leading to tryptophan formation. Applied and Environmental Microbiology 1979, 38:181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han K, Lim HC, Hong J: Acetic Acid Formation in Escherichia coli fermentation. Biotechnology and Bioengineering 1992, 39:663–671. [DOI] [PubMed] [Google Scholar]
- 10.Suarez DC, Kilikian BV: Acetic acid accumulation in aerobic growth of recombinant Escherichia coli. Process Biochemistry 2000, 35:1051–1055. [Google Scholar]
- 11.Phue JN, Lee SJ, Kaufman JB, Negrete A, Shiloach J: Acetate accumulation through alternative metabolic pathways in ackA(-) pta(-) poxB(-) triple mutant in E. coli B (BL21). Biotechnology Letters 2010, 32:1897–1903. 10.1007/s10529-010-0369-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahm DH, Pan J, Rhee JS: Characterization and evaluation of a Pta (Phosphotransacetylase) negative mutant of Escherichia coli Hb101 as production host of foreign lipase. Applied Microbiology and Biotechnology 1994, 42:100–107. [DOI] [PubMed] [Google Scholar]
- 13.Kakuda H, Hosono K, Shiroishi K, Ichihara S: Identification and characterization of the ackA (Acetate Kinase a)-pta (Phosphotransacetylase) Operon and complementation Analysis of Acetate Utilization by an ackA-pta Deletion Mutant of Escherichia coli. Journal of Biochemistry 1994, 116:916–922. [DOI] [PubMed] [Google Scholar]
- 14.Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ: The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. Journal of Bacteriology 2007, 189:5574–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Cheng LK, Wang J, Liu Q, Shen T, Chen N: Genetic engineering of Escherichia coli to enhance production of L-tryptophan. Applied Microbiology and Biotechnology 2013, 97:587–7596. [DOI] [PubMed] [Google Scholar]
- 16.Bauer KA., Ben-Bassat A, Dawson M, de la Puente VT, Neway JO: Improved expression of human interleukin-2 in high-cell-density fermentor cultures of Escherichia coli K-12 by a phosphotransacetylase mutant. Applied and Environmental Microbiology 1990, 56:1296–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang DE, Shin S, Rhee JS, Pan JG: Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. Journal of Bacteriology 1999, 181: 6656–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mccleary WR, Stock JB: Acetyl Phosphate and the Activation of 2-Component Response Regulators. Journal of Biological Chemistry 1994, 269:31567–31572. [PubMed] [Google Scholar]
- 19.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR:Gas chromatography mass spectrometry—based metabolite profiling in plants. Nature Protocols 2006, 1:387–396. [DOI] [PubMed] [Google Scholar]
- 20.Villas-Bôas SG, Bruheim P: Cold glycerol—saline: The promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Analytical Biochemistry 2007, 370:87–97. [DOI] [PubMed] [Google Scholar]
- 21.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology 2006, 2:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madyagol M, Al-Alami H, Levarski Z, Drahovská H, Turňa J, Stuchlík S: Gene replacement techniques for Escherichia coli genome modification. Folia Microbiologica 2011, 56:253–263. 10.1007/s12223-011-0035-z [DOI] [PubMed] [Google Scholar]
- 23.Ramalingam S, Gautam P, Mukherjee KJ, Jayaraman G: Effects of post-induction feed strategies on secretory production of recombinant streptokinase in Escherichia coli. Biochemical Engineering Journal 2007, 33:34–41. [Google Scholar]
- 24.Shimizu M, Suzuki T, Kameda KY, Abiko Y: Phosphotransacetylase of Escherichia coli B, purification and properties. Biochimica et Biophysica Acta 1969, 191:550–558. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 1976, 72:248–254. [DOI] [PubMed] [Google Scholar]
- 26.Mayer MAG, Bronnenmeier K, Schwarz WH, Schertler C, Staudenbauer WL: Isolation and properties of acetate kinase-and phosphotransacetylase-negative mutants of Thermoanaerobacter thermohydrosulfuricus. Microbiology 1995, 141:2891–2896. [Google Scholar]
- 27.Gao X, Pujos-Guillot E, Seébeédio JL: Development of a quantitative metabolomic approach to study clinical human fecal water metabolome based on trimethylsilylation derivatization and GC/MS analysis. Analytical Biochemistry 2010, 82:6447–6456. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda M, Katsumata R: Hyperproduction of Tryptophan by Corynebacterium glutamicum with the Modified Pentose Phosphate Pathway. Applied and Environmental Microbiology 1999, 65:2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu P, Yang F, Kang J, Wang Q, Qi Q: One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microbial Cell Factories 2012, 2;11:30 10.1186/1475-2859-11-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu P, Kang J, Yang F, Wang Q, Liang Q, Qi Q: The improved L-tryptophan production in recombinant Escherichia coli by expressing the polyhydroxybutyrate synthesis pathway. Applied Microbiology and Biotechnology 2013, 97:4121–4127. 10.1007/s00253-012-4665-0 [DOI] [PubMed] [Google Scholar]
- 31.Castano-Cerezo S, Pastor JM, Renilla S, Bernal V, Iborra JL, Canovas M: An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli. Microbial Cell Factories 2009, 8 (54). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid JW, Mauch K, Reuss M, Gilles ED, Kremling A: Metabolic design based on a coupled gene expression-metabolic network model of tryptophan production in Escherichia coli. Metabolic Engineering 2004, 6:364–377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To ensure the accuracy of the data, five biomass samples of each strain were subjected to GC-MS. Most of the samples were within 95% confidence interval, except samples A1 and B1 (date not show). A1 and B1 were judged to be abnormal samples and were omitted from the analysis to ensure the reliability of the results. The rt_mz values express the mass-to-charge ratio of chromatographic retention time.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.