Abstract
Tumor protein p53 (TP53) is the most commonly mutated gene in human cancer. The majority of mutations are missense, and generate a defective protein that is druggable. Yet, for decades, the small-molecule restoration of wild-type (WT) p53 function in mutant p53 tumors (so-called p53 mutant ‘reactivation’) has been elusive to researchers. The p53 protein requires the binding of a single zinc ion for proper folding, and impairing zinc binding is a major mechanism for loss of function in missense mutant p53. Here, we describe recent work defining a new class of drugs termed zinc metallochaperones that restore WT p53 structure and function by restoring Zn2+ to Zn2+-deficient mutant p53.
Introduction
Recent results from The Cancer Genome Atlas program confirm that tumor protein p53 (TP53) is the most commonly mutated gene in human cancer [1–3]. The majority of p53 mutations (>70%) involve the substitution of a single amino acid in its DNA-binding domain (DBD), rendering p53 unable to activate transcription. Mutant p53 consequently accumulates to high levels in cancer cells owing to a lack of MDM2-mediated negative autoregulation [4–7].
Developing drugs that restore wild-type (WT) structure and function to p53 mutants has long been considered a Holy Grail in cancer therapy. Indeed, over the past decades several compounds have been reported to reactivate mutant p53; however, all but one (APR-246/PRIMA-1) have failed to progress to clinical development – previous strategies for mutant p53 reactivation have been thoroughly reviewed [8–10]. APR-246 (PRIMA-1) is a small molecule currently in a Phase Ib/II clinical trial in recurrent platinum-sensitive high-grade serious ovarian cancer (PiSARRO trial, NCT02098343). Clinical evidence that this compound reactivates mutant p53 is still pending because the Phase I dose escalation study recently published did not reveal this information [11].
We and others recently identified a class of small molecules that reactivate p53 mutants that are impaired in their ability to bind zinc. These compounds, termed zinc metallochaperones (ZMCs), share the common characteristic of binding zinc outside the cell and delivering it to mutant p53 to facilitate proper folding. This review will highlight key concepts that define a ZMC and equally importantly identify a class of p53 mutants that are potentially amenable to ZMC treatment. The pharmacologic delivery of a metal ion to correct a defect in protein folding is unprecedented in drug development. This novel mechanism coupled with the identification of a defined patient population for treatment makes ZMCs attractive candidates in the search for p53-targeted drugs.
p53 and zinc
The p53 protein is a 393 residue, zinc-dependent, homotetrameric transcription factor [12]. The monomer comprises the N-terminal transactivation domain, a central DBD and the C-terminal tetra-merization domain. The X-ray crystal structure of the DBD (residues 94–312) reveals a central β-sandwich with a DNA-binding surface consisting of a loop-sheet-helix motif and two large loops (L2 and L3) [13]. These loops are stabilized by the tetrahedral coordination of a single zinc ion by Cys176–His179 of L2 and Cys238–Cys242 of L3. The purified zinc-free (apo)DBD is stable at 10 °C but can no longer discriminate between consensus and non-consensus DNA-binding sequences [14]. At physiologic temperatures apoDBD is predominantly unfolded [14].
Multiple lines of evidence suggest that WT p53 can reversibly transition between a folded ‘WT-like’ and unfolded ‘mutant-like’ conformation under biological conditions, often related to a change in Zn2+-binding status. Hainaut and Milner reported that incubating cells and cell lysates with Zn2+ chelators can starve p53 of Zn2+ and cause an immunophenotype switch from WT to mutant as judged by conformation-specific antibody immunoprecipitation [15]. This conformational change results in a loss of sequence-specific DNA-binding activity, but it can be reversed by adding ZnCl2 or replacing the cell culture media. They also report that metallothionein IIA, a high-affinity endogenous Zn2+-binding protein, can switch the immunophenotype and inhibit p53 function [16]. Other groups have reported immunophenotype switching during the S-phase of the cell cycle upon growth stimulation [17], and after treatment with oxidizing agents [18]. Taken together, these results indicate that the conformation of the p53 protein is flexible and can cycle between folded and unfolded isomers in response to changes in its environment.
Tumorigenic p53 mutations fall into three broad categories: destabilizing, DNA contact and zinc binding [19–21]. Destabilizing mutations are often found in the β-sandwich core of DBD distant from the zinc- and DNA-binding sites and these mutations lower the melting temperature of p53 partially unfolding it at 37 °C. DNA contact mutants are found in the DNA-binding region and disrupt crucial interactions between p53 residues and DNA, lowering the DNA-binding activity but leaving the thermodynamic stability of the protein largely unaffected. Zinc-binding mutants have classically been defined by their proximity to the L2 and L3 loops, and are unique in that they structurally destabilize p53 and abolish its sequence-specific DNA-binding activity [20]. The most-studied zinc-binding mutant is R175H, which is the most frequent p53 mutation in cancer [12] (Fig. 1). R175H DBD readily loses its Zn2+ in the presence of even weak Zn2+ chelators and has a Kd for Zn2+ tenfold weaker than the typical intracellular Zn2+ concentration. The 175 mutant protein probably does not bind Zn2+ in vivo [22].
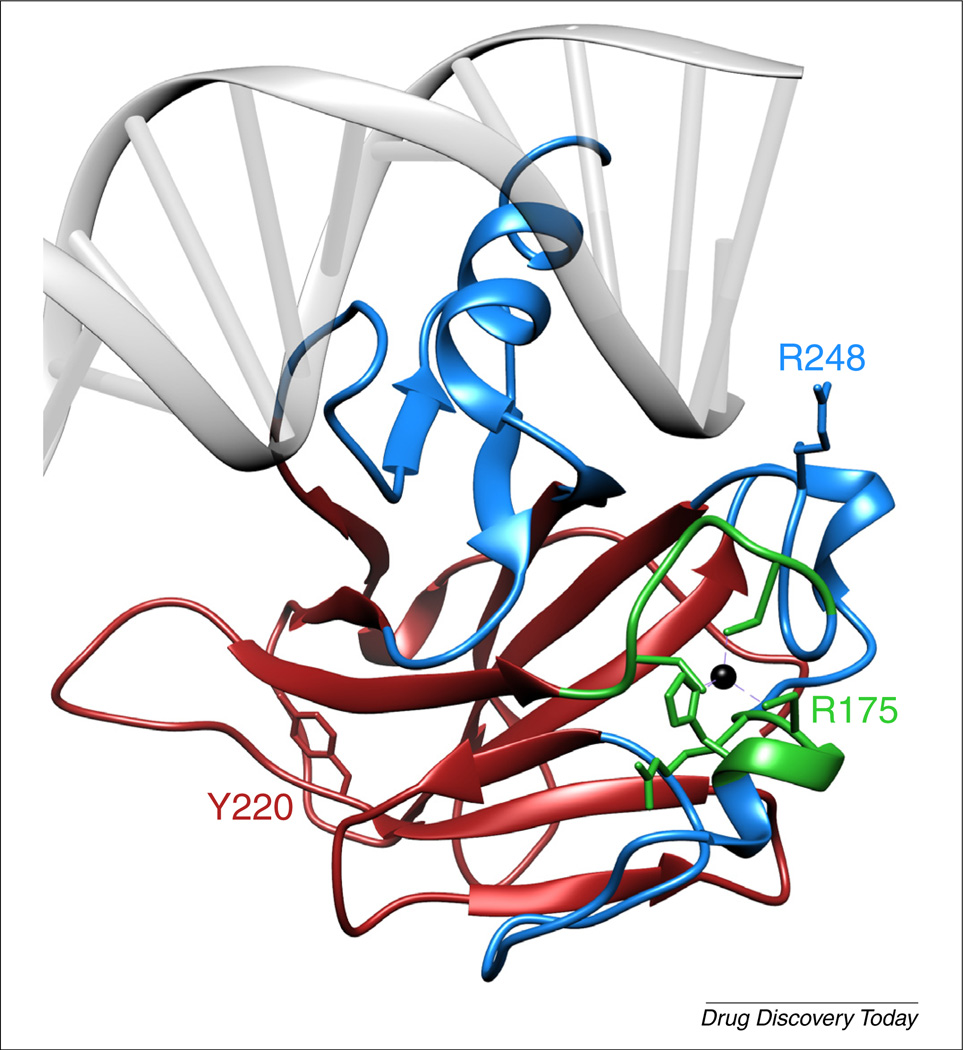
FIGURE 1.
Structure of the wild type p53 DNA-binding domain (DBD) complexed with DNA. Zinc (black sphere) is coordinated by C176, H179, C238 and C242. Mutations in the DNA-binding region (blue; e.g. R248W) impair DNA-binding affinity but leave the protein structure largely intact. Mutations in the β-sandwich domain (red; e.g. Y220C) cause p53 to misfold but maintain a certain amount of DNA-binding affinity. Mutations in the Zn2+-binding region (green; e.g. R175H) cause p53 to misfold and lose its DNA-binding affinity [36]. Structure taken from PDB entry TUP1.
Whereas starving p53 of zinc by chelation or tumorigenic mutation (e.g. R175H) can induce structural changes, too much zinc can do the same. We previously reported that excess zinc induces DBD misfolding, presumably because of interactions of zinc with cysteine and histidine residues located outside of the zinc-binding site of the native protein [for clarity, we define the Kd of the native Zn2+-binding site as Kd1 and that of the non-native zinc-binding site(s) as Kd2] [23]. Importantly, we found that certain zinc chelators can facilitate proper folding of p53 in the presence of excess zinc by functioning as a zinc buffer, providing the chelator has a Kd for Zn2+ between Kd1 and Kd2. This way, the chelator can serve as a source of zinc for the native site and a sink for excess zinc to prevent zinc-induced misfolding. The term given for this type of chelator is zinc metallochaperone.
In principle, it might be possible to correct the folding defect in the p53–R175H mutant by supplying exogenous zinc to tumor cells expressing this mutant and, indeed, evidence for this has been observed in some tumor cells [24]. Moreover, supplemental zinc has also been shown to restore WT p53 structure in certain tumors that overexpress metallothioneins, lending further support to this view [25,26]. However, supplementation with free zinc is not a useful strategy to reactivate mutant p53 pharmacologically because the window of zinc concentrations that leads to activity is relatively narrow, and it is unlikely that Zn2+ supplementation alone could overwhelm the homeostatic mechanisms that govern intracellular Zn2+ levels in humans and maintain it in this range.
NSC319726 (ZMC1) discovery as a lead compound for mutant-p53-targeted drug development
Using in silico screening of the NCI60 anticancer drug screen, we identified three thiosemicarbazones (NSC319725, NSC319726, NSC328784) with predicted high sensitivity in tumor cells expressing mutant p53 but at the same time low sensitivity in cell lines expressing WT p53 [27]. We validated NSC319725 and NSC319726 in cell lines with different TP53 status (WT, null or mutant) and found that the compounds exhibit marked sensitivity in mutant p53 cells, with the maximal sensitivity observed in cells expressing p53–R175H. At doses that induce mutant p53 cell death, the compounds are completely nontoxic to a non-tumor (human fibroblast) cell line.
Using NSC319726 (ZMC1), we demonstrated that the mechanism of cell growth inhibition in p53–R175H mutant cells is through p53-mediated apoptosis resulting from restoration of WT structure and function to p53–R175H [27]. This ‘reactivation’ manifests in mice as acute, mutant-dependent toxicity. ZMC1 is remarkably toxic in mice that have a homozygous germline mutation of p53–R172H (mouse equivalent of human R175H) when compared with WT littermates. Furthermore, when ZMC1 is administered to mice bearing subcutaneous xenograft tumors of different TP53 status, we observe potent antitumor activity only in p53–R175H tumors, with no degree of growth inhibition in tumors with p53–R273H (DNA contact mutant) or WT p53 cells, and no systemic toxicity at the doses that are effective in tumor cell death. This indicates that the antitumor properties of ZMC1 are p53 allele specific.
Thiosemicarbazones (TSCs) like ZMC1 bind divalent metal ions such as Fe2+, Zn2+, Cu2+ and Mn2+ [28]. They have been investigated as anticancer drugs, the most notable being 3-aminopyridine-2-carboxaldehyde (Triapine®) [29,30]. The mechanism of TSCs has been attributed to the inhibition of the iron-dependent enzyme ribonucleotide reductase via iron chelation as well as reactive oxygen species (ROS) generation through Fenton chemistry catalyzed by TSC–Fe3+ complexes [31,32]. At the high doses typically required to kill cancer cells, nonspecific toxicities related to methemoglobinemia and bone marrow suppression have plagued development of TSCs [33,34]. However, ZMC1 is effective at much lower doses, and has a fundamentally different mechanism through mutant p53 reactivation [22,27]. We focused our initial mechanistic work with ZMC1 on its metal-ion-chelating and redox-modulating properties because these are common to TSCs. We found that supplemental zinc increased the apoptotic activity of the compound whereas treatment with the reducing agent, N-acetyl cysteine (NAC), decreased the apoptotic activity.
Mechanistic studies of ZMC1: ZMC1 as a Zn2+ buffer
The selective activity of ZMC1 against the Zn2+-binding mutant p53–R175H combined with the observation that ZMC1 activity is enhanced by supplemental Zn2+ gave clear indications that ZMC1 stabilizes the p53–Zn2+ interaction [27]. However, its mechanism for doing so was unclear. The most direct hypothesis is that ZMC1 binds to the destabilized p53–R175H and forces the protein into a WT-like conformation. Indeed, this is the paradigm for most previous attempts to develop drugs that target p53 mutants. For example, the carbazole derivative PhiKan083 reactivates the core-mutant Y220C by filling a surface cleft left by the removal of the bulky Tyr side chain, thus stabilizing the native conformation [35]. However, this scenario is effectively ruled out for ZMC1 because its Kd for purified DBD–R175H (~ 10−4 M) is 100–1000-fold higher than the EC50 in cells [22]. Therefore, a conventional ligand-binding/stabilization interaction cannot explain its mechanism.
Additional experiments with purified DBD–R175H provide an alternative hypothesis. Although R175H was classified as a Zn2+-binding mutation in 2000 based on its proximity to the native Zn2+-binding site and the resultant structural destabilization of the protein [36], the Zn2+-binding affinity of the native site (Kd1) was not measured until recently. The measured Kd1 value of 2.1 nM is far too weak to result in native Zn2+ binding at concentrations of free zinc, [Zn2+]free, typically present in cells (~ 0.1 nM) [37,38]. However, zinc binding is not abolished, and Kd1 is well separated from the Kd of the non-native sites that lead to misfolding (Kd2 ~ 1 µM) [22]. These data indicate that p53–R175H is nonfunctional in the cell because it is an apoprotein without zinc bound. It stands to reason that to restore WT conformation and activity to p53–R175H one must either decrease its Kd1 below intracellular [Zn2+]free or increase intracellular [Zn2+]free above Kd1. If the latter track is taken then it is essential that [Zn2+]free does not rise above Kd2 to avoid inappropriate zinc binding to p53 and other proteins. This principle was demonstrated in vitro with the Zn2+-binding compound nitrilotriacetate (NTA) (Kd,NTA = 17 nM) and the purified WT DBD [14,39].
The Kd,ZMC1 value of ~30 nM [22] fits well with the above model. It is 15-fold higher than Kd1 of DBD–R175H and 33-fold lower than Kd2, making ZMC1 ideally suited to re-metallate p53–R175H while protecting against improper zinc ligation. Folding studies in vitro with purified DBD protein indicate that ZMC1 can rescue the conformation of DBD misfolded by high Zn2+ concentration, allowing the protein to regain its native structure by chelating the excess Zn2+. Conversely, re-metallation studies indicate that ZMC1 can also rescue Zn2+-deficient DBD, donating Zn2+ to the native Zn2+-binding site and restoring structure- and sequence-specific DNA-binding activity to the protein. It is this ability to chelate Zn2+ when the metal is in excess and donate Zn2+ when it is deficient that separates the ZMC mechanism from that of more-established metal-binding drugs, which typically function only as chelators (e.g. to starve metalloenzymes of the metals on which they depend) [28]. Indeed, the metal ion chelator EDTA, which is used to treat heavy metal poisoning, binds Zn2+ too tightly (Kd,EDTA ~ 10−13 M) to release it to the apo-p53 protein and therefore does not restore WT structure to DBD–R175H.
Requirement for ROS in the ZMC1 mechanism
As previously mentioned, an additional requirement for the mechanism of ZMC1 is ROS generation. This was first indicated by cell killing studies in TOV112D cells, which noted that the oxidizer diamide enhances the effectiveness of ZMC1, whereas the antioxidant NAC decreases it [27]. Follow-up studies demonstrated that ZMC1 treatment generates ROS and induces DNA oxidation as measured by 8-oxy-dGUO staining and qRT-PCR of genes in the antioxidant Nrf2-pathway [9]. Time-course western blots reveal a series of p53 post-translational modifications (PTMs) that correlate with these measurements of ROS levels: phosphorylation at Ser15 and Ser46 and acetylation at Lys120 [22]. These changes also correlate with the intracellular rescue of p53 conformation (measured by PAB240/PAB1620 staining) and p53 function (assayed by p21 and PUMA induction).
Importantly, the ability of ZMC1 to induce cell killing, DNA oxidation, genes in the Nrf2-pathway, p53 function and p53 PTMs are all dramatically reduced upon administration of NAC, indicating that ROS induction is a necessary upstream signal to all of these activities [22,27]. However, ZMC1 retains its ability to rescue p53–R175H conformation in the presence of NAC [22] suggesting that rescue of p53 conformation and the required increase in ROS levels happen through two necessary but separate mechanisms outlined in Fig. 2. It seems that ZMC1 rescues mutant p53 conformation by buffering the intracellular [Zn2+]free to a level such that the defective protein can bind to it, thereby restoring WT-like structure to the protein, and then transactivates the newly rescued p53 post-translationally by ROS signaling or ROS-induced damage.
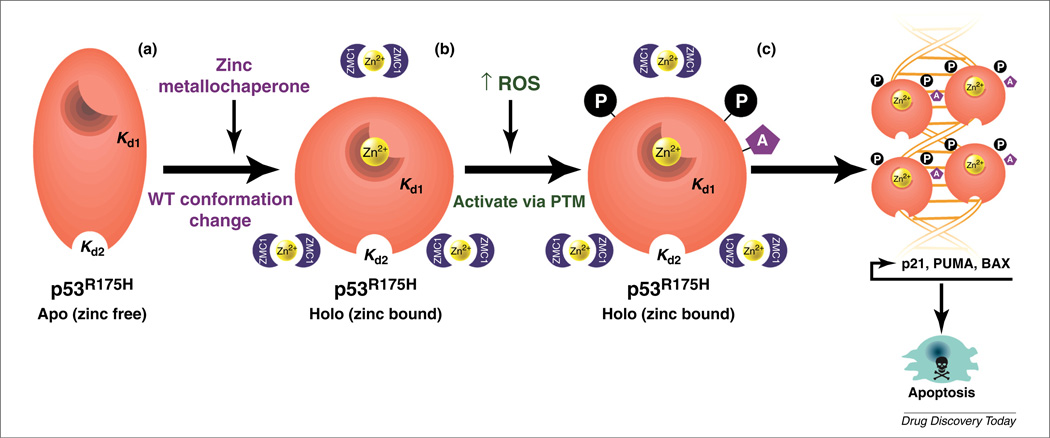
FIGURE 2.
Model of the ZMC1 dual mechanism. (a) The p53–R175H is misfolded owing to impaired zinc binding (apo form) and, as result, no site-specific DNA binding occurs (and no transcription of p53 targets). ZMC1 functions as a zinc metallochaperone (ZMC) by providing a source of zinc to facilitate refolding of the p53–R175H to a WT-like structure (holo form). (b) ZMC1 boosts reactive oxygen species (ROS) levels, which activates a stress response (i.e. ATM) that transactivates the p53–R175H through amino-terminal phosphorylation (P) and acetylation (A) events. (c) The newly conformed and transactivated p53–R175H can now bind DNA in a site-specific manner, causing transcription of apoptotic effectors that leads to tumor cell death. Kd1 = Kd of the native binding site, Kd2 = Kd of the non-native binding site.
This dual mechanism allows ZMC1 to be effective as a monotherapy. It can correct the misfolded conformation of p53–R175H and provide a signal to ‘turn on’ the newly conformed protein. Although these two activities had never been observed together in the same drug previous to ZMC1, the secondary requirement for ROS is consistent with some previous attempts at Zn2+ therapy for p53 Zn2+-binding mutants. D’Orazi and colleagues reported that administration of Zn2+ alone in certain cell lines can restore the WT-like conformation to p53–R175H, but that it is not sufficient to induce cell killing [24]. However, when cytotoxic chemotherapy (a known inducer of ROS and DNA damage) is applied, Zn2+ is an effective adjuvant in susceptible cell lines. Although we do not observe p53–R175H rescue with the application of Zn2+ alone in any of our experimental systems, the point remains that Zn2+ restoration of p53–R175H is necessary but insufficient for functional rescue in cells.
Precisely how ZMC1 induces ROS is unclear, but it probably involves an interaction with one or more redox active transition metals and subsequent Fenton chemistry. Zn2+ itself is redox neutral because it only has one stable oxidation state under biological conditions and therefore cannot generate ROS. However, TSCs are known to interact with a number of metal cations including Fe2+, Cu2+, Mn2+ and Co2+ [24]. Absorbance spectroscopy reveals that ZMC1 interacts with Fe2+, Fe3+ and Cu2+ in addition to Zn2+ [40]. Iron and copper are capable of generating ROS through Fenton chemistry because they can cycle between two oxidation states under biological conditions (Fe2+/Fe3+ and Cu1+/Cu2+) [41]. The source of the redox active metals and how much is needed to induce ROS that we observe for ZMC1 is unclear, but it is probably a very small amount based on the relatively low toxicity seen in p53-WT cell lines and WT mice [27]. We have found that ZMC1 induces some degree of apoptosis in cancer cells (10–20%) that is independent of p53 status and this is probably the result of the ROS induction [27]. However, in a zinc-binding mutant such as the p53–R175H we observed significantly greater degrees of apoptosis (>80%) that is not due to ROS because this could be abrogated using an siRNA to p53 indicating an on-target effect [27].
Identifying ionophore activity
Despite the biophysical, biochemical and biological data all converging on the same Zn2+-buffering/ROS-induction mechanism presented in Fig. 2, a fundamental question remained: where does the Zn2+ come from and how does ZMC1 deliver it to p53 inside the cell? Functional studies in mammalian cells demonstrate that adding supplemental Zn2+ to the cell culture medium enhances the effectiveness of ZMC1 [27], whereas chelating the Zn2+ from the medium decreases it [40]. Additionally, quantitative imaging studies with the fluorescent Zn2+-indicator FluoZin-3 demonstrate that intracellular Zn2+ concentration increases upon ZMC1 treatment when extracellular Zn2+ is present in the imaging medium, but not when it is absent [40]. These findings strongly support an extracellular source for the Zn2+. Because the vast majority of extracellular Zn2+ is present as an albumin-bound complex in serum, albumin is the most likely source of Zn2+ in vivo.
After binding Zn2+ extracellularly, ZMC1 transports the metal across the plasma membrane as an ionophore. Titration and the crystal structure of the ZMC1–Zn2+ complex demonstrate a 2:1 stoichiometry [22,40]. The crystal structure also reveals that the polar pyridine and thiocarbonyl moieties from ZMC1 molecules point inward toward the Zn2+, whereas the hydrophobic aromatic and alkyl moieties point away. This arrangement effectively encapsulates the Zn2+ in a hydrophilic pocket surrounded by a hydrophobic shell – a structural theme shared by valinomycin and other known ionophores. Furthermore, the thiocarbonyl bond length indicates enolization via loss of the α-proton from each ZMC1 molecule, rendering the entire complex neutral [40]. All of these structural features are consistent with a complex that has much higher lipid solubility than the free Zn2+ ion, and would be expected to facilitate Zn2+ diffusion across membranes. Indeed, membrane transport studies with liposomes indicate that ZMC1 dramatically increases the rate of specific Zn2+-transport across synthetic phospholipid bilayers, supporting the ionophore hypothesis [40].
Definition of a ZMC and zinc-binding mutant class
ZMCs represent a fundamental departure from previous attempts to target mutant p53. Virtually all previous attempts to develop p53-reactivating compounds sought to bind or otherwise modify mutant p53 to ‘fix’ the defective protein and make it functional in the cellular environment. Examples of this include PRIMA-1MET (metabolite of PRIMA-1) which was reported to react covalently with thiol groups in mutant p53 [42]. How this binding functions to restore WT structure to zinc-binding mutants like R175H has yet to be reported. PhiKan083 and its analog PK7088 are known to bind and stabilize the Y220C conformational mutant [35,43]. In the case of Zn2+-binding mutants, this would entail developing a small molecule that increases Zn2+ affinity or otherwise stabilizes the WT conformation. The logic behind this approach is clear but it has proven largely unsuccessful with no compounds clinically available despite decades of research. ZMCs by contrast do not try to fix the defective protein but instead change the cellular environment to allow the protein to function despite its defect. For Zn2+-binding mutants this translates to increasing intracellular [Zn2+]free to a set point that is ideally five–tenfold above Kd1 of the p53 mutant, thus allowing the zinc to enter the mutant (defective) binding sites in the protein and restore its ‘native’ conformation. The available evidence suggests that the added zinc in the mutant protein restores some of the native conformation permitting it to bind to DNA specifically and initiate an apoptotic transcriptional program in a cell. This ‘end around’ strategy makes this approach unique compared with conventional pharmacotherapies, and provides a range of possibilities for drug development not available to traditional approaches.
Because no specific binding interaction is required between a ZMC and p53, the potentially functional structures of the ZMC can vary to a much greater extent than those of traditional drugs, which are required to maintain high affinity for their targets. This is an attractive feature of ZMC therapy because it permits the ability to investigate non-thiosemicarbazone zinc chelators for ZMC activity, circumventing TSC toxicity problems related to iron chelation and inhibition of ribonucleotide reductase. Indeed, we have found that the zinc chelator NTA has ZMC activity in vitro. However, NTA is not useful in cells because it has poor ionophore activity, highlighting another requirement of a ZMC. A bipyridine-Zn2+-curcumin complex was recently reported to reactivate p53–R175H and also has the ability to cross the blood–brain barrier [44]. Although the structure bears no resemblance to ZMC1, it is possible that this functions by much the same mechanism given that it binds Zn2+ and transports it across biological membranes. Table 1 illustrates the properties we have identified thus far that are important for ZMC function.
TABLE 1.
Activities necessary for zinc metallochaperone therapy and associated mechanisms
| Activity | Mechanism |
|---|---|
| Increase intracellular [Zn2+]free | Zn2+ ionophore (source: serum albumin) |
| Maintain [Zn2+]free between Kd1 and Kd2 |
Zn2+ buffer (Kd1 < Kd,drug < Kd2)a |
| Activate p53 via post-translational modification |
Reactive oxygen species induction (Fenton reactions) |
Kd1 = Zn2+ Kd of native site, Kd2 = Zn2+ Kd of non-native site.
The lack of a required protein-binding interaction also enables a greater palate of mutants to be reactivated by ZMCs, provided that the mutation compromises Zn2+-binding affinity. The question then becomes: which mutants are impaired for Zn2+-binding? Historically, Zn2+-binding mutants have been classified by their proximity to the Zn2+-binding site and their effect on protein stability and function. This definition encompasses mutations of the four Zn2+-binding residues (C176, C238, C242, H179) and hotspot mutations R175H and M237I. However, in addition to the canonical Zn2+-binding mutants, we observe that ZMC1 reactivates hotspot mutant G245S, which is traditionally defined as a DNA-region mutant. We have previously demonstrated that removal of the Zn2+ from DBD-WT destabilizes the protein by ~3 kcal/mol [14]. Conversely, unfolding DBD destabilizes Zn2+ binding, raising the Kd from <10−10 M to ~10−9 M. Therefore, Zn2+ binding and p53 stability are linked – any mutation that disrupts Zn2+ binding can destabilize p53, and any mutation that destabilizes p53 can impair Zn2+ binding. It is therefore likely that the pool of mutations that exhibit impaired Zn2+ binding and are potentially treatable by ZMCs is much larger than previously appreciated.
It is not yet known how far the range of mutations in the p53 protein that have an altered Zn2+-binding extends. From a direct spatial approach, all mutations tested that lie adjacent to the Zn2+-binding site are reactivated, but this reactivation does not extend to the DNA-interacting surface because R248 mutations are unaffected (Fig. 3). However, a spatial approach might be inadequate because there are a number of mutations in the β-sandwich region distant from the Zn2+-binding site that destabilize the protein to an equal or greater extent than the canonical Zn2+-binding mutants. Because of the link between Zn2+ binding and protein stability, we might expect these mutants to exhibit impaired Zn2+ binding as well. There has never been a rigorous study of which mutants impair zinc binding, and research into ZMC therapy now substantiates the need for one.
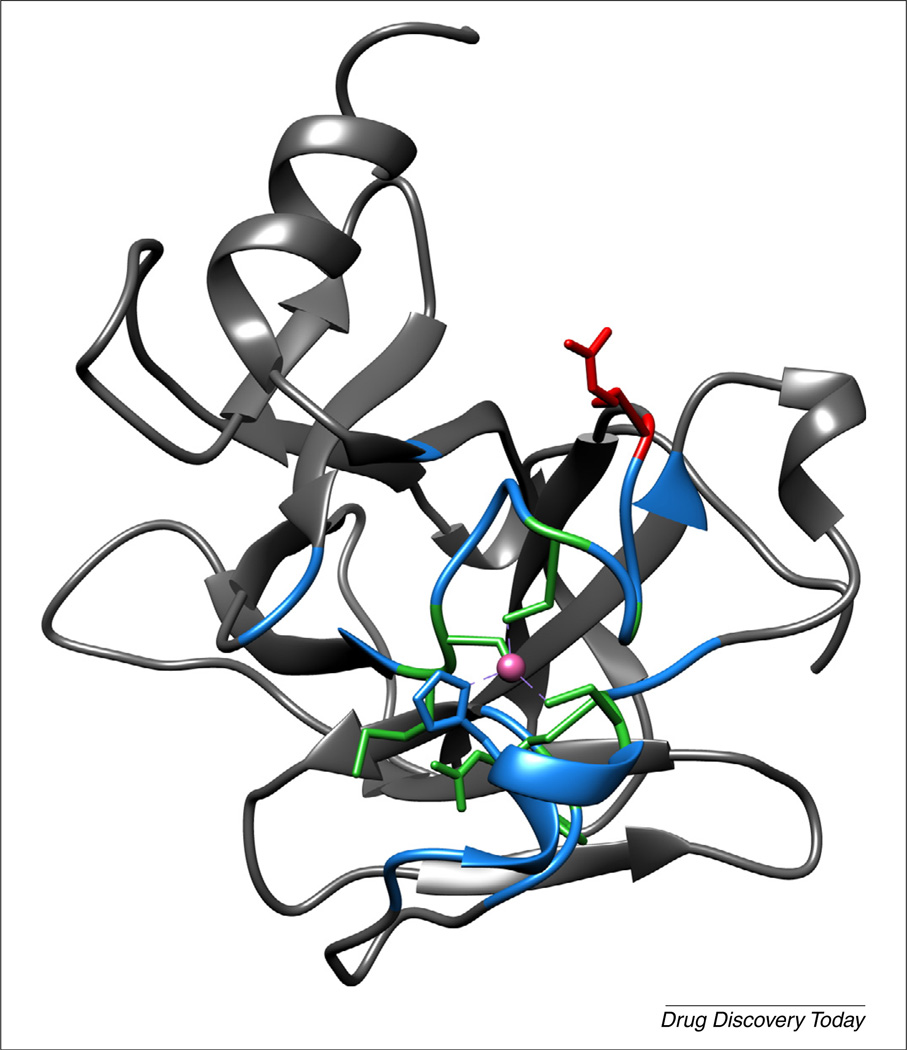
FIGURE 3.
Spatial approach to Zn2+-binding mutations. Residues within 10 Å of the bound Zn2+ ion in the DNA-binding domain (DBD) X-ray crystal structure would be expected to be most probably zinc impaired and (TUP1) are colored. Residues in green have mutations that are confirmed to be reactivated by ZMC1, red are confirmed to be unaffected by ZMC1 and blue have not been investigated. Green residues are R175, C176, C238, C242 and G245 [22], as well as M237 and L194 (unpublished). The residue shown in red is R248.
Another exciting possibility for ZMC therapy is that p53 is not the only protein that can be potentially targeted. In theory, any Zn2+-binding protein that requires Zn2+ to function but has impaired Zn2+-binding affinity could be rescued by the same strategy provided the Zn2+ Kd of the defective binding site and Kd of the putative ZMC are appropriately matched. For example, p53 homolog p63 contains a similar DBD and Zn2+-binding site [45]. Germline mutations of p63 are known to disrupt epidermal and appendage development, and manifest as a number of ectodermal dysplasias [46]. Some of the mutations known to cause these diseases correspond exactly with p53 hotspot mutants [47]. Notably, R204 in p63 corresponds to R175 in p53, so we might expect mutations at those positions to have similar effects on Zn2+ binding in both proteins, and be rescued by similar therapy provided the Kd of the ZMC and Kd of the native ligation site in the R204 mutant of p53 are similarly matched as ZMC1 and the p53–R175H. No zinc-binding studies of any p63 mutants have been published.
The ionophore activity and elevating of intracellular zinc concentrations by a ZMC explain the on-target effect of ZMC1 with p53 mutants with impaired zinc binding, but these activities would also be expected to occur in normal cells and thus there are potential off-target effects of this type of treatment. It is estimated that 3–10% of human genes probably encode for zinc-binding proteins so certainly the question of how ZMC therapy might affect the zinc proteome is relevant [39]. There are some key structural differences between p53 and other classical zinc finger transcription factors that could explain why p53 is particularly vulnerable to ZMC therapy. For instance, classical zinc finger transcription factors have an interior hydrophobic core in which the zinc atom is buried lending to their stability. However, in p53 the zinc atom bridges and stabilizes two large loops of a loop-sheet-helix motif that would otherwise be unstable [48].
Preclinical data suggest however that there is a therapeutic window between toxicity for cancer cells and normal cells in mice but toxic effects of zinc in some selected cell types could be a drawback of this type of treatment [27]. Indeed, in vivo testing of ZMCs at this point has been limited to relatively short intervals (days-to-weeks) and, although well tolerated in WT animals, the long-term effects of modulating intracellular Zn2+ are currently unknown. However, it is likely that the increased intracellular zinc concentrations in cancer and normal cells would be transient because cellular mechanisms that govern zinc homeostasis would be expected to be elicited [37] (also unpublished data). This area of the ZMC mechanism needs to be further studied, but this might explain why these transient increases in intracellular zinc could be tolerated in vivo.
The dual function of ZMC1 (p53 conformational rescue followed by ROS-induced PTM activation) presents a number of potential synergies that might be exploited for cancer treatment. Time-course western blots indicate that p53–R175H levels drop upon ZMC1 treatment in a matter of hours, and that this drop is eliminated by co-treatment with Nutlin-3, presumably by disrupting MDM2-mediated p53 degradation [27]. Combining inhibitors of the p53–MDM2 interaction with a ZMC might potentiate the effectiveness of ZMCs by maintaining the rescued p53 at a higher level. Additionally, many conventional chemotherapy drugs as well as radiation are known to induce ROS, damage DNA and otherwise provide signals that ultimately result in p53 PTM and activation [49,50]. These strategies might synergize with ZMCs by providing additional activating signals in the PTM activation portion of the ZMC1 mechanism. ROS-generating photodynamic therapy might also potentiate ZMC effectiveness for the same reason [51]. Identifying and exploiting potential synergistic effects of ZMCs with established and experimental therapies could greatly enhance the therapeutic potential of this strategy.
Concluding remarks
There is now a new pathway to target mutant p53 using small molecules functioning as ZMCs. The small-molecule delivery of a metal ion to restore the native structure and function of a metalloprotein is unprecedented in drug development. The p53–R175H is the most frequently found p53 mutation in cancer and accounts for an estimated 28 000 new cancer cases in the USA [12]. If one considers the entire class of zinc-binding mutants as we now understand them, this number would increase to over 74 000 annually. This number will probably expand as further zinc-binding impaired p53 mutants and similarly impaired Zn2+-binding proteins involved in other diseases are discovered. Future clinical trials of ZMCs will need to enroll patients specifically with zinc-binding mutations to most effectively exploit the ZMC mechanism.
Acknowledgments
We would like to thank Garrett Lee and Mary Ann Sells for their assistance with the illustration in Fig. 2.
References
- 1.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freed-Pastor WA, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haupt Y, et al. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 7.Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Sun Y. Targeting p53 for novel anticancer therapy. Transl. Oncol. 2010;3:1–12. doi: 10.1593/tlo.09250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu X, et al. Small molecule compounds targeting the p53 pathway: are we finally making progress? Apoptosis. 2014;19:1055–1068. doi: 10.1007/s10495-014-0990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoo KH, et al. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann S, et al. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J. Clin. Oncol. 2012;30:3633–3639. doi: 10.1200/JCO.2011.40.7783. [DOI] [PubMed] [Google Scholar]
- 12.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho Y, et al. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 14.Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–2403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- 15.Hainaut P, Milner J. A structural role for metal ions in the wild-type conformation of the tumor suppressor protein p53. Cancer Res. 1993;53:1739–1742. [PubMed] [Google Scholar]
- 16.Meplan C, et al. Metalloregulation of the tumor suppressor protein p53: zinc mediates the renaturation of p53 after exposure to metal chelators in vitro and in intact cells. Oncogene. 2000;19:5227–5236. doi: 10.1038/sj.onc.1203907. [DOI] [PubMed] [Google Scholar]
- 17.Milner J, Watson JV. Addition of fresh medium induces cell cycle and conformation changes in p53, a tumour suppressor protein. Oncogene. 1990;5:1683–1690. [PubMed] [Google Scholar]
- 18.Hainaut P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993;53:4469–4473. [PubMed] [Google Scholar]
- 19.Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat. Rev. Cancer. 2001;1:68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 20.Joerger AC, Fersht AR. Structure–function–rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 21.Joerger AC, et al. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc. Natl. Acad. Sci. U. S. A. 2006;103:15056–15061. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu X, et al. Small molecule restoration of wildtype structure and function of mutant p53 using a novel zinc-metallochaperone based mechanism. Oncotarget. 2014;5:8879–8892. doi: 10.18632/oncotarget.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler JS, Loh SN. Zn(2+)-dependent misfolding of the p53 DNA binding domain. Biochemistry. 2007;46:2630–2639. doi: 10.1021/bi062106y. [DOI] [PubMed] [Google Scholar]
- 24.Puca R, et al. Restoring p53 active conformation by zinc increases the response of mutant p53 tumor cells to anticancer drugs. Cell Cycle. 2011;10:1679–1689. doi: 10.4161/cc.10.10.15642. [DOI] [PubMed] [Google Scholar]
- 25.Margalit O, et al. Zinc supplementation augments in vivo antitumor effect of chemotherapy by restoring p53 function. Int. J. Cancer. 2012;131:E562–E568. doi: 10.1002/ijc.26441. [DOI] [PubMed] [Google Scholar]
- 26.Puca R, et al. Reversible dysfunction of wild-type p53 following homeodomain-interacting protein kinase-2 knockdown. Cancer Res. 2008;68:3707–3714. doi: 10.1158/0008-5472.CAN-07-6776. [DOI] [PubMed] [Google Scholar]
- 27.Yu X, et al. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Y, et al. Thiosemicarbazones from the old to new: iron chelators that are more than just ribonucleotide reductase inhibitors. J. Med. Chem. 2009;52:5271–5294. doi: 10.1021/jm900552r. [DOI] [PubMed] [Google Scholar]
- 29.Finch RA, et al. Triapine (3-aminopyridine-2-carboxaldehyde-thiosemicarbazone): a potent inhibitor of ribonucleotide reductase activity with broad spectrum antitumor activity. Biochem. Pharmacol. 2000;59:983–991. doi: 10.1016/s0006-2952(99)00419-0. [DOI] [PubMed] [Google Scholar]
- 30.Yu Y, et al. Chelators at the cancer coalface: desferrioxamine to Triapine and beyond. Clin. Cancer Res. 2006;12:6876–6883. doi: 10.1158/1078-0432.CCR-06-1954. [DOI] [PubMed] [Google Scholar]
- 31.Richardson DR, et al. Dipyridyl thiosemicarbazone chelators with potent and selective antitumor activity form iron complexes with redox activity. J. Med. Chem. 2006;49:6510–6521. doi: 10.1021/jm0606342. [DOI] [PubMed] [Google Scholar]
- 32.Shao J, et al. A ferrous-Triapine complex mediates formation of reactive oxygen species that inactivate human ribonucleotide reductase. Mol. Cancer Ther. 2006;5:586–592. doi: 10.1158/1535-7163.MCT-05-0384. [DOI] [PubMed] [Google Scholar]
- 33.Murren J, et al. Phase I and pharmacokinetic study of Triapine, a potent ribonucleotide reductase inhibitor, administered daily for five days in patients with advanced solid tumors. Clin. Cancer Res. 2003;9:4092–4100. [PubMed] [Google Scholar]
- 34.Ma B, et al. A multicenter Phase II trial of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) and gemcitabine in advanced non-small-cell lung cancer with pharmacokinetic evaluation using peripheral blood mononuclear cells. Invest. New Drugs. 2008;26:169–173. doi: 10.1007/s10637-007-9085-0. [DOI] [PubMed] [Google Scholar]
- 35.Boeckler FM, et al. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bullock AN, et al. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene. 2000;19:1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 37.Colvin RA, et al. Cytosolic zinc buffering and muffling: their role in intracellular zinc homeostasis. Metallomics. 2010;2:306–317. doi: 10.1039/b926662c. [DOI] [PubMed] [Google Scholar]
- 38.Vinkenborg JL, et al. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat. Methods. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loh SN. The missing zinc: p53 misfolding and cancer. Metallomics. 2010;2:442–449. doi: 10.1039/c003915b. [DOI] [PubMed] [Google Scholar]
- 40.Blanden AR, et al. Synthetic metallochaperone ZMC1 rescues mutant p53 conformation by transporting zinc into cells as an ionophore. Mol. Pharmacol. 2015;87:825–831. doi: 10.1124/mol.114.097550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bleackley MR, Macgillivray RT. Transition metal homeostasis: from yeast to human disease. Biometals. 2011;24:785–809. doi: 10.1007/s10534-011-9451-4. [DOI] [PubMed] [Google Scholar]
- 42.Lambert JM, et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15:376–388. doi: 10.1016/j.ccr.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, et al. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 2013;41:6034–6044. doi: 10.1093/nar/gkt305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garufi A, et al. A fluorescent curcumin-based Zn(II)-complex reactivates mutant (R175H and R273H) p53 in cancer cells. J. Exp. Clin. Cancer Res. 2013;32:72. doi: 10.1186/1756-9966-32-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C, et al. Structures of p63 DNA binding domain in complexes with half-site and with spacer-containing full response elements. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6456–6461. doi: 10.1073/pnas.1013657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koster MI. p63 in skin development and ectodermal dysplasias. J. Invest. Dermatol. 2010;130:2352–2358. doi: 10.1038/jid.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- 48.Hainaut P, Mann K. Zinc binding and redox control of p53 structure and function. Antioxid. Redox Signal. 2001;3:611–623. doi: 10.1089/15230860152542961. [DOI] [PubMed] [Google Scholar]
- 49.Sykes SM, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solban N, et al. Targeted photodynamic therapy. Lasers Surg. Med. 2006;38:522–531. doi: 10.1002/lsm.20345. [DOI] [PubMed] [Google Scholar]