Abstract
Modular collaboration between iterative fungal polyketide synthases (IPKSs) is an important mechanism for generating structural diversity of polyketide natural products. Inter-PKS communication and substrate channeling are controlled in large by the starter unit acyl carrier protein transacylase (SAT) domain found in the accepting IPKS module. Here, we reconstituted the modular biosynthesis of the benzaldehyde core of the chaetoviridin and chaetomugilin azaphilone natural products using the IPKSs CazF and CazM. Our studies revealed a critical role of CazM's SAT domain in selectively transferring a highly reduced triketide product from CazF. In contrast, a more oxidized triketide that is also produced by CazF and required in later stages of biosynthesis of the final product is not recognized by the SAT domain. The structural basis for the acyl unit selectivity was uncovered by the first X-ray structure of a fungal SAT domain, highlighted by a covalent hexanoyl thioester intermediate in the SAT active site. The crystal structure of SAT domain will enable protein engineering efforts aimed at mixing and matching different IPKS modules for the biosynthesis of new compounds.
INTRODUCTION
The modular, assembly-line biosynthesis of polyketides and nonribosomal peptides exemplifies how Nature assembles complex natural products from simple precursors.1 While this elegant biosynthetic logic is almost universally applicable among bacterial polyketide synthases (PKSs), the eukaryotic fungal iterative polyketide synthases (IPKSs) have evolved to be more simplified and enigmatic.2 Each fungal IPKS consists of a single module, of which the catalytic domains embedded within are used iteratively to construct a finished product. Two major types of chemically distinct fungal IPKS modules have been discovered, including the non-reducing IPKSs (NR-PKSs) that produce aromatic products such as bikaverin;3 and the highly-reducing IPKSs (HR-PKSs) that produce highly reduced polyketides including the cholesterol-lowering lovastatin.4 The HR-PKSs are particularly complex as the domains responsible for α-methylation and β-reduction are used permutatively during each iteration with precise yet unresolved programming rules to yield structurally diverse products.
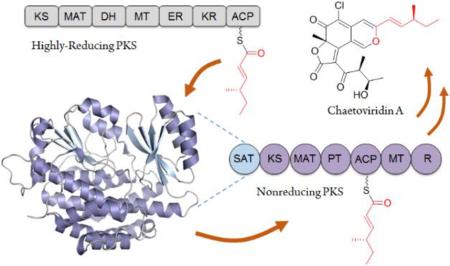
The collaboration between fungal PKS modules is an important mechanism to combine biosynthetic capabilities from different PKSs and represents the fungal equivalent of assembly line biosynthesis.4 The most frequently observed chemical modularity is the combination of an upstream HR-PKS and a downstream NR-PKS, in which a reduced polyketide synthesized by the HR-PKS is transferred to the NR-PKS as a starter unit via the starter-unit acyl carrier protein-transacylase (SAT) domain and further elongated.5 Such distribution of labor leads to a polyketide product that combines two or more chemically distinct substructures. Examples include the biosynthesis of norsolorinic acid (1),5a in which an NR-PKS accepts a hexanoyl chain from an upstream fatty acid synthase (FAS)-like module and synthesizes the anthraquinone core that is eventually morphed into aflatoxin.6 In the biosynthesis of the resorcylic acid lactones (RALs) such as hypothemycin (2),7 radicicol (3)8 and zearalenone (4)9 (Figure 1A), a highly reduced polyketide precursor synthesized by an HR-PKS is transferred to an NR-PKS and further elongated and cyclized by the NR-PKS to yield the RALs. Another large family of fungal polyketides that are derived from the chemically modular synthesis is the azaphilones such as asperfuranone (5)10 and chaetoviridin A (6).11 In these pathways, the NR-PKSs employ a C-terminal reductive (R) domain to release the products as benzylaldehydes, with the benzyl ring typically undergoing further oxidative transformations.
Figure 1.
Chemical modularity in building fungal polyketides. A) Representative molecules. The red portion indicates reduced products of upstream synthases that are transferred to downstream NR-PKSs. B) Proposed biosynthesis of chaetoviridin A (6) and chaetomugilin A (7). Domain abbreviations:ketosynthase (KS), malonyl-CoA:acyl carrier protein acyltransferase (MAT), dehydratase (DH), methyltransferase (MT), enoylreductase (ER), ketoreductase (KR), acyl carrier protein (ACP), starter-unit:ACP-transacylase (SAT), product template (PT), and reductive domain (R). 11-SNAC was synthesized and used to directly load the KS of CazM.
The modularity of the two PKS system is therefore an attractive feature that can be engineered to make new biosynthetic products. Indeed, successful production of analogs of 1, 5 and assorted RALs have been achieved through the mixing-and-matching of different non-reducing and reducing PKS modules, as well as using precursor-directed biosynthesis in which chemically synthesized reduced precursors are used to prime the functions of the NR-PKS module.5b,12–14 Notwithstanding the conceptual simplicity of the collaboration between modules, fundamental insights into how the modules communicate through protein-protein interactions and how substrates are recognized and transferred between modules are needed to enable more rational biosynthetic engineering strategies.
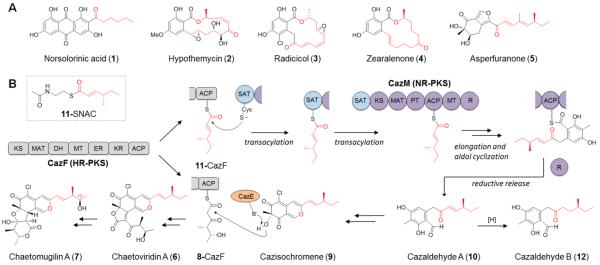
The caz biosynthetic cluster from Chaetomium globosum is responsible for the production of 6 and chaetomugilin A (7), and contains the NR-PKS CazM and HR-PKS CazF.11 In vitro assays confirmed CazF synthesizes the (4S,5R)-5-hydroxy-4-methyl-3-oxohexanoyltriketide 8, which is transferred to cazisochromene 9 by the acyltransferase CazE to yield 6.11 The biosynthesis of 9 is proposed to derive from cazaldehyde A (10), which appears to be the product of a collaborative HR-PKS and NR-PKS interaction. However, the presence of CazF as the lone HR-PKS in the caz gene cluster suggests that CazF may be involved in the biosynthesis of 10 in addition to 8. In this model, CazF is proposed to also synthesize the more reduced (S, E)-4-methyl-hex-2-enoyltriketide 11. The product 11 is attached to the ACP of CazF as a thioester and is transferred to CazM by the SAT domain to serve as the starter unit in the synthesis of 10. The proposed mechanism of the SAT-mediated chain transfer is shown in Figure 1B, in which the acyl chain 11 is first transacylated to the active site cysteine of the SAT, followed by transfer to the phosphopantetheinyl thiol on the ACP domain of CazM. If the collaborative interactions between CazF and CazM in the synthesis of 10 can be established, this would reveal an intriguing example of chemical modularity in fungal PKS programming where an HR-PKS is capable of synthesizing two different reduced portions of a natural product.15 This will also point to an important role of the CazM SAT domain, which must selectively transacylate 11 over the more polar 8 from the CazF ACP to the CazM ACP during the PKS crosstalk. Here, we report the reconstitution of the CazF/CazM interaction in the biosynthesis of 6, as well as the functional and structural characterization of the CazM SAT domain. This first X-ray crystal structure of the SAT domain from a fungal PKS will be highly useful for future biosynthetic engineering efforts.
RESULTS AND DISCUSSION
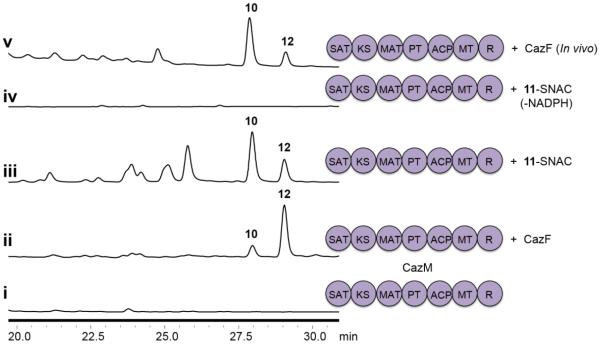
To reconstitute the CazF and CazM interactions, we first aimed to produce CazM recombinantly from Saccharomyces cerevisiae. The entire gene coding sequence of cazM was originally annotated to be three genes: CHGG_07645, CHGG_07646 and CHGG_07647 (Figure S1 and S2). cDNA sequencing revealed the presence of three extra nucleotides in the sequencing data; two in CHGG_07645 and one in CHGG_07646. Removal of these bases yielded one continuous protein-coding sequence. GenBank accession numbers for the corrected gDNA and mRNA sequences are KP764718 and KP771866, respectively. Once the correct uninterrupted coding sequence was in hand, CazM was expressed (299 kDa) as a C-terminal hexahistidine-tagged protein from S.cerevisiae BJ5464-NpgA16,17at 2.2 mg/L. To assay if CazM alone can turnover polyketide products using acetyl-CoA as a starter unit, the protein was incubated with 2 mM each of acetyl-CoA, malonyl-CoA, S-adenosyl-l-methionine (SAM) and NADPH. Because CazM contains a reductive domain at its C-terminus, NADPH was added to the reaction to aid in the reductive offloading of any polyketide product from the megasynthase. After incubation, no products were detected in the organic extract by LC-MS analysis suggesting that CazM requires a priming acyl substrate to initiate its PKS function (Figure 2).
Figure 2.
Modular CazF-CazM interaction and cazaldehyde production. HPLC analysis (λ=290 nM) of polyketide products when i) CazM was incubated with acetyl-CoA, malonyl-CoA, SAM and NADPH; ii) CazM was incubated with CazF, malonyl-CoA, SAM and NADPH; iii) CazM was incubated with 11-SNAC, malonyl-CoA, SAM and NADPH; iv) CazM was incubated with 11-SNAC, malonyl-CoA and SAM; and v) CazM was co-expressed with CazF in S. cerevisiae BJ5464-NpgA. Two benzaldehydes 10 and 12 were observed.
We next examined the modular collaboration between CazF and CazM by mixing equimolar amounts of CazM and CazF, which was previously expressed from S. cerevisiae,11 with 2 mM each of malonyl-CoA, SAM and NADPH. LC-MS analysis of the organic extract showed the formation of two predominant products containing identical UV profiles that are indicative of benzaldehydes. While one of the products had the same mass as the expected 10, the other compound differed in mass (m/z) by + 2 (Figure S8). To elucidate the structures of the two products, CazF and CazM were co-expressed in S. cerevisiae BJ5464-NpgA (Figure S7) and the two compounds were purified in sufficient quantity for complete 1D and 2D NMR spectroscopy (Figures S8–16). The two compounds were confirmed to be 10 (1 mg/L) and cazaldehyde B (12, 0.5 mg/L). While the formation of 10 was expected and confirms the successful collaboration between CazF and CazM, formation of 12, which underwent enoyl reduction in the reduced portion of the molecule, was unexpected. Compound 12 is most likely a shunt product, as no analog of 6 containing the corresponding reduced moiety has been isolated from C. globosum. A variety of reduction assays were performed and verified that the conversion of 10 to 12 is CazM-dependent and not spontaneous (Figure S17).
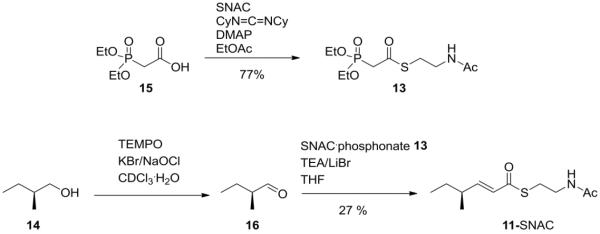
To verify that CazF synthesizes the acyl substrate 11-CazF, we prepared the N-acetyl cysteamine thioester (SNAC) 11 mimic, which is expected to directly prime the ketosynthase (KS) domain of CazM for further chain elongation.8 11-SNAC was prepared from a 3-step sequence using the SNAC-HWE reagent 13 and 2-(S)-methyl-1-butanol 14 (Scheme 1). Thioesterification of commercially available diethylphosphonoacetic acid 15 with SNAC provided the phosphonate thioester 13.18 Oxidation of 14 to the aldehyde 16 followed by a HWE reaction with 13 furnished the trans olefin 11-SNAC. Combining 2 mM 11-SNAC and 25 μM CazM in the presence of 2 mM each of malonyl-CoA, SAM and NADPH similarly led to the synthesis of both 10 and 12 (Figure 2). When NADPH was omitted from the reaction, no production of 10 or 12 could be detected indicating that CazM requires NADPH for the reductive offloading of its polyketide product. Altogether, these results confirm the product of the CazF module to be 11, which is transferred to CazM for four additional rounds of chain elongation, cyclization and reductive release (Figure 1B). The formation of the side product 12 using 11-SNAC also suggests that the extra reduction to yield 12 must take place on CazM, most likely by a previously unknown function of its R domain.
Scheme 1.
Synthesis of 11-SNAC using the SNAC- HWE reagent 13 and 2-(S)-methyl-1-butanol 14.
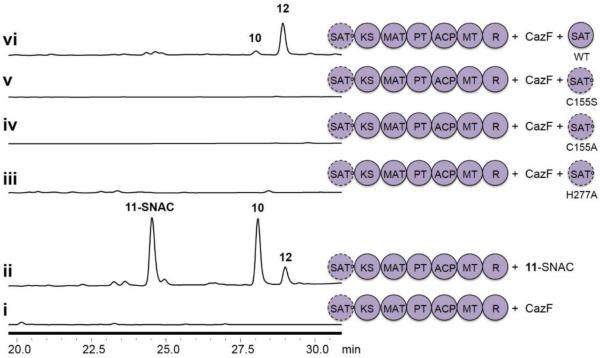
CazF was previously confirmed to also synthesize 8 that is recognized by CazE.11 However, in the presence of CazM, no product derived from 8 can be detected, hence pointing to a strict preference for the more reduced polyketide 11 by the SAT domain of CazM. The CazM SAT domain shows sequence similarity to AT domains from FASs and bacterial modular PKSs.19 The active site Cys155 found within the motif GxCxG and His277 are proposed to form the catalytic dyad5,14,19–21 and were targeted for mutagenesis. While CazM mutants C155A and C155S were not solubly expressed from S. cerevisiae BJ5464-NpgA, the H277A mutant was expressed at a comparable yield to wild-type CazM. In vitro assays with CazM H277A and CazF did not produce 10 and 12 (Figure 3). Precursor feeding of the CazM H227 mutant with 11-SNAC, however, restored the biosynthesis of 10 and 12, thereby confirming the remaining domains of CazM were fully functional. Therefore, a functional SAT domain is absolutely required for the modular synthesis of 10.
Figure 3.
Trans SAT complementation assays. HPLC analysis (λ= 290 nm) of 10 synthesized when i) CazM H277A was incubated with CazF, malonyl-CoA, SAM and NADPH; ii) CazM H277A was incubated with 11-SNAC, malonyl-CoA, SAM and NADPH; iii-vi)CazM H277A was incubated with CazF, malonyl-CoA, SAM, NADPH and standalone SAT domains.
To test if a standalone SAT domain can complement the CazM SAT0 mutant, we solubly expressed and purified the 46 kDa domain containing the linker region between SAT and KS domains from Escherichia coli BL21 (DE3). We reasoned that a functional, free-standing SAT domain will enable facile biochemical assays of mutants, as well as mix-and-match experiments towards analog generation. When equimolar ratios of CazM H277A, CazF and wild type SAT were combined, we detected the restored production of 10 and 12 at levels comparable to that of CazF and wild-type CazM (Figure 3). Using the in trans complementation system, we constructed the active site SAT mutants C155A, C155S and H277A. While it was expected that the C155A and H277A mutants would not complement the CazM SAT0 mutant, the C155S mutant was also surprisingly inactive (Figure 3). Interestingly, the SAT domains of the NR-PKSs Hpm3, Rdc1 and PKS13, which are responsible for the biosynthesis of the RALs 5, 6, and 7, respectively, all contain a GxSxG active site motif. Our complementation assay indicates that the active site thiol is absolutely essential for catalysis by CazM and the labile thioester intermediate is preferred over the more stable oxyester.4 Hence, the catalytic environment in the SAT domain is precisely tuned for the acyl transfer reactions shown in Figure 1B.
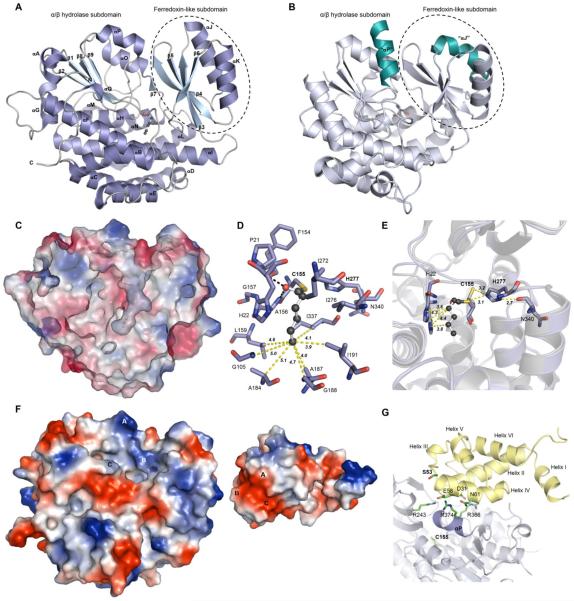
To gain atomic resolution into the acyl transfer step between CazF and CazM; especially the role of the cysteine and the acyl binding pocket, we solved the crystal structure of the standalone SAT domain to 1.6 Å using Single Anomalous Dispersion (SAD) (Table S5). The SAT crystallized in the space group P212121 with one monomer per asymmetric unit. Similar to structures of malonyl-CoA:ACP transacylases (MCAT) from bacterial fatty acid synthases,22–24 both trans-AT25 and cis-AT domains in PKSs19,26–30 (for example, see the avermectin loading AT domainin Figure 4B), the CazM SAT architecture is comprised of a large α/β-hydrolase core and a small ferredoxin-like subdomain containing a four-stranded antiparallel β-sheet and two distal α-helices (Figure 4A).
Figure 4.
Crystal structure of the CazM SAT domain. A) The monomeric SAT domain consists of a large subdomain containing an α/β-hydrolase-like fold and a small subdomain containing a ferredoxin-like fold. B)Structure of the avermectin AT loading domain Ave-AT° (PDB 4RL1).19 The two teal helices indicate a change in orientation compared to αJ and αP in the SAT structure and the active site S120 is shown as a stick. C) Electrostatic surface representation of the SAT domain showing a cross section of the cavity leading to the active site and bound hexanoyl. D) Residues located within 5 Å of bound hexanoyl and distances of residues lining the bottom of the active site cavity to C6 of hexanoyl (yellow dashed lines). Polar contacts are shown as black dashes. E) Active site residue comparison between the apo and hexanoyl-bound SAT. The apo SAT residues are shown in gray, whereas the hexanoyl-bound residues are shown in purple. Bound hexanoyl is shown in ball and stick form. F) Electrostatic surface maps of the apo-SAT (left) and CazF ACP (right) docking interfaces. Colors range from blue (positive) to white to red (negative). The homology model for CazF ACP was generated using I-TASSER34 and docking simulations were performed with PatchDock.35 TheACP is rotated 180° such that A–C on the SAT and ACP interfaces should match up. On the SAT, A is R374, B is R243 and C is R366. On the ACP, A is N61, B is E58 and C is D31.G) The homology model of CazF ACP (colored in yellow) was used as a ligand protein to dock with the apo SAT structure (colored in white). Helix αPof the SAT is highlighted in purple and surface residues that may be responsible for protein-protein interaction are shown as sticks. The active site Ser in the ACP and Cys in the SAT are in bold. All electrostatic surface maps were generated using PyMOL.32
We next performed co-crystallization of the SAT domain with either trans-2-hexenoic acid or hexanoyl-CoA, which served as mimics of the triketide substrate 11. Serendipitously, the co-crystallized structure of the SAT with hexanoyl-CoA showed well-defined electron density for a hexanoyl thioester at the active site Cys and was solved by molecular replacement to 1.4 Å (Figure 4A; Table S5). The active site cavity is located at the α/β-hydrolase and ferredoxin interface and was calculated to have a solvent accessible area of 238.49 Å3 using CASTp31 (Figure 4C). Predominantly hydrophobic amino acids line the cavity consistent with selectivity for a more reduced ligand (Figure 4D). These residues most likely dictate the preference of the SAT for the triketide 11 over the more oxidized 8, both of which can be synthesized by CazF (Figure 1B). Residues G105, L159, A184, G188, A187, I191, and I337 line the bottom of the cavity and are all located within 5 Å of the distal carbon in the hexanoyl chain. These residues therefore limit the length of the starter unit to a triketide and potentially a tetraketide. Additionally, the imidazole side chain of His22 is in close proximity to the β-carbon of the hexanoyl substrate and could further sterically contribute to the observed selectivity towards the reduced β-methylene carbon found in 11 over the more bulky β-ketone present in 8. Hence, capturing the thioester intermediate in the crystal structure not only confirmed the role of the active site cysteine, but also revealed factors that contribute to the SAT acyl substrate selectivity.
Comparison of the apo-SAT and hexanoyl-bound enzyme reveals a subtle conformational change in the active site. After the attachment of hexanoyl, the rotamer conformation of Cys155 located on N-terminus of αH changes and the loop connecting secondary structural elements β1 and αB moves toward the active site (Figure 4E). Catalytic His277 acts as a general acid/base catalyst in the transthioesterification reaction where Nε can deprotonate the thiol in Cys155. The carbonyl backbone of Asn340 helps polarize His277 and has a hydrogen bond distance of 2.7 Å to Nδ of His277 in both apo and hexanoyl-bound structures (Figure 4E). Based on the conserved FGDQ(S/T) sequence in PksA, it was proposed that glutamine is involved in formation of the oxyanion hole.5 The corresponding sequence from the CazM SAT consists of FAPHV and the backbone amides of His22 and Ala156 are most likely responsible for stabilizing the transition state as observed in the crystal structure.
While the majority of amino acids lining the acyl binding site are hydrophobic and select for a more lipid-like ligand, the basic residues R243, R366 and R374 located by the entrance to the channel may be involved in inter-and intramolecular docking with the acidic ACP domains from CazF and CazM, respectively. In the published MCAT and AT structures, positively charged surface residues have been suggested to facilitate ACP binding. To provide further insight into residues responsible for protein-protein interaction, a homology model of the CazF ACP was obtained using I-TASSER.34 The ACP domain from module 2 of 6-deoxyerythronolide B synthase (DEBS) (PDB: 2JU1)36 was selected as the top threading template. The CazF ACP homology model was used as a ligand molecule to dock with the apo SAT structure. The lowest energy docking models placed the ACP in the cleft between the α/β hydrolase and ferredoxin-like subdomains (Figure 4F). This placement is similar to docking models generated for the DEBS AT3 and ACP3,37 the iterative type I enediyne synthase ATDyn10 and ACPDyn,30 and the trans-AT kirromycin synthase KirCII and ACP5Kir.38 The orientation of the ACP homology model places acidic residues D31 and E58 in close proximity to SAT residues R243 and R366 (Figure 4G). R243 is positioned at the entrance to the catalytic cavity and its sidechain could make a salt bridge with the carboxylate group of either an Asp or Glu residue on the ACP. Interestinglyin the SAT structure, the orientation of helix αP is parallel to αO and αJ is parallel to αK. In the majority of MCAT and AT structures, such as the MCATs from Streptomyces coelicolorA3(2),22 Helicobacter pylori,23 and Xanthomanous oryzae pv. oryzae,24 and the ATs from the avermectin synthase,19 DEBS modules 326 and 5,27 the mycolic acid synthase PKS13,28 and zwittermicin synthase ZmaA,29 these helices are perpendicular to each other (Figure 4B and Figure S19). Extensive mutagenesis studies with the KirCII AT and ACP5kir domains performed by Williams and coworkers demonstrated that the perpendicular αP helix is the primary anchoring point for the ACP substrate. In the CazM SAT structure, αP contains Arg374 and the basic residue may aid in the inter- and intramolecular docking with negatively charged residues on ACP domains. The unusual parallel configuration of the helices in the SAT may be important in the recognition of two different ACP partners. Such interactions may be mapped from the covalent ACP trapping experiments that have recently found success in analyzing these weak protein-protein interactions.39
CONCLUSIONS
We reconstituted the modular interaction between a HR-PKS and NR-PKS in the biosynthesis of 6. Biochemical and structural insights into how the SAT domain facilitates module crosstalk and controls substrate selection were obtained. The structure of the first SAT domain can serve as a starting point for rational protein engineering efforts towards biosynthesis of new fungal polyketides.
EXPERIMENTAL SECTION
Synthesis of 11-SNAC
To a solution of 2-(S)-methyl-1-butanol 14 (0.039 mL, 0.34 mmol) in CDCl3 (1 mL) and D2O (1 mL) was added KBr (4.0 mg, 0.04 mmol), TEMPO (~0.6 mg) and NaOCl (0.25 mL of a 1.5 M solution, 0.38 mmol). The reaction was stirred at room temperature for 1 h and the progress of the reaction was monitored by NMR. When completed, the mixture was washed with 1 M HCl (1.0 mL, containing 16.0 mg potassium iodide/mL), 10 % Na2S2O3 (1.0 mL) and water (1.0 mL). The organic layer was dried and filtered to provide the aldehyde 16 as the only product (based on NMR) and was used immediately in the HWE reaction. The SNAC-HWE reagent 13 was prepared as previously reported from diethylphosphonoacetic acid 15.18 Compound 13 (81 mg, 0.30 mmol) was dissolved in dry THF (1.2 mL) and LiBr was added (46 mg, 0.50 mmol). Addition of trimethylamine (0.08 mL, 0.60 mmol) yielded the ylide as a white solid. Aldehyde 16 formed in the first step was added in CDCl3, and the reaction was stirred overnight at room temperature. The solvent was removed, and the residue was purified by silica gel chromatography (7:3, EtOAc-hexanes) to afford 11-SNAC (21.0 mg, 27%, two steps). [α]D; 3.96 (c = 1.45, CHCl3). IR (CHCl3 cast); 3290, 3077, 2963, 2875, 1663, 1632, 1552, 1459, 1438, 1404, 1286, 1052. 1H NMR (500 MHz, CDCl3, ppm); 6.83 (dd, 1H, J = 15.6, 7.8 Hz, =CH), 6.09 (dd, 1H, J = 15.6, 1.2 Hz, =CH), 5.38 (br s, 1H, NH), 3.47 (q, 2H, J = 6.2 Hz, -NHCH2), 3.09 (t, 2H, J = 6.3 Hz, SCH2), 2.25–2.19 (m, 1H, CH), 1.96 (s, 3H, Ac-CH3), 1.44–1.41 (m, 2H, CH3CH2), 1.06 (d, 3H, J = 6.7 Hz, CH-CH3), 0.88 (t, 3H, J = 7.5 Hz, CH3CH2) (Figure S5). 13C NMR (125 MHz, CDCl3, ppm); 191.4, 171.0, 152.5, 127.6, 40.7, 39.0, 29.6, 29.2, 24.1, 19.6, 12.5 (Figure S5). HR-ESIMS (C11H19NO2SNa)+; 252.1029 (calc), 252.1024 (found) (Figure S6).
CazM Reconstitution
To reconstitute an intron-free cazM, five overlapping pieces were constructed (Figure S2). Piece two (2,150 bp) and piece four (3,018 bp) were amplified from genomic DNA using primer pairs CazM_P2_F with CazM_P2_R and CazM_P4_F with CazM_P4_R, respectively (Table S1). Pieces one, three and five were initially annotated to contain introns and were amplified from cDNA. Total RNA was extracted using the RiboPure Yeast kit (Ambion) from a three-d old malt static culture (10 g/L malt extract, 4 g/L yeast extract, 10 g/L D-glucose) incubated at 28°C. Residual genomic DNA was digested with DNase (2 U/μL) (Invitrogen) at 37°C for four h. cDNA was created for pieces one, three and five using the reverse primers CazM_P1_R, CazM_P3_R, and CazM_P5_R, respectively. The cDNA was used as template for PCR and pieces one (1,313 bp), three (1,187 bp) and five (1,733 bp) were amplified using primer pairs CazM_P1_F with CazM_P1_R, CazM_P3_F with CazM_P3_R, and CazM_P5_F with CazM_P5_R, respectively. The five exons were co-transformed with the 2μ URA3 expression plasmid into S. cerevisiae BJ5464-NpgA using an S. c. EasyComp™ Transformation kit (Invitrogen) yielding plasmid pJWT-37. A single transformant was used to inoculate 3 mL SDc media (0.5 g Bacto Technical grade cassamino acids, 2 g Dextrose, and 88 mL Milli-Q water) supplemented with 1 mL adenine (40 mg/20 mL Milli-Q water), 1 mL tryptophan (40 mg/20 mL Milli-Q water) and nitrogen base (1.7 g Nitrogen Base without amino acids and without ammonium sulfate, 5 g ammonium sulfate and 100 mL Milli-Q water) and grown for 72 h with constant shaking at 28°C. A 1 mL aliquot of the seed culture was used to inoculate 1 L of YPD (10 g yeast extract, 20 g peptone and 950 mL Milli-Q water) supplemented with 1% dextrose and the culture was shaken at 28°C for 72 h. The cells were harvested by centrifugation (3,750 rpm at 4°C for 10 min) and the cell pellet was resuspended in 30 mL lysis buffer (50 mM NaH2PO4, 150 mM NaCl, 10 mM imidazole, pH 8.0). The cells were sonicated on ice in one min intervals until homogenous. To remove cellular debris, the homogenous mixture was centrifuged at 17,000 rpm for 1 h at 4°C. The supernatant was passed through a 0.45 μM filter and Ni-NTA agarose resin (2 mL) was added to the cleared lysate. The solution was incubated with gentle rotation at 4°C for 16 h. Soluble CazM was purified by gravity-flow column chromatography using increasing concentrations of imidazole in buffer A (50 mM Tris-HCl, 500 mM NaCl, 20–250 mM imidazole, pH 7.9). Purified protein was concentrated and buffer exchanged into Buffer B (50 mM Tris-HCl, 2 mM EDTA, 100 mM NaCl, pH 7.9) using an Ultracel 100,000 MWCO centrifugal filter (Amicon Inc.) and stored in 10% glycerol. The sequence of an intron-free cazM (Figure S3) was confirmed using primers CazM_seq1–CazM_seq11 and its protein concentration was calculated to be 2.2 mg/L using the Bradford assay with BSA as a standard.
CazM in vitro assays
To assess the activity of CazM and what substrates could prime the enzyme, 25 μM of protein was incubated at room temperature in a 100 μL reactions containing 2 mM S-adenosyl-l-methionine chloride (SAM), 2 mM NADPH, 2 mM malonyl-CoA and either 2 mM acetyl-CoA or 2 mM 11-SNAC in 100 mM phosphate buffer pH 7.4. After 8 h, the reactions were extracted two times with 200 μL 99% ethyl acetate:1% acetic acid and the organic layer was dried by speedvac. The extract was then dissolved in 20 μL MeOH and analyzed on a Shimadzu 2010 EV LC-MS with a Phenomenex Luna 5μ 2.0 × 100 mM C18 column using positive and negative mode electrospray ionization with a linear gradient of 5–95% MeCN:H2O over 30 min followed by 95% MeCN for 15 min and a flow rate of 0.1 mL/min. To assess whether the polyketide product could be released off the ACP without a reducing agent, the assay was repeated using 11-SNAC and omitting NADPH from the reaction. The reaction was incubated for 8 h at room temperature and extracted and analyzed as described above.
CazF was expressed and purified as published previously.11 The activity of CazM with its partnering HR-PKS CazF was analyzed by incubating 25 μM CazM, 25 μM CazF, 2 mM malonyl-CoA, 2 mM NADPH, 2 mM SAM in 100 mM phosphate buffer pH 7.4 in a total volume of 100 μL. The reaction was incubated at room temperature for 12 h and extracted two times with 200 μL 99% ethyl acetate:1% acetic acid. The organic layer was dried using a speedvac and the extract was resuspended in 20 μL MeOH and analyzed by LC-MS as mentioned above. The CazF-CazM activity reaction was repeated but the wild-type CazM was replaced with the CazM H277A mutant (construction described in the supplemental information). The reaction was incubated at room temperature for 12 h and then extracted and analyzed as described previously.
In vivo production of cazaldehyde A (10) and B (12)
Intact cazF was amplified using pJW07638 as template,11 AccuPrimePfx DNA polymerase (Invitrogen) and primers CazF_Trp_F and CazF_Trp_R (Table S1). The 7,863 amplicon was co-transformed with the 2μ Trp expression plasmid into S. cerevisiae BJ5464-NpgA using an S. c. EasyComp™ Transformation kit (Invitrogen) to yield plasmid pJWT-41. pJWT-41 and pJWT-37 were then co-transformed into S. cerevisiae BJ5464-NpgA.16,17 A single transformant was used to inoculate 3 mL SDc media (0.5 g Bacto Technical grade cassamino acids, 2 g Dextrose, and 89 mL Milli-Q water) supplemented with 1 mL adenine (40 mg/20 mL Milli-Q water) and 10 mL nitrogen base (1.7 g Nitrogen Base without amino acids, 5 g ammonium sulfate and 100 mL Milli-Q water) and grown for 72 h with constant shaking at 28°C. A 1 mL aliquot of the seed culture was used to inoculate 100 mL of YPD (1.0 g yeast extract, 2.0 g peptone and 95.0 mL Milli-Q water) supplemented with 1% dextrose and the culture was shaken at 28°C at 250 rpm. A 500 μL aliquot was extracted with ethyl acetate containing 1% acetic acid at d 1, 2, 3 and 7 (Figure S7). The organic layer was dried using a speedvac and the extract was resuspended in 20 μL MeOH. The crude extract was analyzed in positive and negative mode on a Shimadzu 2010 EV Liquid Chromatography Mass Spectrometer (LCMS) (gas flow set to 4.2 mL/min, drying temperature set to 200°C, and capillary voltage set to 2000 V). A Luna C18(2) 100 × 2 mm column was used with a flow rate of 0.1 mL/min and a linear solvent gradient of 5–95% MeCN:H2O containing 0.1% formic acid over a period of 30 min followed by 15 min of 95% MeCH:H2O.
Purification of cazaldehyde A (10) and B (12)
To isolate enough material for NMR characterization, 10 × 100 mL cultures were extracted after 48 h. The mycelium and media were extracted with ethyl acetate containing 1% acetic acid, dried over anhydrous MgSO4 and concentrated in vacuo. The crude extract was fractionated using reverse phase C18 flash column chromatography (Fisher Scientific, PrepSep C18 1 g/6 ml) with 20% MeCN:H2O, 40% MeCN:H2O, 60% MeCN:H2O, 80% MeCN:H2O, and 100% MeCN as the mobile phase. The fractions were analyzed in positive and negative mode on a Shimadzu 2010 EV LCMS using the same parameters as above. Compounds 10 and 12 eluted in the 40% MeCN:H2O fraction and were purified by HPLC (Beckman Coulter) using a Luna C18 column (250 × 10 mm; 5μm particle size) and an isocratic condition of 55% MeCN:H2O containing 0.1% TFA with a flow rate of 2.0 mL/min. The tR of 10 and 12 were 28.5 min and 29.5 min, respectively. Compounds 10 and 12 were isolated at 1 mg/L and 0.5 mg/L, respectively, and their chemical structures were elucidated from the spectroscopic data given in Tables S3 and S4, respectively.
Expression of the SAT domain
The SAT domain of CazM (Figure S3) did not contain any introns and was amplified from C. globosum gDNA using primer pair SAT_pHis8_F and SAT_pHis8_R, which contained EcoRI and NotI restriction sites, respectively. The PCR product was digested with EcoRI and NotI and ligated into the pHis8 expression vector40 yielding expression plasmid pJWT-49, which was transformed into E. coli BL21(DE3) cells. A single colony was used to inoculate 10 mL Luria-Bertani (LB) media supplemented with 35 μg/mL kanamycin and grown overnight with constant shaking at 37°C. A 5 mL aliquot of the seed culture was used to inoculate a 2 L shake flask containing 500 mL of Terrific Broth (per liter: 24 g yeast extract, 12 g tryptone and 4 mL glycerol) supplemented with kanamycin. The cells were shaken at 250 rpm at 37°C to an OD600 of 0.6. To induce protein expression, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM and the culture was shaken at 16°C for 22 h. The cells were harvested by centrifugation (3,750 rpm at 4°C for 25mins) and the cell pellet was resuspended in 25 mL lysis buffer (50 mM Tris-HCl, 2 mM EDTA, 2 mM DTT, 500 mM NaCl, 5 mM imidazole, pH 7.9). The cells were sonicated on ice in 30 s intervals until homogenous. To remove cellular debris, the homogenous mixture was centrifuged at 17,000 rpm for 30 min at 4°C. Ni-NTA agarose resin was added to the supernatant (1 mL) and the solution was incubated at 4°C with gentle agitation for four h. Soluble octahistidyl-tagged SAT was purified by gravity-flow column chromatography using increasing concentrations of imidazole in buffer A (50 mM Tris-HCl, 500 mM NaCl, 20 mM–250 mM imidazole, pH 7.9). Purified SAT was concentrated and buffer exchanged into Buffer B (50 mM Tris-HCl, 2mM EDTA, 100 mM NaCl, pH 7.9) using an Ultracel 30,000 MWCO centrifugal filter (Amicon Inc.) and stored in 10% glycerol. The protein concentration was calculated to be 43 mg/L by Bradford assay using BSA as a standard.
Standalone SAT in vitro studies
To determine if a standalone SAT domain could restore function to an SAT inactive CazM, 25 μM of CazM H277A was incubated with 25 μM of the standalone wild-type SAT domain in a 100 μL reaction containing 2 mM SAM, 2 mM NADPH, 2 mM mal-CoA and 25 μM CazF in 100 mM phosphate buffer pH 7.4. After 8 h, the reaction was extracted two times with 200 μL 99% ethyl acetate:1% acetic acid and the organic layer was dried by speedvac. The extract was then dissolved in 20 μL MeOH and analyzed on a Shimadzu 2010 EV LC-MS with a Phenomenex Luna 5μ 2.0 × 100 mM C18 column using positive and negative mode electrospray ionization with a linear gradient of 5–95% MeCN:H2O over 30 min followed by 95% MeCN for 15min and a flow rate of 0.1 mL/min. The assay was repeated using the standalone SAT C155S, C155A and H277A mutants.
Crystallization of the SAT domain
To obtain selenomethionine-substituted SAT, a single BL21(DE3) colony transformed with pJWT-49 was used to inoculate 5 mL LB media supplemented with kanamycin and grown for nine h with constant shaking at 37°C. A 2 mL aliquot was used to inoculate 50 mL of M9 minimal media in a 250 mL flask. The 50 mL culture was supplemented with kanamycin and shaken overnight at 37°C. A 2 L shake flask containing 500 mL of M9 minimal media was also shaken overnight at 37°C to pre-warm and aerate the media for expression. After overnight incubation, 5 mL of the 50 mL culture was used to inoculate 500 mL of pre-warmed M9 minimal media supplemented with kanamycin. The cells were shaken at 250 rpm at 37 °C to an OD600 of 0.5. To the 500 mL culture, 100 mg Lys, 100 mg Phe, 100 mg Thr, 50 mg Ile, 50 mg Leu, 50 mg Val and 60 mg of selenomethionine (Sigma Aldrich) was added. The culture was shaken at 250 rpm for 15 min at 37°C and then induced with 0.5 mM IPTG. After induction, the culture was shaken at 250 rpm at 16°C for 19 h. The cells were harvested as described above and the lysate was incubated with 1 mL Ni-NTA resin for 4.5 h at 4°C with gently agitation. Soluble selenomethionine-enriched SAT was purified by gravity-flow column chromatography using increasing concentrations of imidazole in buffer A (50 mM Tris-HCl, 500 mM NaCl, 20 mM–250 mM imidazole, pH 7.9).
Thrombin cleavage was used to remove the N-terminal His8-tag from the selenomethionine-labeled protein. To a three inch piece of 6–8 K molecular weight cut-off Spectra/Por dialysis membrane (Spectrum Labs), 72 mg of protein was incubated with 144μL thrombin (0.5 mg/mL, Sigma Aldrich) and incubated overnight at 4°C in 2 L dialysis buffer (50 mM Tris-HCl, 500 mM NaCl, 2 mM DTT, pH 7.9). Thrombin was removed by passing the protein mixture through benzamidine sepharose 4 fast-flow (GE Healthcare). The thrombin-free extract was then passed through Ni-resin to remove any remaining His-tagged SAT. The resulting flow-through was concentrated to 8.5 mg/mL using an Ultracel 30,000 MWCO centrifugal filter (Amicon Inc.) and 100 μL aliquots were dispensed and stored at −80 °C. Purity of the selenomethionine-enriched SAT and efficiency of the cleavage was evaluated by SDS-PAGE (Figure S4).
Crystals for Se-methionine-enriched apo SAT and hexanoyl-bound SAT crystals were obtained using the hanging-drop vapor-diffusion crystallization method. Mixing 1 μL of 10 mg/mL Semethionine-enriched protein with 1 μL crystallization buffer (0.1 M Tris pH 7.0 and 20 % PEG 8,000) and diffusing against a reservoir containing 500 μL of 0.1 M Tris pH 7.0 and 20 % PEG 8,000 produced rectangular plates after five d at 4°C. Initial hits were obtained at the UCLA Crystallization Facility using drops dispensed by a Mosquito liquid handling device (TTP Lab Tech). The rectangular plate crystals belong to the orthorhombic space group P212121 with cell dimensions of a= 43.13, b = 52.16 and c = 163.04 Å. To obtain hexanoyl-bound SAT, hexanoyl-CoA was added to a final concentration of 1.4 mM to a 1 μL:1 μL mixture of Se-methionine-enriched SAT protein and 500 μL reservoir solution (0.1 M Tris pH 7.5 and 20 % PEG 8,000). After seven d at 4°C, rectangular plate crystals were observed. The crystals belong to the orthorhombic space group P212121 with cell dimension of a = 45.67, b = 52.36 and c = 168.03 Å.
Data Collection for the SAT domain
All data was collected at 100K at APS-NECAT beamline 24-ID-C on a DECTRIS-PILATUS 6M pixel detector. Single crystals were mounted on CrystalCap HT Cryoloops (Hampton Research, Aliso Viejo, CA). Se-methionine-enriched SAT crystals were cryoprotected in mother liquor solution containing 33% glycerol and flash frozen in liquid nitrogen prior to data collection.
Data processing, phasing and refinement of the SAT domain
Unit cell dimensions, phasing and refinement statistics are presented in Table S5. Datasets of the Se-methionine Apo-SAT and hexanoyl-SAT crystals were collected at the Advanced Photon Source (APS) at the beamline 24-ID-C (NE-CAT). For the Se-methionine crystal a Single Anomalous Dispersion (SAD) data set was collected at 0.9792 Å. Data was indexed, integrated, and scaled using XDS/XSCALE.41
Phases for the Se-methionine apo SAT protein were obtained using the program HKL2MAP,42 which is the graphic interface for the SHELXC/D/E programs. Seven selenium sites were found by SHELXD.43 SHELXE44 assigned the hand, produced the first set of phases and performed solvent flattening. The final Figure of Merit was 0.817. The new set of phases were fed into the program BUCCANEER,45 which traced 95% of the protein residues. The Se-methionine apo SAT model was refined to 1.6Å. Final iterative rounds of model building and refinement were carried out using Coot46 and PHENIX47 with TLS refinement.48 More than 99.2% of the residues in the model were found in the favored regions of the Ramachandran plot by MOLPROBITY.33 There is one molecule of apo SAT in the asymmetric unit. The apo SAT model has clear electron density for all residues, except for C-terminal residues 397 to 414. Additionally, some side chains with no interpretable electron density were omitted in the final model.
The hexanoyl-bound SAT structure was solved starting with the apo SAT model using rigid body refinement followed by 12 cycles of manual model building and refinement using PHENIX.47 More than 98.7% of the residues in the model were found in the favored regions of the Ramachandran plot by MOLPROBITY.33 There is one molecule of hexanoyl-bound SAT in the asymmetric unit. The hexanoyl-bound SAT model has clear electron density for all residues, except for C-terminal residues 396 to 414.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the US NIH (1R01GM085128 and 1DP1GM106413) to Y.T; NSERC to J.C.V.; and JSPS (No. LS103) to K. W. We thank Mike Collazo at the UCLA-DOE X-ray Crystallization and Crystallography Core Facilities, which are supported by DOE Grant DE-FC02-02ER63421, and M. Cappel, K. Rajashankar, N. Sukumar, J. Schuermann, I. Kourinov and F. Murphy at NECAT beamlines 24-ID at APS, which are supported by grants from the NIH (5P41RR015301-10 and 8 P41 GM103403-10). Use of the APS is supported by the DOE under contract DE-AC02-06CH11357. J.M.W. thanks L'Oréal USA for Women in Science for a postdoctoral fellowship. NMR instrumentation was supported by the NSF equipment grant CHE-1048804.
Footnotes
Supporting Information Spectroscopic information and additional experimental details. This material is available free of charge via the Internet at http://pubs.acs.org. The atomic coordinates and structure factors for apo SAT and hexanoyl-bound SAT have been deposited in the Protein Data Bank under accession codes 4RO5 and 4RPM, respectively.
REFERENCES
- (1).Cane DE, Walsh CT, Khosla C. Science. 1998;282:63. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- (2).Cox RJ. Org. Biomol. Chem. 2007;5:2010. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- (3).Ma SM, Zhan J, Watanabe K, Xie X, Zhang W, Wang CC, Tang Y. J. Am. Chem. Soc. 2007;129:10642. doi: 10.1021/ja074865p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).(a) Chooi YH, Tang Y. J. Org. Chem. 2012;77:9933. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kennedy J, Auclair K, Kendrew SG, Park C, Vederas JC, Hutchinson CR. Science. 1999;284:1368. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- (5).(a) Crawford JM, Dancy BCR, Hill EA, Udwary DW, Townsend CA. Proc. Natl. Acad. Sci. USA. 2006;103:16728. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gao Z, Wang J, Norquay AK, Qiao K, Tang Y, Vederas JC. J. Am. Chem. Soc. 2013;135:1735. doi: 10.1021/ja4001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yu J, Chang P,K, Ehrlich KC, Cary JW, Bhatnager D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW. Appl. Environ. Micro. 2004;70:1253. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhou H, Qiao K, Gao Z, Meehan MJ, Li J, Dorrestein PC, Vederas JC, Tang Y. J. Am. Chem. Soc. 2010;132:4530. doi: 10.1021/ja100060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Zhou H, Qiao K, Gao Z, Vederas JC, Tang Y. J. Biol. Chem. 2010;285:41412. doi: 10.1074/jbc.M110.183574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zhou H, Zhan J, Watanabe K, Xie X, Tang Y. Proc. Nat. Acad. Sci. USA. 2008;105:6249. doi: 10.1073/pnas.0800657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CCC. J. Am. Chem. Soc. 2009;131:2965. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Winter JM, Sato M, Sugimoto S, Chiou G, Garg NK, Tang Y, Watanabe K. J. Am. Chem. Soc. 2012;134:17900. doi: 10.1021/ja3090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Liu T, Chiang Y-M, Somoza AD, Oakley BR, Wang CCC. J. Am. Chem. Soc. 2011;133:13314. doi: 10.1021/ja205780g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Liu T, Sanchez JF, Chiang Y-M, Oakley BR, Wang CCC. Org. Lett. 2014;16:1676. doi: 10.1021/ol5003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Huitt-Roehl CR, Hill EA, Adams MM, Vagstad AL, Li JW, Townsend CA. ACS Chem. Biol. 2015 doi: 10.1021/acschembio.5b00005. DOI: 10.1021/acschembio.5b00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).(a) Antosch J, Schaefers F, Gulder TAM. Angew. Chem., Int. Ed. Engl. 2014;53:3011. doi: 10.1002/anie.201310641. [DOI] [PubMed] [Google Scholar]; (b) Zhang G, Zhang W, Zhang Q, Shi T, Ma L, Zhu Y, Li S, Zhang H, Zhao YL, Shi R, Zhang C. Angew. Chem., Int. Ed. Engl. 2014;53:4840. doi: 10.1002/anie.201402078. [DOI] [PubMed] [Google Scholar]; (c) Li Y, Chen H, Ding Y, Xie Y, Wang H, Cerny RL, Shen Y, Du L. Angew. Chem., Int. Ed. Engl. 2014;53:7524. doi: 10.1002/anie.201403500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lee KK, Da Silva NA, Kealey JT. Anal. Biochem. 2009;394:75. doi: 10.1016/j.ab.2009.07.010. [DOI] [PubMed] [Google Scholar]
- (17).Keszenman-Pereyra D, Lawrence S, Twfieg ME, Price J, Turner G. Curr. Genet. 2003;43:186. doi: 10.1007/s00294-003-0382-7. [DOI] [PubMed] [Google Scholar]
- (18).Zhou H, Gao ZZ, Qiao KJ, Wang JJ, Vederas JC, Tang Y. Nat. Chem. Biol. 2012;8:331. doi: 10.1038/nchembio.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang F, Wang Y, Ji J, Zhou Z, Yu J, Zhu H, Su Z, Zhang L, Zheng J. ACS Chem. Biol. 2015;10:1017. doi: 10.1021/cb500873k. [DOI] [PubMed] [Google Scholar]
- (20).Crawford JM, Vagstad AL, Ehrlich KC, Townsend CA. Bioorg. Chem. 2008;36:16. doi: 10.1016/j.bioorg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Crawford JM, Vagstad AL, Whitworth KP, Ehrlich KC, Townsend CA. ChemBioChem. 2008;9:1019. doi: 10.1002/cbic.200700702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).(a) Keatinge-Clay AT, Shelat AA, Savage DF, Tsai S-C, Miercke LJW, O' Connell JD, III, Khosla C, Stroud RM. Structure. 2003;11:147. doi: 10.1016/s0969-2126(03)00004-2. [DOI] [PubMed] [Google Scholar]; (b) Arthur CJ, Williams C, Pottage K, Ptoskon E, Findlow SC, Burston SG, Simpson TJ, Crump MP, Crosby J. ACS Chem. Biol. 2009;4:625. doi: 10.1021/cb900099e. [DOI] [PubMed] [Google Scholar]
- (23).Zhang L, Liu W, Xiao J, Hu T, Chen J, Chen K, Jiang H, Shen X. Protein Sci. 2007;16:1184. doi: 10.1110/ps.072757307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Natarajan S, Kim J-K, Jung T-K, Doan TTN, Ngo H-P-T, Hong M-K, Kim S, Tan VP, Ahn SJ, Lee SH, Han Y, Ahn Y-J, Kang L-W. Mol. Cells. 2012;33:19. doi: 10.1007/s10059-012-2155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wong FT, Jin X, Mathews II, Cane DE, Khosla C. Biochemistry. 2011;50:6539. doi: 10.1021/bi200632j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tang Y, Chen AY, Kim C-Y, Cane DE, Khosla C. Chem. Biol. 2007;14:931. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tang Y, Kim C-Y, Mathews II, Cane DE, Khosla C. Proc. Natl. Acad. Sci. USA. 2006;103:11124. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Bergeret F, Gavalda S, Chalut C, Malaga W, Quémard A, Pedelacq J-D, Daffé M, Guilhot C, Mourey L, Bon C. J. Biol. Chem. 2012;287:33675. doi: 10.1074/jbc.M111.325639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Park H, Keavany BM, Dyer DH, Thomas MG, Forest KT. PLoS One. 2014;9:e110965. doi: 10.1371/journal.pone.0110965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Liew CW, Nilsson M, Chen MW, Sun H, Cornvik T, Liang Z-X, Lescar J. J. Biol. Chem. 2012;287:23203. doi: 10.1074/jbc.M112.362210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Lliang J. Nucl. Acid.Res. 2006;34:W116. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).DeLano WL. The PyMOL User's Manual. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- (33).Chen VBW, Arendall B, Headd JF, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. Acta Cryst. 2010;D66:12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).(a) Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. Nat. Methods. 2015;12:7. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Roy A, Kucukural A, Zhang Y. Nat. Protoc. 2010;5:725. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhang Y. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. Nucl. Acids.Res. 2005;33:W363. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Alekseyev VY, Liu CW, Cane DE, Puglisi JD, Khosla C. Protein Sci. 2007;16:2093. doi: 10.1110/ps.073011407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wong FT, Chen AY, Cane D,E, Khosla C. Biochemistry. 2010;49:95. doi: 10.1021/bi901826g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ye Z, Musiol EM, Weber T, Williams GJ. Chem. Biol. 2014;21:636. doi: 10.1016/j.chembiol.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Nguyen C, Haushalter RW, Lee DJ, Markwick PR, Bruegger J, Caldara-Festin G, Finzel K, Jackson DR, Ishikawa F, O'Dowd B, McGammon A, Opella SJ, Tsai S-C, Burkart MD. Nature. 2013;505:427. doi: 10.1038/nature12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. Biochemistry. 2000;39:890. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- (41).Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Pape T, Schneider TR. J. Appl. Crystallogr. 2004;37:844. [Google Scholar]
- (43).Uson I, Sheldrick G. Curr. Opin. Struct. Biol. 1999;9:643. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- (44).Sheldrick GM. Z.Kristallogr. 2002;217:644. [Google Scholar]
- (45).Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Acta. Crystallogr. D Biol. Crystallogr. 2011;67:235. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Emsley P, Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- (47).Adams PD, Grosse-Kunstleve RW, Hung LW, Loerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- (48).Painter J, Merritt EA. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.