Abstract
Glioblastomas co-opt stem cell regulatory pathways to maintain brain tumor–initiating cells (BTICs), also known as cancer stem cells. NOTCH signaling has been a molecular target in BTICs, but NOTCH antagonists have demonstrated limited efficacy in clinical trials. Recombining binding protein suppressor of hairless (RBPJ) is considered a central transcriptional mediator of NOTCH activity. Here, we report that pharmacologic NOTCH inhibitors were less effective than targeting RBPJ in suppressing tumor growth. While NOTCH inhibitors decreased canonical NOTCH gene expression, RBPJ regulated a distinct profile of genes critical to BTIC stemness and cell cycle progression. RBPJ was preferentially expressed by BTICs and required for BTIC self-renewal and tumor growth. MYC, a key BTIC regulator, bound the RBPJ promoter and treatment with a bromodomain and extraterminal domain (BET) family bromodomain inhibitor decreased MYC and RBPJ expression. Proteomic studies demonstrated that RBPJ binds CDK9, a component of positive transcription elongation factor b (P-TEFb), to target gene promoters, enhancing transcriptional elongation. Collectively, RBPJ links MYC and transcriptional control through CDK9, providing potential nodes of fragility for therapeutic intervention, potentially distinct from NOTCH.
Introduction
Glioblastoma (World Health Organization grade IV glioma) represents the most prevalent and malignant primary intrinsic brain tumor. Current therapy for glioblastoma includes maximal surgical resection followed by chemoradiation, and then adjuvant chemotherapy with the oral methylator, temozolomide (1). Despite aggressive treatment, glioblastoma remains universally fatal. The myriad drivers of glioblastoma malignancy include co-option of stem cell pathways through a combination of hardwired genetic lesions and epigenetic dysregulation. Resident stem cells contribute to normal organ development and wound responses. Genetic lesions in glioblastomas target developmental regulation, and cancer is “the wound that does not heal.” Concordantly, the regenerative potential mediated by stem cell transcriptional programs has proven critical in tumor initiation, therapeutic resistance, and regrowth after cancer therapy (2–4).
Based on this background, novel targeted therapeutics against core regulatory pathways in embryonic and tissue-specific stem cells have been developed as cancer therapeutics. A therapeutic index of these agents may be achievable if dependence on these pathways is relatively less critical in normal tissues, due to completion of development. In the adult brain, neurogenesis is far less active than in early development, and neural stem cells (NSCs) are quiescent (5), both of which are important distinctions from brain tumors. In most organs, the NOTCH pathway serves multiple roles in cell fate and lineage commitment, as it mediates interactions between heterologous cell types (6). Upon engagement of NOTCH receptors to their ligands, the extracellular NOTCH region is cleaved by a disintegrin and metalloproteinase (ADAM) family of metalloproteases followed by the rate-limiting step of liberation of the NOTCH intracellular domain (NICD) by the γ-secretase complex. The NICD translocates to the nucleus and binds to transcriptional regulators to direct gene regulation, including target genes that, in turn, have strong transcriptional regulatory function (e.g., the HES and HEY transcription factors). NOTCH signaling is complex due to the differential expression of 4 receptors and 5 ligands, but NOTCH converges on a limited repertoire of transcriptional regulators, prominently RBPJ (recombining binding protein suppressor of hairless or recombination signal-binding protein for immunoglobulin kappa j region; also known as CBF1 [mammalian C promoter–binding factor 1]) or CSL (for CBF1, suppressor of hairless [fly], Lag2 [worm]). In the absence of NOTCH activation, RBPJ recruits histone deacetylases and corepressor components. Nuclear localization of NICD leads to binding to and displacement of an RBPJ transcriptional corepressor complex to act as a transcriptional activator by the recruitment of histone acetylases to activate transcription of NOTCH target genes (7).
NOTCH signaling is highly context dependent with strong differences in the biologic outcome of NOTCH activation or disruption based on developmental stage, tissue type, and cell type (6). As brain tumors, including gliomas, have stem-like features, the role of NOTCH signaling has been the focus of many studies in neuro-oncology. Gliomas express NOTCH ligands (8). γ-Secretase inhibitors (GSIs) that block the liberation of NICD have efficacy in preclinical studies (9, 10), but GSIs have greater effect in disrupting the resistance of brain tumor–initiating cells (BTICs) to ionizing radiation (11) and chemotherapy (12). Translation of NOTCH targeting into clinical trials for glioma treatment has shown transient efficacy (13), suggesting that the current treatment paradigms against NOTCH will require modification. Here, we examined the role of RBPJ given the proposed convergence of NOTCH signaling on RBPJ transcriptional control.
Results
Targeting NOTCH does not inhibit BTIC proliferation.
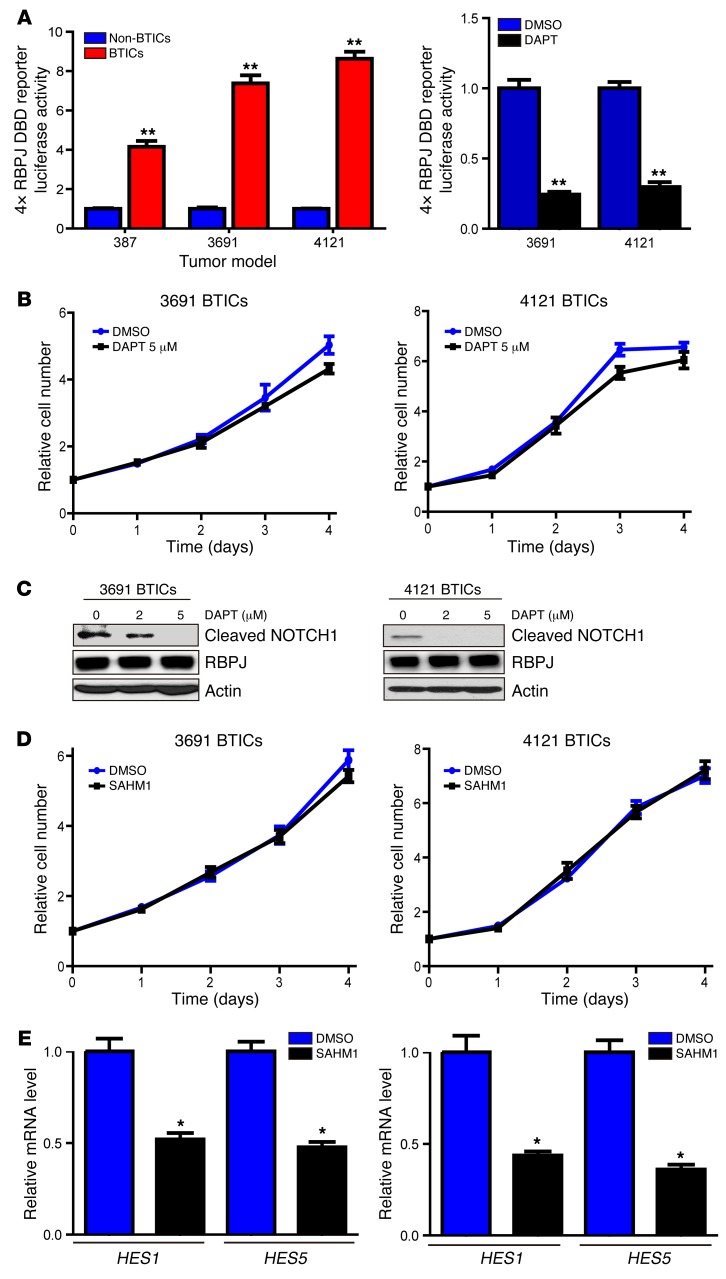
While phase I clinical trials showed transient responses in treating gliomas with NOTCH antagonists (13), other clinical trials have shown more limited activity. As NOTCH is hypothesized to function primarily in BTICs, we first examined a surrogate of NOTCH activity through use of a 4× RBPJ-binding reporter — a common method of measuring NOTCH activity (Figure 1A) — in matched BTICs and non-BTICs derived from human glioblastoma patients, which were functionally validated as previously described (data not shown). In each model tested, BTICs displayed strong preferential activation (4- to 9-fold greater) of RBPJ (Figure 1A). Based on activation of a pathway downstream of NOTCH, we tested the dependence of BTICs on NOTCH signaling by treating BTICs with the GSI, DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester), a widely employed inhibitor of NOTCH signaling. We previously found that clinically achievable concentrations of DAPT and other GSIs did not inhibit BTIC proliferation as monotherapy, but synergized with radiation (11). Concordant with these results, NOTCH inhibition by DAPT had a very weak effect on BTICs (Figure 1B). The inhibitory effect of DAPT on NOTCH signaling was confirmed by immunoblot of cleaved (activated) NOTCH, with NICD levels abolished at 5 μM (Figure 1C; see complete unedited blots in the supplemental material; supplemental material available online with this article; doi:10.1172/JCI86114DS1). To validate these results, we used another NOTCH antagonist from another drug class. SAHM1 is a synthetic, cell-permeant, stabilized α-helical peptide that targets a critical protein–protein interface in the NOTCH-RBPJ-MAML1 transcription activator complex (14). Similar to the effects of DAPT, inhibiting NOTCH signaling by SAHM1 did not reduce proliferation of BTICs (Figure 1D). Successful targeting of NOTCH signaling by SAHM1 was confirmed by real-time PCR (qPCR) of canonical downstream targets, HES1 and HES5 (Figure 1E).
Figure 1. NOTCH inhibition does not attenuate cell autonomous BTIC growth.
(A) Left: Matched sets of BTICs and non-BTICs (387, 3691, and 4121) were transfected with a 4× RBPJ luciferase reporter together with a tk-renilla reporter. Data are displayed as mean ± SEM for the ratio of firefly-to-renilla luciferase (t test, **P < 0.01, n = 3). Right: 3691 and 4121 BTICs were transfected with a 4× RBPJ luciferase reporter together with a tk-renilla reporter, and then treated with the NOTCH antagonist, DAPT (5 μM), or vehicle control (DMSO). Data are displayed as mean ± SEM for the ratio of firefly-to-renilla luciferase (Student’s t test, **P < 0.01, n = 3). (B) Effects of DAPT or vehicle control (DMSO) on cell proliferation were tested in two BTIC models (3691 and 4121). Data are displayed as the mean values for each time point. (C) Cleaved NOTCH1 (NOTCH intracellular domain [NICD]) levels were analyzed by immunoblot after treatment with vehicle control or DAPT in two BTIC models (3691 and 4121). (D) Effects of the stapled peptide NOTCH inhibitor, SAHM1, or vehicle control (DMSO) on cell proliferation were tested in two BTIC models (3691 and 4121). Data are displayed as the mean values for each time point. (E) Effects of SAHM1 treatment on downstream NOTCH target gene expression (HES1 and HES5) were tested in two BTIC models (3691 and 4121). Cells were treated with SAHM1 for two days. Total RNA was isolated and cDNA was synthesized by reverse transcription. The mRNA levels of indicated genes were detected by qPCR (t test, *P < 0.05, n = 3).
BTICs preferentially express RBPJ without differential NOTCH activation.
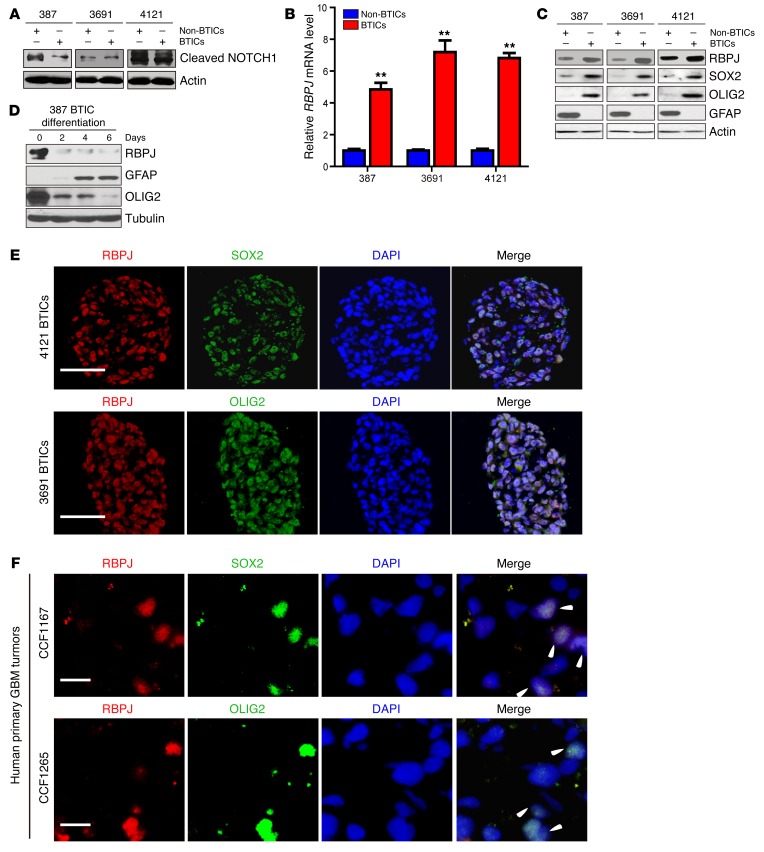
Based on the weak antitumor effects of NOTCH inhibitors, we sought potential explanations for this therapeutic failure. Surprisingly, multiple patient–derived matched BTICs and differentiated progeny (non-BTICs) revealed no significant differences in NOTCH activation, as measured by NICD levels (Figure 2A). While NOTCH signaling can alter transcription through RBPJ regulation, recent studies have demonstrated NOTCH-independent activities of RBPJ (15). To determine if RBPJ might be differentially expressed in BTICs, which would explain the preferential RBPJ transcriptional activation, we examined RBPJ levels in matched, functionally validated BTICs and non-BTICs from patient-derived xenografts by qPCR and immunoblotting. In each matched set of BTICs and non-BTICs that we tested, BTICs displayed strikingly elevated RBPJ mRNA and protein levels relative to matched non-BTICs (Figure 2, B and C). To determine the relationship between cellular differentiation and RBPJ regulation, we induced differentiation in BTICs and found a marked decrease of RBPJ protein levels after inducing BTIC differentiation (Figure 2D), strongly indicating dynamic regulation of RBPJ associated with BTIC self-renewal and differentiation. We next confirmed coexpression of RBPJ with BTIC markers, SOX2 and OLIG2, by immunofluorescent staining of cultured BTIC tumorspheres and human primary glioblastoma tissue sections (Figure 2, E and F).
Figure 2. RBPJ is highly expressed in BTICs.
(A) Cleaved NOTCH1 levels were assayed by immunoblot in matched BTICs and non-BTICs isolated from patient-derived xenografts (387, 3691, and 4121). (B) RBPJ mRNA levels in BTICs and non-BTICs were detected by qPCR. Data are displayed as mean ± SEM (t test, **P < 0.01, n = 3). (C) RBPJ protein levels were assayed in matched BTICs and non-BTICs isolated from patient-derived xenografts (387, 3691, and 4121). (D) RBPJ protein levels were assayed by immunoblotting during a time course of BTIC differentiation induced by 10% serum. (E) Coexpression of RBPJ and BTIC markers, SOX2 and OLIG2, was assayed by immunofluorescence in tumorspheres isolated from patient-derived xenografts (3691 and 4121). Nuclei were visualized by DAPI staining. Scale bars: 50 μm. (F) Coexpression of RBPJ and BTIC markers, SOX2 and OLIG2, was assayed by immunofluorescence in two primary human glioblastoma (GBM) specimens (CCF1167 and CCF1265). Nuclei were visualized by DAPI staining. Scale bars: 10 μm.
To interrogate the preferential expression of RBPJ in BTICs in the absence of culture, we performed an in silico analysis of RBPJ expression in glioma patients in The Cancer Genome Atlas (TCGA). RBPJ mRNA expression correlated with BTIC markers — SOX2, SOX4, OLIG2, and CD133 — in the TCGA glioblastoma data set (Supplemental Figure 1, A–D). Glioblastomas have been classified into different subtypes based on their transcriptional profiles (16). RBPJ was expressed in all glioblastoma subtypes, but most highly in the Proneural group, especially in the glioma-CpG island methylator phenotype (G-CIMP) group (Supplemental Figure 1E). Collectively, these results suggest that BTICs express elevated levels of RBPJ compared with differentiated progeny, even in the absence of preferential NOTCH activation.
RBPJ is necessary for BTIC maintenance.
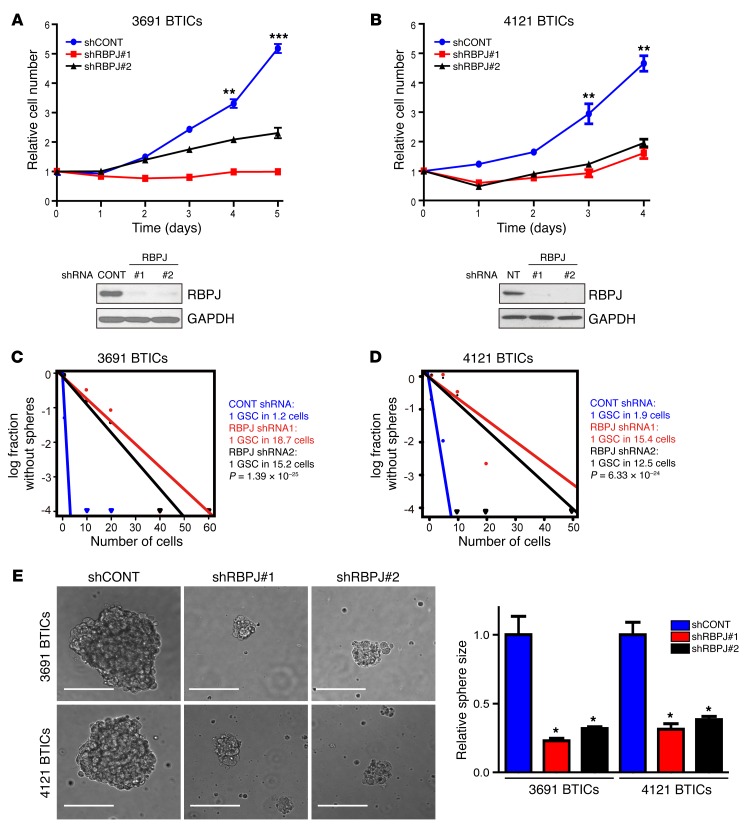
The concordant expression of RBPJ and tumor cell differentiation suggests a potential functional role of RBPJ in BTIC biology, prompting us to interrogate the functional requirement for RBPJ in BTIC maintenance. We developed 2 independent, nonoverlapping shRNA lentiviral constructs to knock down RBPJ and compared their effects to a nontargeting control sequence shRNA insert (shCONT), which does not target any known genes from any species. RBPJ shRNAs significantly reduced RBPJ protein expression levels on immunoblot (Figure 3, A and B). We then examined the phenotypic consequences of shRNA-mediated reduction of RBPJ expression. Silencing RBPJ strikingly decreased the growth of BTICs (Figure 3, A and B) and induced apoptosis (Supplemental Figure 2, A and B) without changes in the proliferation of non-BTICs (Supplemental Figure 3), supporting a specific requirement of RBPJ for BTIC growth. To test whether targeting RBPJ influences tumorsphere formation, a surrogate marker of self-renewal, we performed in vitro limiting dilution assays. RBPJ knockdown resulted in a more than 6-fold decrease in the frequency of sphere formation and a greater than 2-fold decrease in the sphere size (Figure 3, C–E). The most important property of BTICs is their potent ability to form tumors in vivo. To address the requirement for RBPJ in maintaining the tumorigenic potential of BTICs, BTICs transduced with either of 2 nonoverlapping RBPJ-targeting shRNAs or a control, nontargeting shRNA were transplanted into the brains of immunocompromised mice. Consistent with the in vitro results, RBPJ knockdown strikingly impaired tumor growth, as shown by quantitative bioluminescent imaging (Figure 4A), and significantly extended survival of tumor-bearing hosts (Figure 4B). Taken together, our findings demonstrate that RBPJ is necessary for BTIC phenotypes including proliferation, self-renewal, and tumor formation.
Figure 3. Targeting RBPJ decreases BTIC growth and self-renewal.
(A and B) Effects of shRNA against RBPJ were tested in two BTIC models (3691 and 4121). Top: BTICs were transduced with a control, nontargeting shRNA sequence (shCONT) or one of two nonoverlapping shRBPJ sequences. Proliferation was measured by CellTiter-Glo (2-way ANOVA: **P < 0.01, ***P < 0.001, n = 4). Bottom: RBPJ protein levels were assayed by immunoblot following transduction with shCONT or shRBPJ. (C and D) Tumorsphere formation efficiency was measured by extreme in vitro limiting dilution assays in two BTIC models (3691 and 4121) after transduction with shCONT or shRBPJ. (E) Left: Representative images of tumorspheres derived from two BTIC lines transduced with shCONT or shRBPJ are shown. Scale bars: 100 μm. Right: Quantification of tumorsphere size is displayed as mean ± SEM (2-way ANOVA, *P < 0.05, n = 3).
Figure 4. Targeting RBPJ decreases BTIC tumor formation.
(A) Tumor size of orthotopic glioblastoma xenografts derived from luciferase-expressing BTICs transduced with shCONT or shRBPJ was tracked by bioluminescence over a time course. (B) Survival of immunocompromised mice bearing intracranial 3691 or 4121 BTICs transduced with shCONT or shRBPJ is displayed by the Kaplan-Meier method (log-rank analysis, **P < 0.01, n = 5).
NOTCH and RBPJ regulate different transcriptional programs.
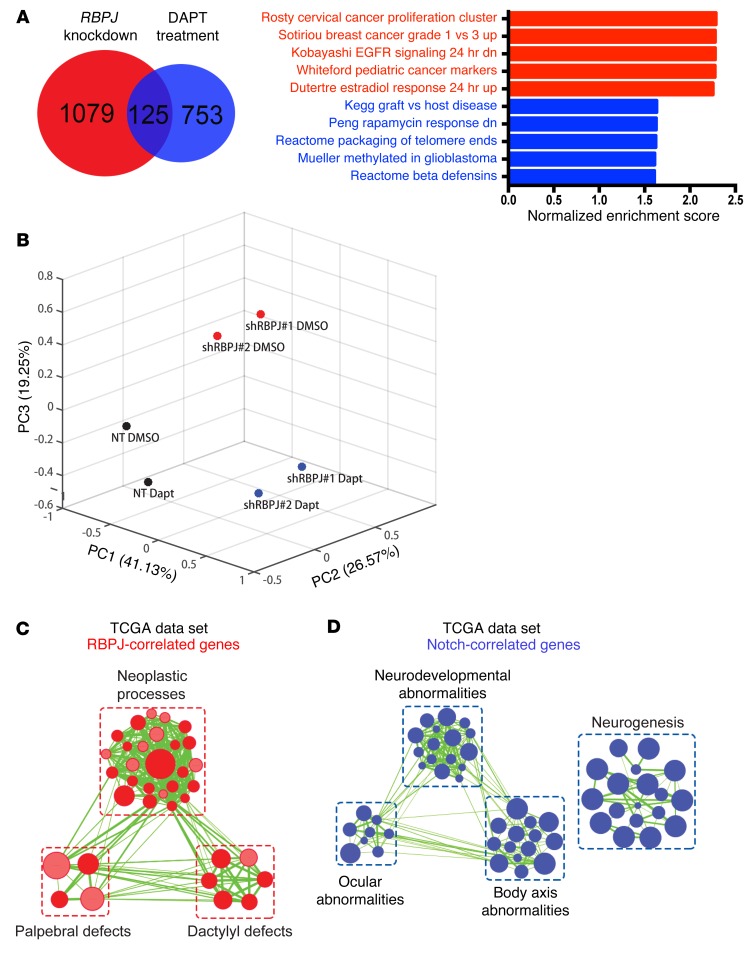
The discordant results of targeting RBPJ and pharmacologic inhibition of NOTCH prompted us to interrogate the global effects of these approaches on the transcriptional landscape by RNA sequencing (RNA-seq). We compared 3691 BTICs treated with NOTCH antagonists to vehicle control (DMSO vs. DAPT at 5 or 10 μM) and RBPJ RNA interference (shRBPJ) relative to a control, nontargeting shRNA (shCONT vs. 2 independent shRBPJ sequences). As expected, GSI treatment inhibited the expression of downstream effectors of NOTCH signaling (Supplemental Figure 4A). However, GSI treatment had no effect or even a paradoxical effect on the expression of key BTIC effectors or cell cycle regulators that have been found to be important in 3691 BTIC growth in previous reports (Supplemental Figure 4A). In contrast, targeting RBPJ had limited effects on the expression of canonical NOTCH downstream targets (HES1, HES5, et al.), but significantly reduced the expression of a series of key genes regulating BTIC stemness and cell cycle progression. Compared with respective controls, RBPJ knockdown in the 3691 BTIC model by itself resulted in at least a 1.5-fold reduction in 1,079 genes that were mutually exclusive from 753 genes decreased by DAPT treatment alone (Figure 5A). Only 125 genes were commonly downregulated by either treatment. Gene set enrichment analysis (GSEA) of RBPJ–knocked down 3691 BTICs versus nontargeting control or DAPT treatment alone versus vehicle control indicated reduced protumorigenic signatures upon RBPJ knockdown, while DAPT treatment alone in BTICs enriched for stress response signatures. We independently confirmed these results by testing the expression of individual genes using qPCR (Supplemental Figure 4, B and C). In each case, NOTCH antagonists and RBPJ ablation controlled the transcription of different gene sets.
Figure 5. RBPJ induces transcriptional profiles in BTICs distinct from NOTCH activation.
(A) GSEA results from ranked genes in 3691 BTICs in which RBPJ was knocked down or treated with DAPT alone. Genes that exhibited at least a 1.5-fold decrease upon RBPJ knockdown or DAPT treatment compared to respective controls (nontargeting vs. DMSO). (B) The first three principal components and their loadings for 4121 BTICs based on RNA sequencing after transduction with either shCONT or shRBPJ and treatment with either vehicle control (DMSO) or DAPT treatment (5 μM). (C and D) Gene signature enrichment was analyzed using gProfiler (34) with genes whose RNA expression was most significantly correlated with (P < 0.001, r > 0.3) and mutually exclusive for (C) RBPJ or (D) NOTCH1 in the TCGA data set. Enriched gene sets for either gene were visualized via Enrichment Map on Cytoscape (35) for signatures with FDR < 0.001 and P < 0.005.
To further confirm our findings, we performed RNA sequencing in a second BTIC model (4121) with RBPJ knockdown in the presence or absence of DAPT. PCA analysis of this second BTIC model revealed that DAPT treatment in addition to RBPJ knockdown accounted for 19.25% of variance in gene expression (Figure 5B). The 4121 BTICs in which RBPJ was knocked down alone (cluster 1) or the same BTICs with both RBPJ knockdown and DAPT treatment (cluster 2) segregated along the principal component 3. Genes most significantly correlated with the BTICs in cluster 1 suggested enriched programs in vesicle trafficking and semaphorin signaling (Supplemental Figure 5), which we have recently shown regulates BTICs (17). In the BTIC cluster 2, the significantly correlated genes suggested increased activity in translation, ribosomal biogenesis, and intrinsic apoptotic pathway. As additional confirmation to rule out potential effects of cell culture, we interrogated transcriptional profiling of glioblastomas in the TCGA data set. RBPJ and NOTCH expression (measured by NOTCH1 expression, as NICD is not assayable by RNA) shared some gene expression targets, but the majority of genes were distinct (Figure 5, C and D). RBPJ transcriptional programs included neoplastic and developmental programs, whereas NOTCH expression correlated with distinct programs of neurogenesis and development of the eye and brain.
Key promoters regulated by RBPJ in our RNA-seq studies include FOXM1, cyclin A2 (CCNA2), and KRAS (data not shown). Using RBPJ ChIP followed by qPCR (ChIP-qPCR), we confirmed that RBPJ preferentially bound to the FOXM1 promoter in BTICs compared to non-BTICs (Supplemental Figure 6A). To find additional links between RBPJ and a stem-like state in glioblastoma, we interrogated RBPJ binding to stem cell genes described as potential reprogramming factors in acquisition of the BTIC state (18). We found that stemness genes expressed by 3691 BTICs were significantly more likely to be bound by RBPJ than putative stemness genes that were not expressed (Supplemental Figure 6B). We confirmed that RBPJ bound one of these key factors, OLIG2, in BTICs by ChIP-qPCR (Supplemental Figure 6C). Stunnenberg and co-workers demonstrated that RBPJ binding at enhancers could be at either NOTCH-dependent or NOTCH-independent constant sites (19). To confirm the NOTCH independence of key binding sites, we confirmed that RBPJ binding to the FOXM1 and OLIG2 promoters in BTICs was not dependent on NOTCH activity, using DAPT treatment, followed by ChIP-qPCR (Supplemental Figure 6D). Collectively, our data demonstrate a potentially novel RBPJ function in BTICs, which may be independent of the canonical NOTCH pathway.
RBPJ binds to CDK9 to regulate transcription.
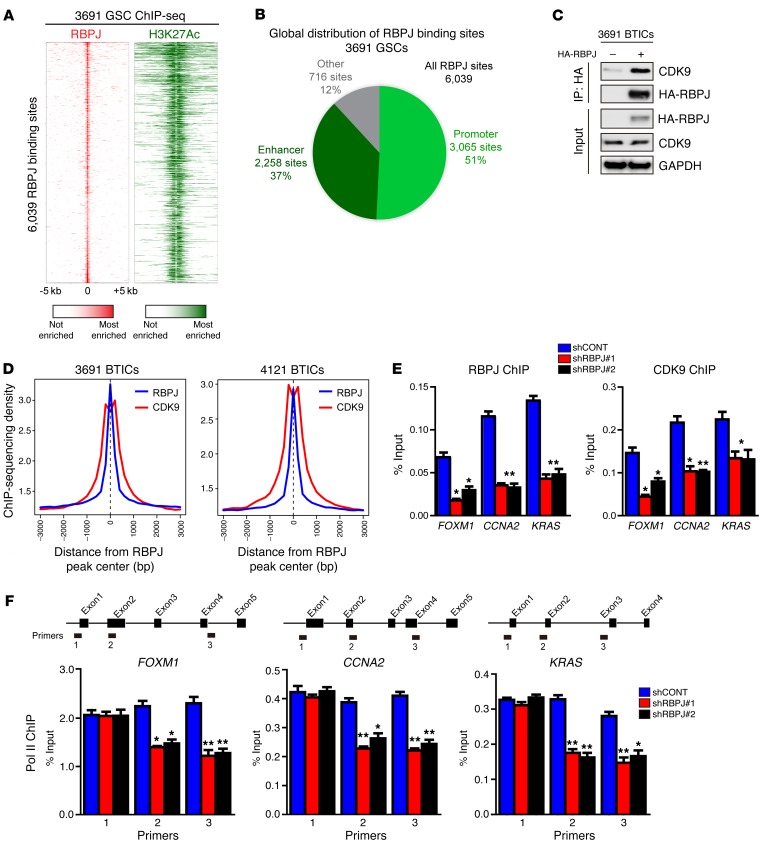
To determine the global landscape of RBPJ binding at regulatory elements, including active enhancers, we performed ChIP followed by deep sequencing (ChIP-seq) for histone 3 lysine 27 acetylation (H3K27Ac) in 3691 BTICs to directly call active enhancers and promoters, and then overlapped this information with RBPJ binding from RBPJ ChIP-seq in 3691 BTICs (Figure 6A). We found striking support for a role for RBPJ as a transcriptional enhancer. Analysis of the global landscape of RBPJ revealed that 88% of RBPJ binding sites fell within an active promoter or enhancer (Figure 6B). This is quite remarkable, as active promoter and enhancer regions only constitute 2% of the genome, demonstrating an extreme enrichment at these sites and providing evidence that RBPJ binding is specific to this site. To identify potential novel downstream effectors of RBPJ, we performed proteomic analysis after transducing 3691 BTICs with HA-tagged RBPJ and then immunoprecipitated RBPJ through HA binding. One hundred thirty-eight proteins were identified exclusively in the RBPJ sample compared to the IgG control, including the RBPJ bait. Prominently, 10 peptides from cyclin-dependent kinase 9 (CDK9) were identified (an example is displayed in Supplemental Figure 7A). We validated direct RBPJ binding to CDK9 by immunoprecipitation (Figure 6C). CDK9 is a core component of positive transcription elongation factor b (P-TEFb), which regulates RNA polymerase II (POLR2L) to release transcriptional pausing and promote transcriptional elongation (20) and has been shown to be essential for somatic cell reprogramming (21). We hypothesized that RBPJ may function by recruiting CDK9 to key promoter regions. Therefore, we performed an in silico analysis of RBPJ and CDK9 ChIP data for the average enrichment of RBPJ and CDK9 around promoter regions genome wide. Promoters displayed high levels of co-occupancy by RBPJ and CDK9, with CDK9 binding in the center of broader RBPJ peaks (Figure 6D).
Figure 6. RBPJ binds to CDK9 to promote target gene transcription elongation.
(A) Global analysis of the RBPJ landscape reveals that RBPJ binds nearly exclusively to active promoters and enhancers. ChIP-seq was conducted in 3691 GSCs for both RBPJ and H3K27Ac, a histone mark of active promoters and enhancers. Binding heatmaps of RBPJ (red) and H3K27Ac (green) centered on the center of RBPJ binding sites. Nearly all RBPJ sites are surrounded by H3K27Ac. (B) RBPJ global localization. Active promoters and enhancers were called using H3K27Ac sites. H3K27Ac peaks within 1 kb upstream or downstream of a transcription start site of an expressed gene were considered active promoter sites. All other H3K27Ac sites were considered active enhancer sites. (C) 3691 BTICs cells were transfected with HA-RBPJ plasmid or control vector. Anti-HA immunoprecipitates were immunoblotted with either an anti-CDK9 or anti-HA antibody. Input controls were immunoblotted with indicated antibodies. (D) Aggregate plots of RBPJ and CDK9 ChIP-seq peak intensity centered on RBPJ-bound loci in BTICs. CDK9 ChIP-seq data from GSE51633. (E) Cross-linked chromatin was prepared from 3691 BTICs expressing shCONT, shRBPJ-1, and shRBPJ-2, and then immunoprecipitated with an anti-CDK9 antibody or IgG control, followed by qPCR using primers specific for FOXM1, CCNA2, and KRAS promoters. Knockdown of RBPJ significantly decreased CDK9 recruitment to relevant promoters (2-way ANOVA, *P < 0.05, **P < 0.01, n = 3). (F) Cross-linked chromatin was prepared from 3691 BTICs transduced with shCONT, shRBPJ-1, and shRBPJ-2, and then immunoprecipitated with an anti-POL2 antibody or IgG control followed by qPCR using primers specific for the indicated regions of FOXM1, CCNA2, and KRAS (2-way ANOVA, *P < 0.05, **P < 0.01, n = 3).
To further link RBPJ and CDK9, we performed ChIP-qPCR for CDK9 and confirmed that CDK9 bound shared promoters (Figure 6E). To determine the potential functional contribution of RBPJ to CDK9 binding to target gene promoters, we targeted RBPJ using shRNA. As expected, shRBPJ attenuated promoter occupancy of RBPJ at key target gene promoters, but inhibition of RBPJ expression also attenuated promoter occupancy of CDK9 at these sites (Figure 6E), suggesting that RBPJ recruits CDK9 to key target genes. Based on the function of CDK9 as a component of P-TEFb to regulate POLR2L function, we interrogated the dependence on RBPJ of POLR2L function for CDK9 target genes. Targeting RBPJ by RNA interference did not alter POLR2L binding at the start of each gene, but knockdown of RBPJ attenuated POLR2L binding at downstream sites, concordant with regulation of transcriptional elongation (Figure 6F). These results support a potentially novel function of RBPJ to regulate the expression of key oncogenes not only as a transcriptional regulator, but also through coordination of transcriptional elongation. It should be noted that RBPJ likely regulates transcription through other mechanisms beyond CDK9, as we find that RBPJ can bind other relevant transcriptional regulators, including the lysine-specific demethylase LSD1 (KDM1) (Supplemental Figure 7B).
CDK9 targeting decreases BTIC growth and self-renewal.
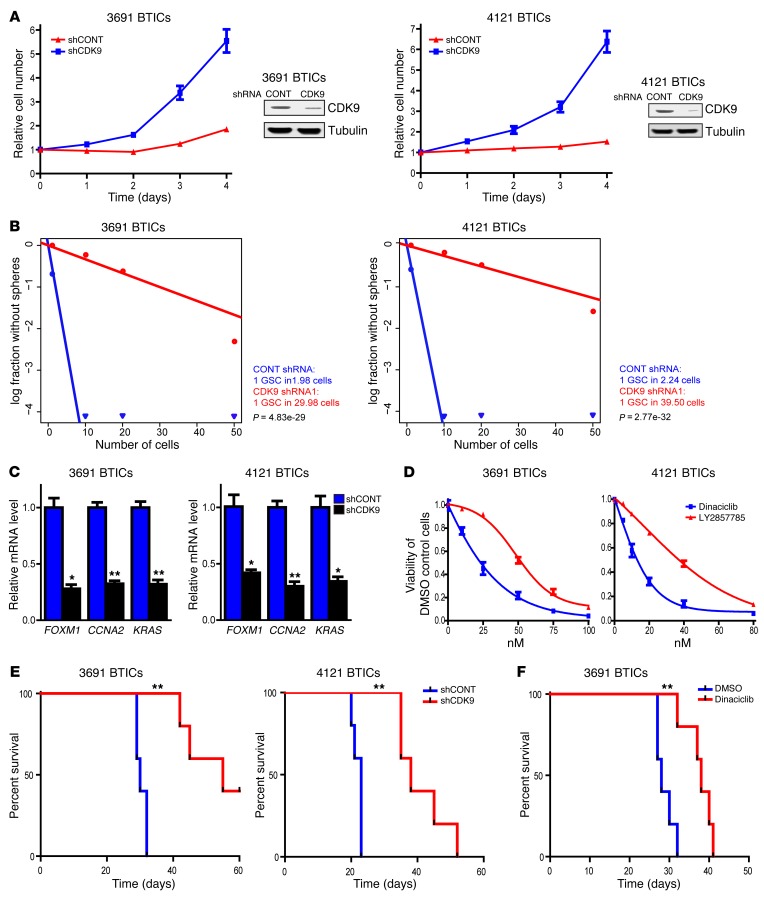
Transcriptional elongation may represent a therapeutic target in cancer, with CDK9 as a central effector. We hypothesized that CDK9, like RBPJ, may be critical in the maintenance of BTICs. As a proof of principle, we targeted CDK9 expression using RNA interference. We developed an shRNA lentiviral construct to knock down CDK9, which we confirmed by immunoblot (Figure 7A). We then examined the phenotypic consequences of shRNA-mediated reduction of CDK9 expression. Silencing CDK9 significantly decreased the growth of two BTIC models (Figure 7A). As sphere formation is a surrogate of self-renewal, we performed an in vitro limiting dilution assay in BTICs expressing nontargeting control shRNA or CDK9-directed shRNA. Targeting CDK9 resulted in a more than 10-fold decrease in the frequency of sphere formation (Figure 7B), indicating an important role of CDK9 signaling in BTIC maintenance. Inhibiting CDK9 expression attenuated the gene expression of key RBPJ-regulated genes (Figure 7C), supporting its role in regulating key genes downstream of RBPJ.
Figure 7. Targeting CDK9 decreases BTIC growth and self-renewal.
(A) Two BTIC models (3691, 4121) were transduced with shCONT or shCDK9 by lentivirus. Right: Decreased CDK9 levels were measured after shRNA transduction by immunoblotting. Left: Cellular proliferation was measured by CellTiter-Glo after shRNA transduction over a time course. (B) In vitro extreme limiting dilution assays demonstrated that shRNA-mediated knockdown of CDK9 in 2 BTIC models (3691 and 4121) decreased the frequency of tumorsphere formation. (C) 3691 and 4121 BTICs were transduced with shCONT or shCDK9 for 2 days. Total RNA was isolated and cDNA was synthesized by reverse transcription. mRNA levels of indicated genes were detected by qPCR (t test, *P < 0.05, **P < 0.01, n = 3). (D) 3691 and 4121 BTICs were treated with DMSO or two different CDK9 inhibitors, dinaciclib and LY2857785, over a concentration range. Cellular viability was tested at 48 hours. (E) Survival of immunocompromised mice bearing intracranial 3691 or 4121 BTICs transduced with shCONT or shCDK9 is displayed by the Kaplan-Meier method (log-rank analysis, **P < 0.01, n = 5). (F) Kaplan–Meier survival curves of immunocompromised mice bearing orthotopic 3691 BTICs. Seven days after tumor implantation, mice were treated with dinaciclib (30 mg/kg) or DMSO vehicle control for 2 weeks (3 times per week, **P < 0.01, n = 5).
CDK9 mediates its function in association with its kinase activity, leading to the development of several CDK9 pharmacologic inhibitors. To support a therapeutic paradigm, we tested the efficacy of 2 different CDK9 inhibitors, dinaciclib and LY2857785, against BTICs, finding significant activity as monotherapy in culture (Figure 7D). Finally, we investigated the role of CDK9 in the most critical functional assay of BTICs, in vivo tumor growth. In orthotopic studies, both RNA interference against CDK9 and a pharmacologic inhibitor of CDK9 attenuated tumor growth and extended the survival of tumor-bearing mice (Figure 7, E and F). Taken together, our findings demonstrate that CDK9 downregulation attenuates BTIC phenotypes, including proliferation and self-renewal.
Myc regulates RBPJ expression in BTICs.
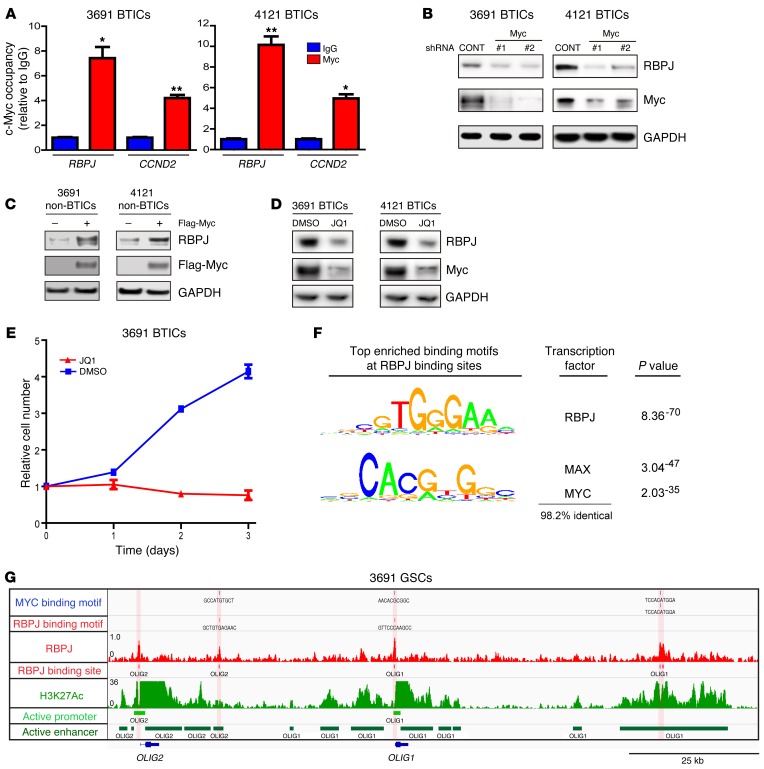
Elevated RBPJ mRNA expression in BTICs suggested that stem cell transcription factors could serve as upstream drivers of RBPJ. Therefore, we interrogated the publicly available ChIP-seq database (WashU EpiGenome Browser; http://epigenomegateway.wustl.edu/) with the human embryonic stem cell (ESC) line H1. MYC, a central regulator of ESCs and BTICs, was highly enriched in a promoter region of RBPJ and coenriched with H3K27ac, marking transcriptional activation, which strongly indicated that MYC may control RBPJ transcription in BTICs (Supplemental Figure 8). To directly test this potential regulation using functionally validated BTICs, we performed ChIP for MYC in BTICs and confirmed binding of MYC to the RBPJ promoter in BTICs by ChIP-qPCR (Figure 8A). MYC binding to cyclin D2 was used as a positive control. In loss-of-function studies, silencing MYC expression in BTICs by 2 independent shRNAs significantly reduced RBPJ expression (Figure 8B). In gain-of-function studies in non-BTICs, MYC overexpression induced RBPJ expression (Figure 8C). JQ1, a selective BET bromodomain inhibitor, specifically targets BRD4, a transcriptional activator of MYC. JQ1 effectively inhibits the expression and function of MYC in several cancers, including myeloma and leukemia (22, 23). Treating BTICs with JQ1 significantly reduced the expression of MYC and RBPJ (Figure 8D). In concordance with effects targeting RBPJ, JQ1 treatment also potently decreased BTIC growth (Figure 8E). To further address the connection between RBPJ and MYC, we performed transcription factor binding motif enrichment analysis to see if MYC binding motifs were enriched under RBPJ ChIP-seq peaks at enhancers and/or promoters (Figure 8F). We found that MYC binding sites were very highly enriched under RBPJ peaks at both enhancers and promoters. MYC is the third most highly enriched known binding motif, only after the RBPJ binding motif and the MAX binding motif, which shares greater than 98% conservation with the MYC binding motif. While this does not settle which factor initiates pause-release, it does suggest that there is a functional relationship between MYC and RBPJ. We then used our RBPJ ChIP-seq data and found that both core neural transcription factors, OLIG2 and OLIG1, have very strong RBPJ binding sites at their promoters and in their enhancers (Figure 8G), concordant with our findings that RBPJ bound to the OLIG2 promoter by ChIP-qPCR (Supplemental Figure 6C). Collectively, our data demonstrate that MYC directly activates RBPJ to facilitate transcription in BTICs, consistent with its role as an upstream regulator of RBPJ.
Figure 8. c-MYC is an upstream inducer of RBPJ expression in BTICs.
(A) Cross-linked chromatin was prepared from 2 BTIC models (3691 and 4121) and then immunoprecipitated with an anti-MYC antibody or IgG control, followed by qPCR using primers specific for the RBPJ promoter. CCND2 was used as a positive control (t test, *P < 0.05, **P < 0.01, n = 3). (B) Lysates of 3691 and 4121 BTICs expressing shCONT, shMYC-1, or shMYC-2 were immunoblotted with the indicated antibodies. shRNA-mediated knockdown of MYC decreased RBPJ levels. (C) 3691 and 4121 non-BTICs were transduced with either MYC or vector control, and then lysates were prepared and immunoblotted with the indicated antibodies. MYC expression in non-BTICs induced increased RBPJ levels. (D) 3691 and 4121 BTICs were treated with the BET domain inhibitor, JQ1 (1 μM), or vehicle control (DMSO). Lysates were immunoblotted with the indicated antibodies. JQ1 treatment decreased MYC and RBPJ levels. (E) 3691 BTICs were treated with JQ1 (1 μM) or DMSO and cellular proliferation was measured sequentially with CellTiter-Glo. (F) Global analysis of the RBPJ landscape reveals that RBPJ binds nearly exclusively to active promoters and enhancers. Transcription factor motif enrichment analysis found that RBPJ binding sites were the most enriched transcription factor–binding motif found under RBPJ ChIP-seq peaks, followed by MYC binding motifs, suggesting an interaction between these two transcription factors. (G) An example of RBPJ binding at enhancers and promoters of OLIG2 and OLIG1. The bottom two rows demonstrate called promoters and enhancers.
RBPJ and CDK9 inform patient prognosis.
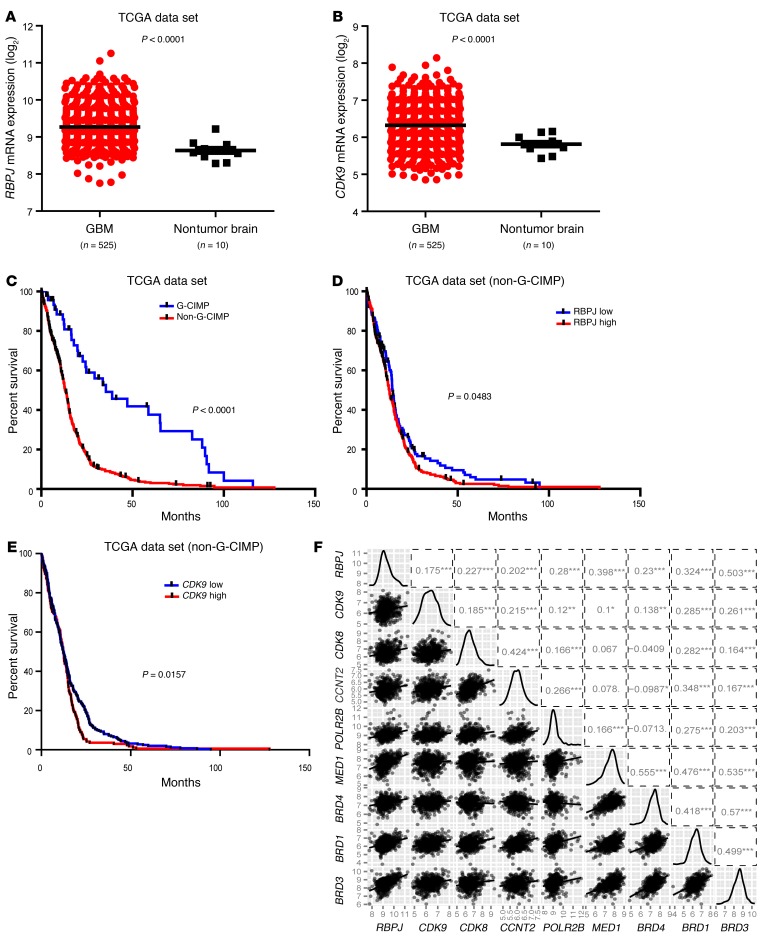
To determine the clinical relevance of these findings, we performed in silico studies of the TCGA glioblastoma data set. Both RBPJ and CDK9 were highly expressed in glioblastomas relative to nontumor brain (Figure 9, A and B). While most glioblastoma patients are diagnosed de novo, a minority of patients are diagnosed with glioblastomas after a preceding diagnosis of a lower grade glioma (secondary glioblastoma). It is now recognized that gliomas can be divided into 2 groups: G-CIMP associated with mutations in isocitrate dehydrogenase 1 or 2 (IDH1/2), and non-CIMP tumors, which do not harbor IDH mutations (24). G-CIMP glioblastomas are more common in the secondary glioblastoma fraction and share transcriptional and epigenetic landscapes with low-grade gliomas and are associated with increased survival relative to non-CIMP tumors (Figure 9C). Therefore, we segregated our analysis of the prognostic significance of target genes based on the non-CIMP, more aggressive phenotype. Concordant with the roles of RBPJ and CDK9 in maintenance of tumor growth, high expression of RBPJ and CDK9 informed poor prognosis (Figure 9, D and E). We further investigated the relationship between RBPJ mRNA expression and key regulators of transcriptional elongation through in silico studies, revealing positive correlations with multiple transcriptional elongation mediators (Figure 9F). These data support a clinically important role for the RBPJ-CDK9 transcriptional elongation-signaling axis in glioblastoma.
Figure 9. RBPJ and CDK9 regulation informs patient prognosis.
(A and B) Relative mRNA expression levels of (A) RBPJ and (B) CDK9 in nontumor brain and glioblastoma were determined in TCGA data set. (C) Analysis of TCGA data indicates that non–G-CIMP glioblastoma (GBM) patients have much poorer survival (P < 0.0001 by log-rank analysis). (D) Analysis of TCGA data indicates that higher RBPJ mRNA expression informs poor prognosis of non–G-CIMP patients (P = 0.0483 by log-rank analysis). (E) Analysis of TCGA data indicates that higher CDK9 mRNA expression informs poor prognosis of non–G-CIMP patients (P = 0.0157 by log-rank analysis). (F) Pairwise correlation analysis of RBPJ and transcriptional elongation–related genes was performed in the TCGA glioblastoma data set. Plots indicate expression data from TCGA patients for indicated genes, and numbers represent correlation coefficient (r) values.
Discussion
Signal transduction pathways are commonly viewed as linked sequential nodes, but each component in a pathway may have complex interactions with distinct pathways and molecules. NOTCH signaling is remarkably complex due to the context-specific activation of multiple receptors by distinct ligands, with associated diversity of enzymatic activity regulating receptor cleavage and liberation of the intracellular domain. Due to the challenges of measuring the activation of different receptors and the diversity of downstream transcriptional targets, many studies have used reporter systems with RBPJ binding sites as a surrogate of NOTCH activation. Our results suggest that RBPJ may have critical functions beyond simply mediating NOTCH signaling, suggesting that studies with RBPJ activity measurement will need reinterpretation. Further complicating the role of NOTCH signaling in cancer is the dual role of NOTCH as a tumor suppressor in some tumors and activator in others, not unlike TGF-β. Disruption of NOTCH signaling may be important in altering the balance of differentiation and fate decisions at tumor initiation, and then serve a different function in supporting the cellular hierarchy of established tumors. These results are important, as NOTCH targeting agents have entered clinical trials for cancers with amplification or mutations of NOTCH components and others based on targeting TICs. Antibodies targeting NOTCH receptors and ligands have undergone evaluation in systemic cancers, but limitations on drug delivery have limited NOTCH inhibition to GSI treatments (13). Few clinical trials have been reported for brain tumors, but clinical activity has been limited, despite evidence of successful inhibition of NOTCH activity.
Our studies may provide a partial explanation for divergent results in preclinical studies and clinical efficacy. RBPJ activity has been used as a reporter as a surrogate for NOTCH activity, but RBPJ may serve important NOTCH-independent roles in the maintenance of BTIC self-renewal and tumorigenic potential. RBPJ may differentially regulate gene expression in the presence and absence of NOTCH ligand-receptor binding (19). For example, RBPJ can suppress HES and HEY family expression in the basal state, so knockdown of RBPJ may derepress the expression of some targets in the canonical NOTCH pathway. RBPJ is tightly localized to gene promoters and enhancers, accounting for 88% of binding sites (Figure 6, A and B). Our results suggest that the majority of transcriptional targets downstream of RBPJ do not overlap with NOTCH-regulated pathways. For example, RBPJ binding to 2 prominent targets (FOXM1 and OLIG2) was resistant to GSI treatment, suggesting independence from NOTCH signaling (Supplemental Figure 6D). We performed a pathway analysis of our RNA-seq data using the Ingenuity Pathway Analysis tools to identify potential new signaling or interactome with RBPJ that may diverge between normal stem cells and BTICs (Supplemental Figure 9). Although RBPJ was involved with a core SOX family of transcription factors shared in both BTIC and normal neural stem cell biology, SOX2 and RBPJ interactions with CHD7 were unique to BTIC and brain tumor pathways, suggesting the importance of enhancer interaction in BTICs compared with normal developmental processes in neural stem cells. Thus, the relatively higher proliferation of BTICs may offer therapeutic opportunities to exploit compared to the more quiescent neural stem and progenitor cells.
The identification of a potentially novel MYC-RBPJ-CDK9 pathway (modeled in Supplemental Figure 10) supports an oncogenic linkage between stem cell regulation and transcriptional control. A recent RNA interference screen in a MYC-driven hepatocellular carcinoma found CDK9 as a target and showed that CDK9 inhibitors blocked proliferation of MYC-amplified tumors (25). Treatment of mixed-lineage leukemias with CDK9 inhibitors reduced MYC expression (26), suggesting that MYC is a target of CDK9-mediated transcriptional control. CDK9 regulation of MYC was recently demonstrated to be mediated by BRD4, the target of JQ1 (27). Our results provide an explanation for these observations and suggest that this pathway may be a feed-forward loop to amplify oncogene expression. Activation of CDK9-mediated transcriptional elongation of oncogenes may be controlled by RBPJ direction of the P-TEFb complex binding. Lu and co-workers found that neither BRD4 inhibition nor CDK9 inhibition alone was sufficient to abrogate proliferation in their system, but rather required a combination approach (27). As MYC can bind only to open chromatin (28), RBPJ may serve to specify cis-regulatory elements near critical BTIC genes to promote pause release. This model may be similar to a relationship between MYC and IRF4 in multiple myeloma, where there is a positive feedback loop to coordinately regulate coexpression to drive a prosurvival gene expression program (29). Thus, we would suggest that BRD4 inhibition of MYC functions in part to break the feedback loop and decrease RBPJ, while CDK9 inhibition could attenuate the enzymatic activity. In future studies, we propose to examine the interplay between RBPJ and CDK9 in the control of BTIC growth and resistance to cytotoxic therapies. The interaction with canonical NOTCH signaling may provide a further node of therapeutic benefit.
Transcription factors are generally considered undruggable, although pharmacologic inhibitors may block interactions of transcription factors with essential binding partners. In response, therapeutic strategies for disrupting the oncogenic function of transcription factors have often been upstream with kinases that modify the transcription factors (e.g., JAK inhibitors with STATs) or the primary point of initial activation (e.g., NOTCH). Our findings suggest that caution must be exercised, as transcriptional regulators may be an intersection point for different activities. Targeting NOTCH activation may reduce downstream functional activity, but these approaches may not inform the full role of a signaling pathway. The regulation of transcription by RBPJ, especially in the Proneural glioblastoma subgroup (Supplemental Figure 1E), at multiple levels supports its potent role in BTIC biology and importantly could be a potential therapeutic target even if anti-NOTCH studies fail in clinical trial.
Methods
Isolation and culture of cells.
To prevent culture-induced drift, patient-derived xenografts were generated and maintained as a recurrent source of tumor cells for study. Immediately upon xenograft removal, a Papain Dissociation System (Worthington Biochemical) was used to dissociate tumors according to the manufacturer’s instructions (detailed protocol: http://www.worthington-biochem.com/PDS/default.html). Cells were then cultured in Neurobasal medium supplemented with B27, L-glutamine, sodium pyruvate (Invitrogen), 10 ng/ml basic FGF (bFGF), and 10 ng/ml EGF (R&D Systems) for at least 6 hours to recover surface antigens. DAPT (LY-374973) was purchased from Sigma-Aldrich.
All specimens were verified by comparison of short tandem repeat (STR) analysis. No marker is uniformly informative for BTICs; therefore, we used a combination of functional criteria to validate BTICs. Where indicated, BTICs and non-BTICs were derived immediately after dissociation or after transient xenograft passage in immunocompromised mice using prospective sorting followed by assays to confirm stem cell marker expression, sphere formation, and secondary tumor initiation. Although CD133 is controversial, in the models used in these studies, CD133 has previously identified functional BTICs (2, 30). Therefore, in experiments with matched TIC and non-TIC cultures, we segregated AC133 marker–positive and marker–negative populations using an APC-conjugated anti-CD133/2 antibody (293C3, Miltenyi Biotech) by FACS or magnetic bead separation (Miltenyi Biotech), as previously described (2, 30). The TIC phenotype of these cells was validated by stem cell marker expression (CD133, OLIG2, and SOX2), functional assays of self-renewal (serial tumorsphere passage), and tumor propagation by in vivo limiting dilution.
Proliferation and neurosphere formation assay.
Cell proliferation was measured using Cell-Titer Glow (Promega). Neurosphere formation was measured by in vitro limiting dilution as previously described (2). All data were normalized to day 0 and presented as mean ± SEM.
Vectors and lentiviral transfection.
The 4XRBPJ-Luc reporter plasmid was a gift from Takahiro Maeda (Harvard University), and the tk-renilla reporter plasmid was a gift from Zengqiang Yuan (Institute of Biophysics, Chinese Academy of Sciences, Beijing, China). HA-tagged RBPJ (HA-RBPJ) was generated by cloning the human RBPJ open reading frame with the N-terminal HA sequence into the pCDH-MCS-T2A-Puro-MSCV vector (System Biosciences). Flag-tagged c-Myc (Flag-Myc) was generated by subcloning the human c-Myc open reading frame with the N-terminal Flag sequence into the pCDH-MCS-T2A-Puro-MSCV vector. Lentiviral clones to express shRNA directed against RBPJ (TRCN0000016203, TRCN000016204), MYC (TRCN0000010389, TRCN0000010390), CDK9 (TRCN0000199187), or a control shRNA insert that does not target human and mouse genes (shCONT, SHC002) were obtained from Sigma-Aldrich. Lentiviral particles were generated in 293FT cells in stem cell media with cotransfection with the packaging vectors pCMV-dR8.2 dvpr and pCI-VSVG (Addgene) by Lipofectamine 2000 (Invitrogen).
Western blotting.
Cells were collected and lysed in hypotonic buffer with nonionic detergent (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.5% NP-40; 50 mM NaF with protease inhibitors), incubated on ice for 15 minutes, and cleared by centrifugation at 10,000 g at 4°C for 10 minutes. Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories). Equal amounts of protein were mixed with reducing Laemmli loading buffer, boiled, and electrophoresed in NuPAGE Gels (Invitrogen), and then transferred to PVDF membranes (Millipore). Blocking was performed for 30 minutes with 5% nonfat dry milk in TBST and blotting performed with primary antibodies for 16 hours at 4°C. Antibodies included RBPJ (Cell Signaling Technology, 5313), SOX2 (R&D Systems, AF2018), OLIG2 (R&D Systems, AF2418), GFAP (Covance, PRB-571), MYC (Santa Cruz Biotechnology, sc-764), HA (Santa Cruz, sc-7392), CDK9 (Santa Cruz Biotechnology, sc-484), cleaved NOTCH1 (Cell Signaling Technology, 4147), ACTIN (Sigma-Aldrich, A5441), and GAPDH (Sigma-Aldrich, SAB1405848).
In vitro limiting dilution assay.
For in vitro limiting dilution assays, decreasing numbers of cells per well (20, 10, 5, and 1) were plated in 96-well plates. Ten days after plating, the presence and number of neurospheres in each well was quantified. Extreme limiting dilution analysis was performed using software from the Walter and Eliza Hall Institute (http://bioinf.wehi.edu.au/software/elda), as previously described.
Immunofluorescent staining.
Cells or 10-μm-thick slices of xenografted brain tissue were fixed in 4% paraformaldehyde and immunolabeled using the following antibodies: RBPJ (Lifespan Biosciences, LS-C158830), OLIG2, and SOX2. Primary antibodies were incubated overnight at 4°C, followed by species-appropriate secondary antibodies (Alexa 488 and 568; Invitrogen Molecular Probes) with incubation for 1 hour. Nuclei were stained with DAPI, and slides were then mounted using Fluoromount (Calbiochem). Images were obtained using a Leica DM4000 upright microscope.
ChIP assay.
4 × 106 cells per condition were plated, and ChIP was performed with a ChIP assay kit (Invitrogen, 49-2024) following the manufacturer’s protocol. Briefly, 5 μg MYC antibody (Santa Cruz Biotechnology, sc-764), RBPJ antibody (Abcam, 25949), anti-HA antibody (Santa Cruz, sc-7392), CDK9 antibody (Santa Cruz Biotechnology, sc-484), POL2 antibody (Santa Cruz Biotechnology, sc-899), or rabbit IgG was used for the immunoprecipitation of the DNA–protein immunocomplexes. Crosslinking was reversed by heating for 6 hours at 65°C, followed by digestion with proteinase K. The purified DNA was analyzed by qPCR with the indicated primers as follows: RBPJ forward: TTCTCGCGAGGTTTAGGAAA, RBPJ backward: CTCACGCTACACCTCCTTCC. CCND2 forward: TCAGTAAATGGCCACACATGTG, CCND2 backward: GGAGCTCTCGACGTGGTCAA, FOXM1 P1 forward: GGCCTTGTCTCGGCATTC, FOXM1 P1 backward: CCACTTCTTCCCCCACAAG. FOXM1 P2 forward: CCTAGCTGCAGGTTTTGGTC, FOXM1 P2 backward: CAACTCAGCCTCCAGGACTC. FOXM1 P3 forward: GGAAAGTGGTCCTCAATCCA, FOXM1 P3 backward: AGCGGCCACCCTACTCTTAC. CCNA2 P1 forward: ATCCCGCGACTATTGAAATG, CCNA2 P1 backward: CCCCAGCCAGTTTGTTTCT. CCNA2 P2 forward: GGGCATCTTCACGCTCTATT, CCNA2 P2 backward: CCTCCTTGGAAAGCAAACAG. CCNA2 P3 forward: TCACTGACTCTGCCTGGTGT, CCNA2 P3 backward: AACTTCAGCTTGTGGGCACT. KRAS P1 forward: GAGCACACCGATGAGTTCG, KRAS P1 backward: CTCTTCCCTCTTCCCACACC. KRAS P2 forward: AACTCCTCCATCGACGCTTA, KRAS P2 backward: GCAGAACAGCAGTCTGGCTA. KRAS P3 forward: TCCTGAGAAGGGAGAAACACA, KRAS P3 backward: TCAAGTCCTTTGCCCATTTT, OLIG2 forward: AGTCCCTCCCCAAGAATCTC, OLIG2 backward: GATGTCATCCGGGCTAATTC.
ChIP-seq.
ChIP was performed as previously described (18). Briefly, formaldehyde-fixed cells were lysed and sheared (Branson S220) on wet ice. The sheared chromatin was cleared and incubated overnight at 4°C with RBPJ antibody (Abcam, 25949) or H3K27ac antibody (Abcam, 4729). Antibody-chromatin complexes were immunoprecipitated with protein G magnetic Dynal beads (Life Technologies), washed, eluted, reverse crosslinked, and treated with RNAse A followed by proteinase K. ChIP DNA was purified using Ampure XP beads (Beckmann Coulter) and then used to prepare sequencing libraries for sequencing with the Next-Seq Illumina genome analyzer.
Peak calling.
Reads were aligned to hg19 using Burrows-Wheeler Aligner (BWA) (31), and identical ChIP-seq sequence reads were collapsed to a single read to avoid PCR duplicates. Peaks were called using HOMER v4.6 (32) using matched inputs with the parameter, –factor. ChIP-seq data for CDK9 in HEK293T were used from GSE51633 (33) and analyzed in the same way.
Intracranial tumor formation and in vivo bioluminescence imaging.
BTICs were transduced with lentiviral vectors expressing RBPJ or a nontargeting, control (shCONT) shRNA for the knockdown experiments. Thirty-six hours after infection, viable cells were counted and engrafted intracranially into NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, The Jackson Laboratory) mice. In parallel survival experiments, animals were monitored until they developed neurological signs. To monitor tumor growth in living animals, BTICs used for the animal studies were transduced with firefly luciferase through lentiviral infection. BTICs expressing firefly luciferase were then transduced with shCONT or shRBPJ. Thirty-six hours after infection, viable cells were counted and engrafted intracranially into NSG mice. For the imaging analysis, animals were administered D-luciferin intraperitoneally and anesthetized with isoflurane. Tumor luciferase images were captured by using the IVIS imaging system (Xenogen-100). All animal procedures conformed to the Cleveland Clinic IACUC-approved protocol.
RNA sequencing.
Total RNA was isolated from cells using the RNeasy Kit (QIAGEN). Strand-specific libraries were generated using the Illumina TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Gold. Paired-end 125-bp reads were generated on an Illumina HiSeq 2500 instrument at the Case Western Reserve University Genomics Core Facility. Reads were aligned to the hg19 genome using TopHat v2.0.6 with the library type option set to first strand. FPKMs of known genes were calculated using Cufflinks v2.0.2 with the “--GTF” option set.
Accession numbers.
All original microarray data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE79736).
Statistics.
All grouped data are presented as mean ± SEM. Data distribution was assumed to be normal, but this was not formally tested. Differences between groups were assessed by 2-tailed Student’s t test or 2-way ANOVA using InStat software (GraphPad Software). Kaplan-Meier curves were generated and log-rank analysis was performed using MedCalc software. A P value less than 0.05 was considered significant.
Study approval.
The Cleveland Clinic IRB approved studies using human tissue samples. Glioblastoma tissues were obtained from excess surgical materials from patients at the Cleveland Clinic after neuropathologist review in accordance with an IRB-approved protocol. Written informed consent was received from all patients. All studies were performed in the Biological Resources Unit (BRU) at the Lerner Research Institute of the Cleveland Clinic under an Institutional Animal Care and Use Committee–approved protocol specific for this study.
Author contributions
QX and JNR designed the experiments, analyzed data, and prepared the manuscript. QX, QW, TEM, BBL, KY, DCF, XF, ZH, WZ, KA, and XW performed the experiments. LK, TEM, SCM, and DCF performed database analyses. BEB and SB provided scientific input and helped to edit the manuscript.
Supplementary Material
Acknowledgments
We appreciate the kind provision of the 4XRBPJ-Luc reporter plasmid from Takahiro Maeda (Harvard University) and the tk-renilla reporter plasmid from Zengqiang Yuan (Institute of Biophysics, Chinese Academy of Sciences). Mary McGraw and the Cleveland Clinic Foundation Tissue Procurement Service provided primary brain tumor tissues. We would like to thank the Cleveland Clinic Lerner Research Institute imaging core and proteomics core for technical assistance and the members of the Rich and Bao laboratories for critical reading of the manuscript and helpful discussions. Finally, we would like to thank our funding sources: The NIH (grants CA197718, CA154130, CA171652, CA169117, CA198892, NS087913, NS089272 [to J.N. Rich], NS070315 [to S. Bao], and HG006991 [to B.E. Bernstein]), Research Programs Committees of Cleveland Clinic (to J.N. Rich), James S. McDonnell Foundation (to J.N. Rich), and the Howard Hughes Medical Institute (to B.E. Bernstein).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(7):2757–2772. doi:10.1172/JCI86114.
See the related Commentary beginning on page 2415.
Contributor Information
Qi Xie, Email: xieqi724@gmail.com.
Qiulian Wu, Email: wuq2@ccf.org.
Leo Kim, Email: jyk29@case.edu.
Kailin Yang, Email: yangk2@ccf.org.
Zhi Huang, Email: huangz@ccf.org.
Wenchao Zhou, Email: zhouw@ccf.org.
Kareem Alazem, Email: alazemk@ccf.org.
Xiuxing Wang, Email: xiuxing81@gmail.com.
Shideng Bao, Email: Baos@ccf.org.
References
- 1.Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 5.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97(6):703–716. doi: 10.1016/S0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 6.Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131(5):965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 7.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13(9):654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purow BW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65(6):2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 9.Fan X, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28(1):5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, et al. Inhibition of NOTCH signaling blocks growth of glioblastoma cell lines and tumor neurospheres. Genes Cancer. 2010;1(8):822–835. doi: 10.1177/1947601910383564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28(1):17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert CA, Daou MC, Moser RP, Ross AH. Gamma-secretase inhibitors enhance temozolomide treatment of human gliomas by inhibiting neurosphere repopulation and xenograft recurrence. Cancer Res. 2010;70(17):6870–6879. doi: 10.1158/0008-5472.CAN-10-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling — are we there yet? Nat Rev Drug Discov. 2014;13(5):357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 14.Moellering RE, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hori K, et al. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22(2):166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Man J, et al. Sema3C promotes the survival and tumorigenicity of glioma stem cells through Rac1 activation. Cell Rep. 2014;9(5):1812–1826. doi: 10.1016/j.celrep.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suva ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castel D, Mourikis P, Bartels SJ, Brinkman AB, Tajbakhsh S, Stunnenberg HG. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27(9):1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20(8):2629–2634. doi: 10.1128/MCB.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu L, et al. Transcriptional pause release is a rate-limiting step for somatic cell reprogramming. Cell Stem Cell. 2014;15(5):574–588. doi: 10.1016/j.stem.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Ott CJ, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120(14):2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang CH, et al. CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma. Genes Dev. 2014;28(16):1800–1814. doi: 10.1101/gad.244368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia-Cuellar MP, et al. Efficacy of cyclin-dependent-kinase 9 inhibitors in a murine model of mixed-lineage leukemia. Leukemia. 2014;28(7):1427–1435. doi: 10.1038/leu.2014.40. [DOI] [PubMed] [Google Scholar]
- 27.Lu H, et al. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. Elife. 2015;4: doi: 10.7554/eLife.06535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guccione E, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8(7):764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 29.Shaffer AL, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454(7201):226–231. doi: 10.1038/nature07064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Q, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci. 2015;18(4):501–510. doi: 10.1038/nn.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155(7):1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler — a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5(11): doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.