Abstract
Targeting glioblastoma stem cells with γ-secretase inhibitors (GSIs) disrupts the Notch pathway and has shown some benefit in both pre-clinical models and in patients during phase I/II clinical trials. However, it is largely unknown why some glioblastoma (GBM) does not respond to GSI treatment. In this issue of the JCI, Xie et al. determined that GSI-resistant brain tumor–initiating cells (BTICs) from GBM express a higher level of the gene RBPJ, which encodes a mediator of canonical Notch signaling, compared to non-BTICs. Knockdown of RBPJ in BTICs decreased propagation in vitro and in vivo by inducing apoptosis. Interestingly, RBPJ was shown to regulate a different transcription program than Notch in BTICs by binding CDK9, thereby affecting Pol II–regulated transcript elongation. Targeting CDK9 or c-MYC, an upstream regulator of RBPJ, with small molecules also decreased BTIC propagation, and prolonged survival in mice bearing orthotopic GBM xenografts. This study not only provides a mechanism for GSI treatment resistance, but also identifies two potential therapeutic strategies to target GSI-resistant BTICs.
Challenges of targeting brain tumor stem cells by Notch pathway blockade
Glioblastoma (GBM) is the most common malignant brain tumor in humans and has a median survival of only 14 months; therefore, new treatment strategies for this devastating disease are desperately needed (1–3). Brain tumor stem cells, also known as brain tumor–initiating cells (BTICs), have been prospectively isolated by several research groups (4–7) and have been shown to be resistant to conventional radiation therapy and chemotherapy (8, 9). Targeting brain tumor stem cells, by blocking Notch signaling with a γ-secretase inhibitor (GSI) or by inducing activation of the bone morphogenetic protein (BMP) pathway with BMP4, has shown some efficacy in preclinical studies (10, 11), bringing hope to improving brain tumor treatment based on targeting cancer stem cells.
The Notch pathway is a developmental signaling pathway that regulates cell fate decisions and stem cell self-renewal in multiple organs of almost all species, including neural stem cells of the mammalian CNS (12–15). Dysregulation of Notch signaling has been observed in many types of neoplasm, including GBM (7, 16–19). Several reports have shown that Notch pathway blockade by GSI inhibits BTIC propagation and prolongs survival in mice bearing intracranial xenografts (20, 21). Moreover, in a recent Phase I clinical trial, 24% of patients with malignant glioma (a total of 44) responded to GSI treatment and had stabilized disease for more than four months (22). Although some malignant glioma patients benefited from GSI treatment, most GBM patients did not respond to GSI treatment, and the mechanism of GSI resistance in GBM cells is largely unknown.
GBMs are molecularly divided into proneural, proliferative, and mesenchymal subclasses, according to an initial study based on gene expression profiling (23). It was immediately speculated that the proneural subgroup would be sensitive to GSI therapy, as this population of GBMs exhibits a higher level of Notch pathway activation than the other subgroups (23). Indeed, a recent study showed that GBM neurosphere cells with a strong proneural gene signature respond to GSI treatment (24). Although no stratified clinical trial to assess GSI for the treatment of proneural GBMs has been carried out, identification of the molecular mechanism of GSI resistance in GBM has the potential to help develop novel therapies for this deadly disease. In the current issue, Xie et al. elegantly demonstrate that a mediator of Notch signaling, RBPJ, is overexpressed in GSI-resistant BTICs preferentially in proneural GBMs and is required for BTIC propagation both in vitro and in vivo (25). The results of this study are of substantial clinical relevance, because a mechanism of GSI resistance in BTICs has been identified, thereby providing a new approach to target GSI-resistant tumor cells in general.
The Notch signaling pathway and RBPJ
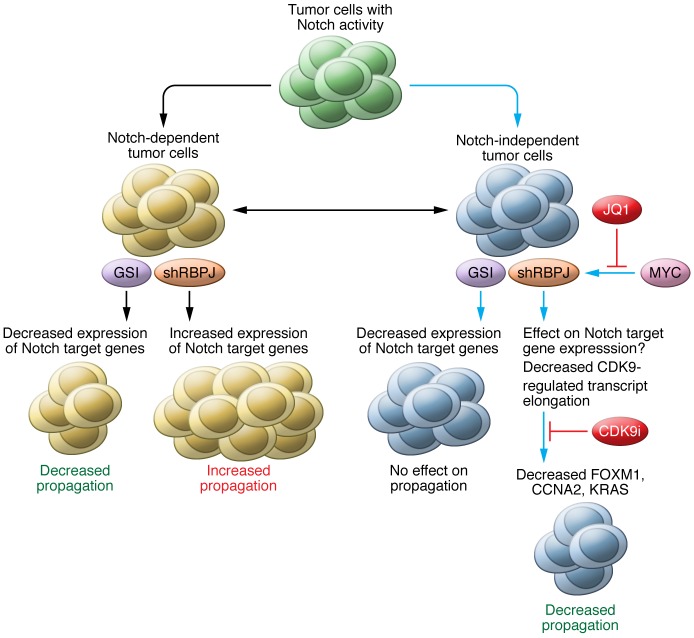
Canonical Notch signaling is initiated when a Notch ligand, such as JAGGED (JAG1 or JAG2) or DELTA (DLL1, 3, or 4), binds to a NOTCH receptor (NOTCH1, 2, 3, or 4) on adjacent cells. In turn, ligand binding to the NOTCH receptor induces γ-secretase–mediated proteolytic cleavage of the NOTCH receptor at the transmembrane domain to release NOTCH intracellular domain (NICD), which translocates into the nucleus, binds RBPJ protein complexes situated on DNA, recruits co-activators, and removes co-repressors to activate downstream target genes, such as HES and HEY family genes (26–28). In the absence of NICD, RBPJ associates with histone deacetylase–containing co-repressors, including SMRT, SHARP, CtBP, SKIP, and CIR, in closed chromatin to suppress gene transcription (26–28). Binding of NICD to RBPJ dissociates the co-repressor complex from RBPJ, and recruits mastermind-like proteins (MAMLs) and histone acetyltransferases to the NICD-RBPJ complex, thereby remodeling chromatin to activate transcription of target genes (26–28). Active NOTCH1 mutations have been found in about 60% of T cell acute lymphoblastic leukemias (T-ALLs) that respond to GSI treatment (29, 30). Subsequently, it has been shown that genetic and epigenetic alterations of tumor cells contribute to GSI resistance in T-ALL (31–33). However, GSI resistance in GBMs has not been carefully investigated. Interestingly, Xie et al. found that GSI-resistant BTICs have elevated RBPJ expression compared to non-BTICs and that BTICs lose RBPJ expression and stem cell markers upon their differentiation (ref. 25 and Figure 1). In addition, knockdown of RBPJ expression by shRNA decreased BTIC propagation in vitro and in vivo by inducing apoptosis (25). This reduction in BTIC propagation prolonged survival in mice bearing intracranial xenografts (25). Together, these results demonstrate that RBPJ is required for GSI-resistant BTIC propagation.
Figure 1. Targeting tumor cells with elevated level Notch activity with γ-secretase inhibitors (GSIs) or shRBPJ.
Tumor cells with elevated levels of Notch activity can be divided into Notch signaling–dependent and –independent classes, which are based on their genetic or epigenetic background. For Notch-dependent tumor cells, GSI treatment can reduce Notch target gene expression and decrease propagation. However, shRPBJ in these same cells will release RBPJ-mediated repression of gene transcription, induce expression of Notch target genes, and increase tumor cell propagation. In contrast, in Notch-independent tumor cells, or GSI-resistant cells, GSI treatment still can block NICD formation and decrease Notch target gene expression. However, GSI treatment has no effect on propagation, because growth of Notch-independent tumor cells depends on genes that are not Notch targets. In this issue, Xie et al. demonstrate that knockdown of RBPJ in Notch-independent cells downregulates expression of genes, including FOXM1, CCNA2, and KRAS, that control brain tumor–initiating cell (BTIC) self-renewal and proliferation, and thereby decreases tumor growth through partnering with CDK9, which regulates elongation of RBPJ target genes. Furthermore, Xie et al. found that RBPJ is regulated by MYC and that blocking MYC expression, upstream of RBPJ, with JQ1 or blocking activity of CDK9 also decreases Notch-independent tumor cell propagation. Solid blue lines indicate results from Xie et al. Solid black lines indicate results from other reports.
While most studies have shown that canonical Notch signaling activates target genes through the DNA-binding RBPJ protein complex, Rbpj knockout mice do not have the same phenotype as Notch knockout mice (34). This discrepancy suggests that RBPJ can function in both Notch-dependent and -independent manners (refs. 35, 36, and Figure 1). In addition, RBPJ generally represses target gene expression in the absence of NICD; therefore, it is believed that knockout or knockdown of RBPJ could result in derepression of target genes (26, 36). Indeed, loss of RBPJ has been shown to induce expression of several Notch target genes, either in the presence or absence of NICD, and increase tumorigenesis in breast cancer and Burkitt lymphoma cells (37). In contrast, Xie et al. show that knockdown of RBPJ only derepresses a few Notch target genes, such as HES5, but induces apoptosis in BTICs (25). One possible reason for these opposite effects from knockdown of RBPJ in tumor cells is that the breast cancer cells and Burkitt lymphoma cells used were dependent on Notch signaling to grow (37), whereas the BTICs used by Xie et al. were already independent of Notch signaling for their growth (GSI-resistant cells) (Figure 1). Another possibility is that there could be different RBPJ binding landscapes across the genome and different Notch-dependent target genes in different types of tumor cells.
RBPJ target genes
Xie et al. performed RNA-seq to examine changes in the transcriptional profile of BTICs in response to RBPJ knockdown or Notch signaling blockade with GSI (25). Only 10% to 15% of genes were commonly regulated by both RBPJ and Notch signaling, suggesting that RBPJ regulates BTIC propagation mostly through the regulation of Notch-independent genes, an observation supported by the fact that propagation of GSI-treated BTICs is independent of Notch signaling. Xie et al. determined that tumorigenesis-associated genes FOXM1, CCNA2 (cyclin A2), and KRAS are not only exclusively regulated by RBPJ at the transcriptional level but also contain RBPJ binding sites at their promoter regions, suggesting that these genes are possible direct targets of RBPJ in BTICs (25). ChIP-PCR analysis confirmed that FOXM1, CCNA2, and KRAS are indeed direct targets of RBPJ and independent of Notch regulation in BTICs, suggesting that RBPJ regulates propagation of GSI-resistant BTICs at least in part through direct regulation of FOXM1, CCNA2, and KRAS expression.
Furthermore, Xie et al. transduced HA-tagged RBPJ into BTICs and performed immunoprecipitation using an anti-HA antibody and carried out proteomic analysis of RBPJ binding proteins to identify RBPJ co-factors (25). CDK9 tightly bound to RPPJ and regulated transcription of RBPJ target genes, including FOXM1, CCNA2, and KRAS, through transcription elongation, a result that is consistent with previous studies showing that CDK9, unlike CDK8, is involved in RBPJ-regulated transcription independent of NICD (38). The study by Xie et al. indicates that CDK9 is only involved in transcription elongation; therefore, it remains unknown how RBPJ modulates the changes in chromatin conformation that are required to initiate transcription, as RBPJ generally binds to closed chromatin in its default state. Interestingly, Xie et al. indeed found that the lysine-specific demethylase LSD1 is also associated with RBPJ and may be involved in transcription initiation (25). Further investigation of the mechanism of RBPJ-regulated, Notch-independent transcription initiation will be very interesting. Nevertheless, Xie et al. have now shown that CDK9 is also required for propagation of GBM BTICs through regulation of FOX1, CCNA2, and KRAS, and that pharmacological inhibition of CDK9 activity with the CDK9 inhibitor dinaciclib (or LY2857785) inhibits BTIC growth and prolongs survival in mice bearing intracranial GBM xenografts (ref. 25 and Figure 1). Together, these results of Xie and colleagues indicate that interfering with RBPJ-regulated gene transcription with CDK9 inhibitors has potential as a clinically relevant therapeutic approach to treat GSI-resistant BTICs.
Regulation of RBPJ expression
RBPJ has been shown to function as a housekeeping gene in cells from different organs (39, 40). It will be interesting to know how RBPJ expression is elevated in BTICs compared to non-BTICs. Xie et al. explored a publicly available ChIP-seq data set and found c-MYC binding sites located in the promoter region of RBPJ (25). Moreover, they confirmed that overexpression of c-MYC induces transcription of RBPJ, as detected by a luciferase reporter assay, and demonstrated direct c-MYC binding at the RBPJ promoter using ChIP-PCR. In addition, knockdown of MYC decreased RBPJ expression at the protein level. Taken together, these results demonstrate that c-MYC is one of the upstream regulators of RBPJ that directly regulates RBPJ expression at the transcriptional level. However, MYC has been shown to be a canonical Notch target in previous studies (41–43). Notch regulates c-MYC expression through RBPJ binding to both promoter and super-enhancer regions of MYC (41–43). As the BTICs used by Xie et al. have canonical Notch activity, albeit they were not dependent on Notch signaling to grow, it is unclear if canonical Notch signaling contributes to the expression of c-MYC in these cells. While GSI treatment indeed blocked NICD1 formation in BTICs (25), the effects of GSI on c-MYC expression are not known. Nevertheless, Xie et al. found that blocking c-MYC expression with the selective bromodomain inhibitor JQ1 decreases BTIC propagation in vitro and in vivo, providing another potential therapeutic strategy for treating GSI-resistant GBMs (Figure 1).
Indication and future directions
Xie and colleagues have discovered that the MYC/RBPJ/CDK9 pathway is critical for BTIC self-renewal (25), results with clinical implications not only for GBM treatment, but also for cancer-targeting therapies in general, particularly for GSI-resistant tumors (Figure 1). Based on previous reports and the current study by Xie et al., tumor cells with Notch activity can be roughly divided into Notch-dependent and Notch-independent classes (Figure 1). Notch-dependent tumor cells most likely will respond to GSI treatment; however, knockdown of RBPJ in Notch-dependent tumor cells will release repression of Notch target genes and may promote tumor growth. In contrast, for Notch-independent tumor cells, although GSI treatment can still block Notch signaling, this strategy will not inhibit growth of these cells. Therefore, Notch-independent tumor cells are considered to be GSI resistant. Knockdown of RBPJ expression in these GSI-resistant tumor cells will decrease propagation by reducing expression of genes that are regulated exclusively by RBPJ, but not Notch, including FOXM1, CCNA2, and KRAS. In addition, the results of Xie et al. demonstrated that these GSI-resistant tumor cells can be treated with the c-MYC inhibitor JQ1 or the CDK9 inhibitor dinaciclib (ref. 25 and Figure 1). As RBPJ is considered to be a housekeeping gene and is expressed in most cells in the body (39, 40), the potential toxicity of these approaches in normal cells will need to be closely watched during future studies aimed at evaluating the clinical application of RBPJ-targeting therapies.
Acknowledgments
This work was funded by grants from the NIH: R01CA148621 (to X. Fan) and R01CA163737 (to X. Fan).
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Reference information:J Clin Invest. 2016;126(7):2415–2418. doi:10.1172/JCI88619.
See the related article beginning on page 2757.
References
- 1.Louis D, Ohgaki H, Wiestler O, Cavenee W. WHO Classification of Tumours of the Central Nervous System. Lyon, France: IARC Press; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Furnari FB, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 4.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100(25):15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 6.Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101(3):781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli R, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 8.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 9.Liu G, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5: doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piccirillo SG, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 11.Fan X, et al. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 2006;66(15):7445–7452. doi: 10.1158/0008-5472.CAN-06-0858. [DOI] [PubMed] [Google Scholar]
- 12.Shen Q, et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304(5675):1338–1340. doi: 10.1126/science.1095505. [DOI] [PubMed] [Google Scholar]
- 13.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8(6):709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 14.Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7(2):93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- 15.Androutsellis-Theotokis A, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442(7104):823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 16.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39(3):193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 17.Purow BW, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65(6):2353–2363. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat Rev Cancer. 2011;11(5):338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 20.Fan X, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28(1):5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, et al. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28(1):17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krop I, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 2012;30(19):2307–2313. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 23.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Saito N, et al. A high Notch pathway activation predicts response to γ secretase inhibitors in proneural subtype of glioma tumor-initiating cells. Stem Cells. 2014;32(1):301–312. doi: 10.1002/stem.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Q, et al. RBPJ maintains brain tumor–initiating cells through CDK9-mediated transcriptional elongation. J Clin Invest. 2016;126(7):2757–2772. doi: 10.1172/JCI86114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 27.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124(5):973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 29.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 30.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herranz D, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med. 2015;21(10):1182–1189. doi: 10.1038/nm.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knoechel B, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46(4):364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakhai H, et al. Conditional ablation of Notch signaling in pancreatic development. Development. 2008;135(16):2757–2765. doi: 10.1242/dev.013722. [DOI] [PubMed] [Google Scholar]
- 35.Fujimoto M, et al. RBP-J promotes neuronal differentiation and inhibits oligodendroglial development in adult neurogenesis. Dev Biol. 2009;332(2):339–350. doi: 10.1016/j.ydbio.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Tanigaki K, Honjo T. Two opposing roles of RBP-J in Notch signaling. Curr Top Dev Biol. 2010;92:231–252. doi: 10.1016/S0070-2153(10)92007-3. [DOI] [PubMed] [Google Scholar]
- 37.Kulic I, et al. Loss of the Notch effector RBPJ promotes tumorigenesis. J Exp Med. 2015;212(1):37–52. doi: 10.1084/jem.20121192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell. 2004;16(4):509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 39.Hamaguchi Y, et al. Biochemical and immunological characterization of the DNA binding protein (RBP-Jκ) to mouse J κ recombination signal sequence. J Biochem. 1992;112(3):314–320. doi: 10.1093/oxfordjournals.jbchem.a123898. [DOI] [PubMed] [Google Scholar]
- 40.Kawaichi M, et al. Genomic organization of mouse J κ recombination signal binding protein (RBP-Jκ) gene. J Biol Chem. 1992;267(6):4016–4022. [PubMed] [Google Scholar]
- 41.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herranz D, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20(10):1130–1137. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yashiro-Ohtani Y, et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc Natl Acad Sci U S A. 2014;111(46):E4946–E4953. doi: 10.1073/pnas.1407079111. [DOI] [PMC free article] [PubMed] [Google Scholar]