Abstract
The immune microenvironment of tumor plays a critical role in therapeutic responses to chemotherapy. Cancer tissues are composed of a complex network between anti-tumor and pro-tumor immune cells and molecules; therefore a comprehensive analysis of the tumor immune condition is imperative for better understanding of the roles of the immune microenvironment in anticancer treatment response. In this study, we performed T cell receptor (TCR) repertoire analysis of tumor infiltrating T cells (TILs) in cancer tissues of pre- and post-neoadjuvant chemotherapy (NAC) from 19 breast cancer patients; five cases showed CR (complete response), ten showed PR (partial response), and four showed SD/PD (stable disease/progressive disease) to the treatment. From the TCR sequencing results, we calculated the diversity index of the TCRβ chain and found that clonal expansion of TILs could be detected in patients who showed CR or PR to NAC. Noteworthy, the diversity of TCR was further reduced in the post-NAC tumors of CR patients. Our quantitative RT-PCR also showed that expression ratio of CD8/Foxp3 was significantly elevated in the post-NAC tumors of CR cases (p=0.0032), indicating that antitumor T cells were activated and enriched in these tumors. Collectively, our findings suggest that the clonal expansion of antitumor T cells may be a critical factor associated with response to chemotherapy and that their TCR sequences might be applicable for the development of TCR-engineered T cells treatment for individual breast cancer patients when their tumors relapse.
Keywords: T cell receptor, tumor-infiltrating T cell, tumor immunity, breast cancer, biomarker
Introduction
Breast cancer is the most common female cancer in the United States (1). The mortality of breast cancer has been decreasing over the last two decades due to early detection and incremental improvements in systemic therapy. Adjuvant chemotherapy reduces the risk of recurrence and prolongs survival (2). Increasingly, these same chemotherapy regimens are used in the neoadjuvant setting for the patients with locally advanced breast cancer or high-risk biology (3–5). Regarding the clinical response to chemotherapy, the presence of pathological complete response (pCR) (defined as no cancer cells in the breast and axilla lymph nodes at the time of surgical resection) is associated with improved disease-free and overall survival (4,6). Conversely, patients with residual disease post-chemotherapy have a significantly higher risk of disease recurrence (6).
The presence of immune cells in tumor tissues has repeatedly been associated with response to chemotherapy (7). In particular, tumor-infiltrating T lymphocytes (TILs) are often observed in solid tumors and are associated with improved survival of cancer patients (8). However, the tumor immunity is governed in a complex network between antitumoral and pro-tumoral immune cells and molecules. For example, TILs consist of various lymphocyte subtypes with different roles such as elimination of cancer cells (e.g. CD8+ cytotoxic T cell (CTL) and CD4+ helper T cells) or protection of cancer cells from host immune attack [e.g. CD4+FoxP3+ regulatory T cell (Treg)]. In general, a higher level of CD8+ T cell infiltration is thought to contribute to better prognoses of patients with solid tumors (8). A meta-analysis focusing on breast cancer showed that higher TIL numbers in pre-treatment biopsies was associated with higher CR rates to neoadjuvant chemotherapy (NAC), while more FoxP3+ Treg cells in post-treatment breast tissue were associated with lower CR rates (9). However, most studies have simply quantified the number of T lymphocytes in tumor tissues, but their clonality and temporal changes in T cell repertoire during the treatment have not been well investigated. In this regard, a recent study revealed that clonal expansion of TILs was associated with better response to a PD-1 (programmed death-1) blockade (pembrolizumab) treatment in human melanoma patients (10). Therefore comprehensive characterization of TILs during cancer treatment is imperative to identify a predictive immune biomarker.
However, T lymphocytes are an extremely heterogeneous immune cell population, due to the molecular diversity in the T cell receptor (TCR) (11). Most T cells express TCR consisting of a heterodimer of α (TCRA) and β (TCRB) chains. Both TCRA and TCRB are generated through genetic rearrangement between V-J (for TCRA) and V-D-J (for TCRB) segments. In addition, since nucleotides are randomly deleted from V, (D), J exons and/or inserted between a V-J junction (for TCRA), or V-D and D-J junctions (for TCRB) during recombination (11–14), a huge variety in TCRs has hindered detailed characterization of the T cell-mediated adaptive immunity.
For the comprehensive analysis of immune functions of TILs, we have established TCR repertoire analysis employing next-generation sequencing technologies (15–20). In this study, we investigated expression levels of immune-related genes as well as the TCR repertoire of TILs in breast tumors in order to characterize the T-cell mediated antitumor immunity that is associated with clinical response to NAC.
Materials and methods
Clinical specimens
This study includes breast cancer patients who had been treated with neoadjuvant chemotherapy. Fresh tumor biopsy samples and surgically resected tumors were collected at Mayo Clinic (MC-1 to MC-9) as previously described (21), Tokushima Breast Care Clinic (TK-1 to TK-6), and Hyogo College of Medicine (HG-1 to HG-4). In case of patients who showed CR to chemotherapy, the tumor bed where tumor had been detected was collected. All human clinical specimens were obtained according to a study protocol and informed consent procedures approved by Institutional Review Board of each institution. Tumor samples were transferred to the University of Chicago and subsequently TCR repertoire was analyzed under the University of Chicago IRB protocol 13-0797. Detailed clinical information is described in Table I.
Table I.
Clinical information of 19 female breast cancer patients.
| Patient | Response | Age | Subtype | Ki67 (%) | Chemotherapy | |
|---|---|---|---|---|---|---|
| MC-1 | SD/PD | 59 | TNBC | 43 | Paclitaxel → Adriamycin/Cyclophosphamide | |
| MC-2 | PR | 35 | ER−/HER2+ | ND | Paclitaxel + Trastuzumab → Adriamycin/Cyclophosphamide | |
| MC-3 | SD/PD | 60 | Luminal B | 5 | Paclitaxel → Adriamycin/Cyclophosphamide | |
| MC-4 | PR | 48 | TNBC | 6 | Paclitaxel → Adriamycin/Cyclophosphamide | |
| MC-5 | SD/PD | 35 | ER+/HER2+ | 11 | Paclitaxel + Trastuzumab → Adriamycin/Cyclophosphamide | |
| MC-6 | CR | 70 | TNBC | ND | Paclitaxel → Adriamycin/Cyclophosphamide | |
| MC-7 | PR | 38 | TNBC | 75 | Paclitaxel → Adriamycin/Cyclophosphamide | |
| MC-8 | CR | 54 | TNBC | ND | Paclitaxel → Adriamycin/Cyclophosphamide | |
| MC-9 | PR | 53 | TNBC | 6 | Paclitaxel → Adriamycin/Cyclophosphamide | |
| Patient | Response | Age | ER (%) | PgR (%) | HER2 | Chemotherapy |
| TK-1 | SD/PD | 67 | 80 | 0 | 2+ | Epirubicin/Cyclophosphamide → Docetaxel |
| TK-2 | PR | 63 | 20 | 0 | 3+ | Epirubicin/Cyclophosphamide → Docetaxel + Trastuzumab |
| TK-3 | CR | 64 | 0 | 0 | 2+ | Epirubicin/Cyclophosphamide → Paclitaxel + Trastuzumab |
| TK-4 | PR | 56 | 1 | <1 | 2+ | Epirubicin/Cyclophosphamide → Docetaxel |
| TK-5 | PR | 46 | <1 | <1 | 3+ | Epirubicin/Cyclophosphamide → Paclitaxel + Trastuzumab |
| TK-6 | CR | 64 | 0 | 0 | 3+ | Epirubicin/Cyclophosphamide → Paclitaxel + Trastuzumab |
| HG-1 | PR | 48 | 2 | 1 | - | Epirubicin/Cyclophosphamide + Capecitabine → Docetaxel + Capecitabine |
| HG-2 | PR | 57 | 0 | 0 | - | Epirubicin/Cyclophosphamide + Capecitabine → Docetaxel + Capecitabine |
| HG-3 | PR | 71 | 90 | 10 | - | 5-Fluorouracil/Epirubicin/Cyclophosphamide → Docetaxel |
| HG-4 | CR | 40 | 0 | 0 | - | Epirubicin/Cyclophosphamide + Capecitabine → Docetaxel + Capecitabine |
CR, complete response; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; ND, not done; PgR, progesterone receptor; PR, partial response; SD/PD, stable disease/progressive disease; TNBC, triple-negative breast cancer.
RNA isolation and cDNA library preparation
The next generation sequencing technology-based TCR repertoire analysis was done as previously described (16,17). Briefly, total RNA was extracted from tumor sample using RNeasy mini kit (Qiagen, Valencia, CA, USA), and cDNA was synthesized by SMART (Switching Mechanism At 5′ end of RNA Transcript) cDNA library construction kit (Clontech, Mountain View, CA, USA). PCR reactions were performed to amplify compatible amplicon libraries of TCRβ with the Ion Torrent sequencing platform (Life Technologies, Carlsbad, CA, USA). A forward primer (5′-CCTCTCTATGGGCAGTCGGTGATTATCAACGCAGAGTGGCCAT-3′) was designed for the sequence of the SMART IV adaptor and the P1 adaptor. A reverse primer (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGCAGAAGGAACGATTCTGATGGCTCAAACACAGC-3′) was designed for the constant region of TCRβ, including the A1 adaptor sequence. Underline indicates the IonXpress barcode sequences. PCR reaction was performed as follows: 3 min at 94°C; 40 cycles of 30 sec at 94°C, 30 sec at 65°C, and 35 sec at 68°C. Amplified PCR products were purified using AMPure XP reagent (Beckman Coulter, Brea, CA, USA) and products of 300–900 bp were selected by Pippin Prep system (Sage Science, Beverly, MA, USA). Finally, we measured the concentration of size-selected PCR products by Agilent 2200 TapeStation System (Agilent, Santa Clara, CA, USA).
TCR repertoire sequencing
The sequencing libraries for TCRβ were amplified onto the proprietary Ion Sphere particles (ISP) by emulsion PCR using the Ion PGM™ Template OT2 400 kit and the OneTouch2 system (Life Technologies), as previously described (16,17). After enrichment and purification of the template-bound ISPs, the final ISP products were sequenced using the Ion 314v2 Chip, Ion Sequencing 400 kit, and Ion PGM Sequencer (Life Technologies), according to the manufacturer's instructions.
TCR repertoire analysis
Sequencing reads of TCRβ were produced in the FASTQ format and mapped to the reference sequences derived from the IMGT/GENE-DB (http://www.imgt.org), using the Bowtie2 aligner (Version 2.1.0) (22,23). After decomposition of V, D, and J segments, the CDR3 region was defined from a conserved cysteine encoded in the 3′ portion of the V segment to a conserved phenylalanine encoded in the 5′ portion of the J segment. The nucleotide sequences between the conserved TCR-V cysteine and TCR-J phenylalanine were extracted to determine the amino acid sequence of CDR3 region (24). Using the identified CDR3 clonotypes, we calculated the Simpson's diversity index to quantify the clonality of the TCRβ repertoires, as we previously described (15,18). For data visualization, we used the Excel program (Microsoft, Redmond, WA, USA) to generate bar graphs and pie charts of TCR repertoire.
Quantitative RT-PCR
To examine expression levels of immune signature genes, we conducted quantitative RT-PCR from the tumor and tumor bed cDNAs using TaqMan Gene Expression assays (Thermo Fisher Scientific, Carlsbad, CA, USA) and ABI ViiA 7 system (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions. The TaqMan probes for CD4, CD8, FoxP3, IDO1, PD-1 and PD-L1 genes were purchased from Life Technologies. Samples were run in triplicate reactions using one FAM and VIC-labeled probes per reaction. A probe for GAPDH was used for data normalization.
Statistical analysis
To compare patient groups of CR, PR, and SD/PD, we conducted the Mann Whitney test to examine any difference in the expression levels of immune-related genes. A p-value of <0.05 was considered statistically signifi-cant. Analysis was carried out using the Prism 6 software (GraphPad, San Diego, CA, USA).
Results
Antitumoral immune microenvironment in the tumors of CR patients
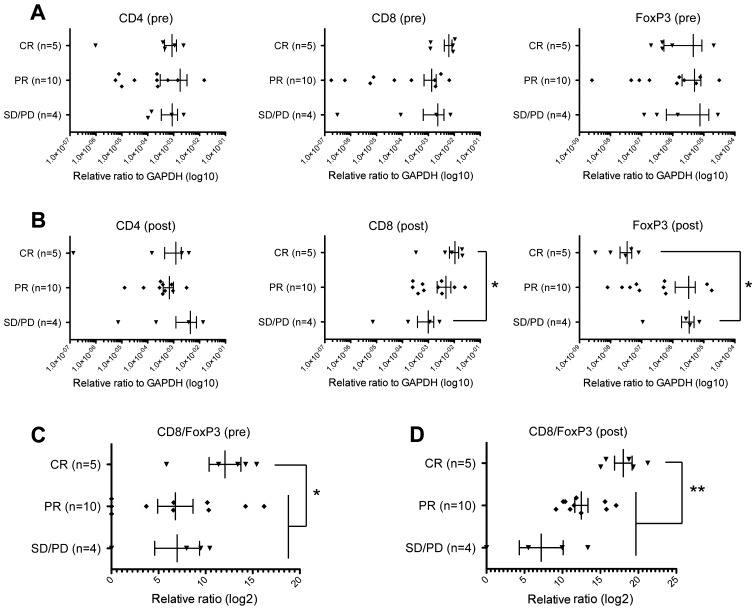
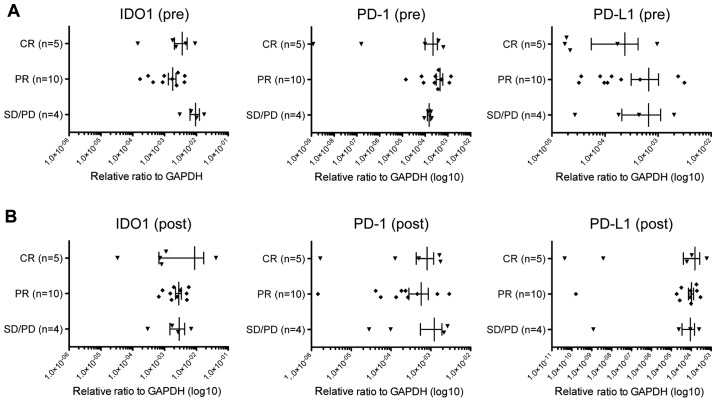
In this study, we obtained pre- and post-treatment cancer tissues from 19 breast cancer patients treated with NAC. Five cases showed complete response (CR), ten cases showed partial response (PR), and four had stable disease/progressive disease (SD/PD) to NAC (Table I). In the case of patients who showed CR, we collected tissue samples derived from the tumor bed. At first, we examined chemotherapy-induced changes of the immune microenvironment based on expression levels of CD4, CD8, and Foxp3. In pre-NAC tumors, we found no significant differences comparing CD4, CD8, and FoxP3 expression levels among the three patient groups (Fig. 1A). CD8 expression levels in the post-NAC tumor tissues of CR cases were significantly higher than those in SD/PD cases (p=0.0317, Mann Whitney test) (Fig. 1B), indicating that stronger infiltration of CD8+ CTLs during NAC contributed to effective elimination of cancer cells. On the contrary, expression levels of Foxp3 in post-NAC tissues were significantly lower in CR cases than in SD/PD cases (p=0.0159, Mann Whitney test) (Fig. 1B). Concordantly, the CD8/FoxP3 expression ratios in the tumors of CR cases were higher than those of PR or SD/PD cases before NAC (p=0.0349, Mann Whitney test) (Fig. 1C) as well as after NAC (p=0.0032, Mann Whitney test) (Fig. 1D). These results indicated that reduction of FoxP3+ Tregs by a certain mechanism might also contribute to achieving complete elimination of tumors and supporting the idea that immune responses in tumor tissues are likely to play critical roles even in chemotherapy response (25). We could not observe any difference in expression levels of other immune suppressive molecules, IDO1, PD-1 and PD-L1 among CR, PR, and SD/PD tumors (Fig. 2).
Figure 1.
Expression changes of CD4, CD8, and FoxP3 in tumors. Expression levels of CD4, CD8, and FoxP3 were graphed according to tumor samples of pre-chemotherapy (A) or post-chemotherapy (B). The x-axis indicates expression level of each gene relative to that of GAPDH. The expression ratios of CD8 and Foxp3 were graphed in the tumor samples of pre-chemotherapy (C) or post-chemotherapy (D). Vertical lines represent the means ± standard deviations, and asterisks indicate *p<0.05 or **p<0.01.
Figure 2.
Expression of IDO1, PD-1, and PD-L1 in tumors. Expression levels of IDO1, PD-1, and PD-L1 were graphed in the tumor samples of pre-chemotherapy (A) or post-chemotherapy (B). The x-axis indicates expression level of each gene relative to that of GAPDH. Vertical lines represent the means ± standard deviations.
Chemotherapy induces changes in TCR repertoire
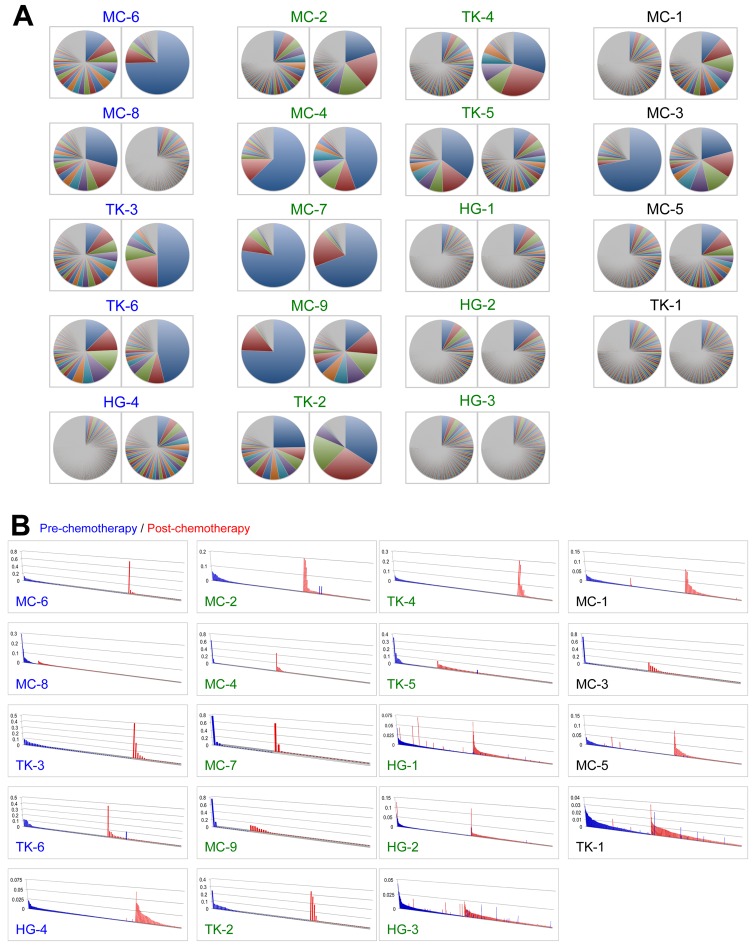
To further characterize infiltrated T cells in breast tumors, we performed TCR repertoire analysis using the next generation sequencing technologies. We isolated RNA from tumor tissue samples, generated TCRB cDNA, performed PCR amplification, and then sequenced with the Ion Torrent PGM sequencer. Subsequently, we performed quantitative analysis for the frequency of individual TCRB clonotypes with a unique V-D-J combination and CDR3 sequences, and compared TCR repertoire in tumors of pre- and post-NAC. Of note, the TCRB repertoire was markedly changed during NAC and we observed in some cases significant expansion of certain clonotypes through the NAC (Fig. 3A). Next, we focused on unique clonotypes with the read frequency of >0.001 and found that TILs with some unique clonotypes were significantly increased or decreased during the NAC (Fig. 3B), implying that cancer cell death by the neoadjuvant chemotherapy might influence the T cell population in tumors.
Figure 3.
The clonality of T lymphocytes in tumors of 19 breast cancer patients. (A) All identified CDR3 clonotypes of TCR β chain are presented in pie charts (left, pre-chemotherapy; right, post-chemotherapy). Since each pie chart was separately colored according to the CDR3 frequency ranks, the same color did not represent an identical CDR3 clonotype. (B) Comparison of TCRB clonotypes identified in tumors. Vertical axis indicates each of CDR3 clonotype (frequency >0.001) of TCRB in tumors of pre-chemotherapy (blue) and post-chemotherapy (red). CDR3 clonotypes are sorted by their frequency in tumors (y-axis).
Clonal expansion of antitumor T cells in the tumors of CR patients
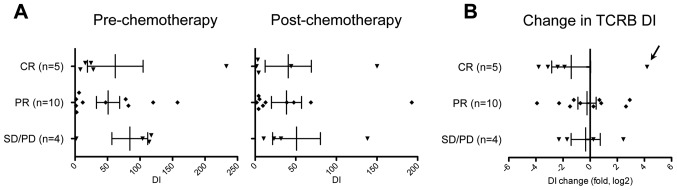
To assess the chemotherapy-induced changes in TIL population, we employed a statistical approach to measure significance of the temporal clonal changes in the identified TCR sequences. We calculated the diversity index (DI) of TCRB repertoire to represent clonality of T cells in cancer tissues (15). The DI values are reduced when T cell clones with certain TCRB are expanded while they are elevated when TCRB clonotypes are evenly distributed. The mean DI value for TCRB was 60.8 (range of 1.65–233) in tumor samples of pre-treatment. The mean DI was decreased to 41.8 (range of 1.77–193) by NAC. We subsequently compared DI values of TCRB according to NAC response (Fig. 4A), and found that DI values of TCRB seemed to be more strikingly decreased in CR cases than in PR or SD/PD cases (Fig. 4B) while one exceptional CR case, MC-8, showed an 18.2-fold increase (from 8.18 to 150) in DI values of TCRB by NAC treatment (possibly by antigen spreading). Our findings suggest that a certain population of TIL was expanded in the tumors showing strong response by NAC. Moreover, considering the high expression ratio of CD8/FoxP3 in CR tumors (Fig. 1D), these results collectively suggest that antitumor T cells were likely to be clonally expanded in the tumors of CR patients by the chemotherapy treatment.
Figure 4.
Changes in the diversity of TCRB by chemotherapy. (A) The diversity indexes (DIs) of TCRB were graphed according to patient's response to chemotherapy (left, pre-chemotherapy; right, post-chemotherapy). (B) Fold changes in the DI of TCRB were calculated and graphed according to the patient's response. Vertical lines represent the means ± standard deviations. Arrow indicates an exceptional case of CR, MC-8 sample.
Discussion
A tumor consists of a mixture of cancer cells and noncancerous cells, of which immune cells are considered to play critical roles in tumor progression/metastasis as well as in response to cancer treatment (25). For example, infiltration of T cells is frequently observed in breast cancer and generally a higher number of T cell populations (particularly CD8+ T cells) in tumors are associated with better response to chemotherapy and better prognosis (9). Accumulating evidence has shown that effective chemotherapy induces cancer cell death and in turn enhances tumor-specific immune responses (26). For example, dead cancer cells are recognized and phagocytosed by antigen presenting cells such as dendritic cells (DCs) which subsequently present tumor-specific antigens. Such chemotherapy-induced cancer cell death is very immunogenic because dead cancer cells release high-mobility group box 1 protein (HMGB1) that promotes the cross-presentation of tumor-derived antigens to T cells (27). The activated DCs migrate to the draining lymph node, where they activate naïve T cells to memory T cells or cytotoxic T cells acting on tumor antigens on cancer cells. These activated T cells are released to blood circulation, infiltrate into tumor area by chemotaxis, and then kill cancer cells that will become additional source of tumor antigens. In addition, chemotherapy was shown to induce intratumoral expression of certain chemokines (e.g. CCL2, CCL5, CXCL8, CXCL9, and CXCL10), allowing more T cells to infiltrate the tumor lesion (28).
In this study, we conducted expression analysis of immune-related molecules including CD4, CD8, and FoxP3, followed by detailed analysis of clonotypes of TILs by the deep TCR sequencing. At first, we examined whether infiltration of CD8+ T cells and FoxP3+ Treg cells in tumor was associated with clinical response to NAC. In concordance with a previous report (9), we found that the CD8/FoxP3 expression ratios in CR cases were higher than those in PR or SD/PD cases. This relationship was more obvious comparing the tumor bed and residual tumors post-NAC. The tumor tissues of CR cases showed the highest CD8/FoxP3 expression ratio with very low FoxP3 expression, implying that NAC shifted the anti-tumor/pro-tumor T cell balance to generate strong antitumor immunity. It might be caused by the release of higher levels of tumor antigens from dead cancer cells and by the secretion of certain chemokines/cytokines that preferentially expanded CD8+ antitumor T cells in the CR cases. It was also suggested that NAC might decrease systemic Tregs and then restore antitumor immunity of the pre-existing CTLs leading to high CR rate (29,30). Breast cancer patients with CR showed that the number of CD8+ T cells remained high in the tumor beds but that of FoxP3+ Treg cells significantly decreased after chemotherapy (31).
Secondly, our results revealed that NAC induced expansion of a certain population of TILs strongly in the tumor beds that displayed CR. This finding might be concordant to a recent paper reporting that significant clonal expansion of TILs was observed in human melanoma patients who showed better response to PD-1 blockade (pembrolizumab) treatment (10). When we consider these results with our observations in the gene expression analysis, chemotherapy might induce selective expansion of CD8+ antitumor CTLs in the tumor beds of CR cases. Moreover, since such a low TCR DI and high CD8/Foxp3 expression was associated with better relapse-free survival of muscle-invasive bladder cancer patients (20), the oligoclonal expansion of CD8+ antitumor TILs may contribute to long-lasting disease-free survival of breast cancer patients. However, at present, we are unable to validate the antitumor activity of individual TIL subclones that we have identified. Further study of TIL subclones using a single cell analysis is expected to identify the TCR α and β pairing and to assess their killing effects against target cancer cells (32).
Although a massive amount of genetic information has been accumulated in cancer research, it is critically necessary to understand patient's systemic and local immune conditions to better understand the detailed molecular mechanism in cancer therapy, not only immunotherapy, but also in radiation therapy, chemotherapy, and molecular-targeted therapy (33). Integration of whole exome/genome sequencing, transcriptome analysis, antigen presentation machinery, and comprehensive characterization of the immune environment will improve our understanding of cancer treatment and should also improve the quality of cancer treatment (33). Our previous TCR repertoire analyses have shown that the TCR sequencing is a rational approach to examine T cell-mediated immune response in a wide range of cancers (15–20). As shown by our results, we successfully isolated potent TCR sequences of antitumor T cells that may have high affinity to oncoantigens (oncogenic proteins with high immunogenicity) or neoantigens (derived from non-synonymous mutation in cancer cells). These TCR sequences can be applied to generate genetically-engineered T cells for adoptive T cell therapy, as examplified by a recent trial of the NY-ESO-1-specific TCR-engineered T cells that led to favorable clinical responses in 16 out of 20 multiple myeloma patients (34).
In conclusion, this study offers renewed evidence that clonal expansion of antitumor T cells is associated with better response to chemotherapy in breast cancer. Future studies in functional validation of the enriched antitumor TILs as well as identification of cancer-derived antigens corresponding to their TCRs will accelerate cancer immunotherapies, such as adoptive T cell therapy.
Acknowledgements
We thank Drs Rui Yamaguchi, Seiya Imoto, and Satoru Miyano of The University of Tokyo for developing the algorithm of TCR repertoire analysis and helpful support in data management. The super-computing resource (http://sc.hgc.jp/shirokane.html) was provided by Human Genome Center, the Institute of Medical Science, The University of Tokyo. This work was also supported by the following grants: Mayo Clinic Center for Individualized Medicine, Nadia's Gift Foundation, John P. Guider, The Eveleigh Family, Pharmacogenomics Research Network (U10GM 61388-15) (R.W., L.W., M.G.), R01 GM28157 (R.W.), R01 CA196648 (L.W.), CA 15083-40A2 (M.G.), George M. Eisenberg Foundation for Charities, and Mayo Clinic Breast SPORE P50CA 116201-9 (K.K., V.S., M.G.).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: An overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 3.Kaufmann M, Hortobagyi GN, Goldhirsch A, Scholl S, Makris A, Valagussa P, Blohmer JU, Eiermann W, Jackesz R, Jonat W, et al. Recommendations from an international expert panel on the use of neoadjuvant (primary) systemic treatment of operable breast cancer: An update. J Clin Oncol. 2006;24:1940–1949. doi: 10.1200/JCO.2005.02.6187. [DOI] [PubMed] [Google Scholar]
- 4.von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 5.Printz C. I-SPY 2 may change how clinical trials are conducted: Researchers aim to accelerate approvals of cancer drugs. Cancer. 2013;119:1925–1927. doi: 10.1002/cncr.28172. [DOI] [PubMed] [Google Scholar]
- 6.Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L, Valagussa P, et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 7.Denkert C, Loibl S, Noske A, Roller M, Müller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R, Hanusch C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 8.Jochems C, Schlom J. Tumor-infiltrating immune cells and prognosis: The potential link between conventional cancer therapy and immunity. Exp Biol Med (Maywood) 2011;236:567–579. doi: 10.1258/ebm.2011.011007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao Y, Qu Q, Zhang Y, Liu J, Chen X, Shen K. The value of tumor infiltrating lymphocytes (TILs) for predicting response to neoadjuvant chemotherapy in breast cancer: A systematic review and meta-analysis. PLoS One. 2014;9:e115103. doi: 10.1371/journal.pone.0115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi Q, Liu Y, Cheng Y, Glanville J, Zhang D, Lee JY, Olshen RA, Weyand CM, Boyd SD, Goronzy JJ. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scaviner D, Lefranc MP. The human T cell receptor alpha variable (TRAV) genes. Exp Clin Immunogenet. 2000;17:83–96. doi: 10.1159/000019128. [DOI] [PubMed] [Google Scholar]
- 13.Folch G, Lefranc MP. The human T cell receptor beta variable (TRBV) genes. Exp Clin Immunogenet. 2000;17:42–54. doi: 10.1159/000019123. [DOI] [PubMed] [Google Scholar]
- 14.Clambey ET, Davenport B, Kappler JW, Marrack P, Homann D. Molecules in medicine mini review: The αβ T cell receptor. J Mol Med Berl. 2014;92:735–741. doi: 10.1007/s00109-014-1145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang H, Yamaguchi R, Liu X, Daigo Y, Yew PY, Tanikawa C, Matsuda K, Imoto S, Miyano S, Nakamura Y. Quantitative T cell repertoire analysis by deep cDNA sequencing of T cell receptor α and β chains using next-generation sequencing (NGS) Oncoimmunology. 2015;3:e968467. doi: 10.4161/21624011.2014.968467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Venkataraman G, Lin J, Kiyotani K, Smith S, Montoya M, Nakamura Y, Kline J. Highly clonal regulatory T-cell population in follicular lymphoma - inverse correlation with the diversity of CD8(+) T cells. Oncoimmunology. 2015;4:e1002728. doi: 10.1080/2162402X.2014.1002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang M, Yew PY, Hasegawa K, Ikeda Y, Fujiwara K, Fleming GF, Nakamura Y, Park JH. Characterization of T cell repertoire of blood, tumor, and ascites in ovarian cancer patients using next generation sequencing. Oncoimmunology. 2015;4:e1030561. doi: 10.1080/2162402X.2015.1030561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yew PY, Alachkar H, Yamaguchi R, Kiyotani K, Fang H, Yap KL, Liu HT, Wickrema A, Artz A, van Besien K, et al. Quantitative characterization of T-cell repertoire in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2015;50:1227–1234. doi: 10.1038/bmt.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K, Hazama S, Yamaguchi R, Imoto S, Takenouchi H, Inoue Y, et al. Characterization of T cell repertoire in tumor tissues and blood in advanced colorectal cancers through deep T cell receptor sequencing. Oncol Lett. doi: 10.3892/ol.2016.4465. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhury NJ, Kiyotani K, Yap KL, Campanile A, Antic T, Yew PY, Steinberg G, Park JH, Nakamura Y, O'Donnell PH. Low T cell receptor diversity, high somatic mutation burden, and high neoantigen load as predictors of clinical outcome in muscle-invasive bladder cancer. Eur Urol Focus. 2015 Oct 8; doi: 10.1016/j.euf.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Ellingson MS, Hart SN, Kalari KR, Suman V, Schahl KA, Dockter TJ, Felten SJ, Sinnwell JP, Thompson KJ, Tang X, et al. Exome sequencing reveals frequent deleterious germline variants in cancer susceptibility genes in women with invasive breast cancer undergoing neoadjuvant chemotherapy. Breast Cancer Res Treat. 2015;153:435–443. doi: 10.1007/s10549-015-3545-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: A comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33:D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolotin DA, Shugay M, Mamedov IZ, Putintseva EV, Turchaninova MA, Zvyagin IV, Britanova OV, Chudakov DM. MiTCR: Software for T-cell receptor sequencing data analysis. Nat Methods. 2013;10:813–814. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 25.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 26.Park JH. Immune microenvironment to predict response of cancer chemotherapy and radiotherapy. In: Nakamura Y, editor. Immunopharmacogenomics. Springer; Tokyo: 2015. pp. 143–155. [DOI] [Google Scholar]
- 27.Gebremeskel S, Johnston B. Concepts and mechanisms underlying chemotherapy induced immunogenic cell death: Impact on clinical studies and considerations for combined therapies. Oncotarget. 2015;6:41600–41619. doi: 10.18632/oncotarget.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong M, Puaux A-L, Huang C, Loumagne L, Tow C, Mackay C, Kato M, Prévost-Blondel A, Avril MF, Nardin A, et al. Chemotherapy induces intratumoral expression of chemokines in cutaneous melanoma, favoring T-cell infiltration and tumor control. Cancer Res. 2011;71:6997–7009. doi: 10.1158/0008-5472.CAN-11-1466. [DOI] [PubMed] [Google Scholar]
- 29.Oda N, Shimazu K, Naoi Y, Morimoto K, Shimomura A, Shimoda M, Kagara N, Maruyama N, Kim SJ, Noguchi S. Intratumoral regulatory T cells as an independent predictive factor for pathological complete response to neoadjuvant paclitaxel followed by 5-FU/epirubicin/cyclophosphamide in breast cancer patients. Breast Cancer Res Treat. 2012;136:107–116. doi: 10.1007/s10549-012-2245-8. [DOI] [PubMed] [Google Scholar]
- 30.Matsushita N, Pilon-Thomas SA, Martin LM, Riker AI. Comparative methodologies of regulatory T cell depletion in a murine melanoma model. J Immunol Methods. 2008;333:167–179. doi: 10.1016/j.jim.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladoire S, Arnould L, Apetoh L, Coudert B, Martin F, Chauffert B, Fumoleau P, Ghiringhelli F. Pathologic complete response to neoadjuvant chemotherapy of breast carcinoma is associated with the disappearance of tumor-infiltrating foxp3+ regulatory T cells. Clin Cancer Res. 2008;14:2413–2420. doi: 10.1158/1078-0432.CCR-07-4491. [DOI] [PubMed] [Google Scholar]
- 32.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura Y. Challenges and future directions of immunopharmacogenomics. In: Nakamura Y, editor. Immunopharmacogenomics. Springer; Tokyo: 2015. pp. 159–162. [DOI] [Google Scholar]
- 34.Rapoport AP, Stadtmauer EA, Binder-Scholl GK, Goloubeva O, Vogl DT, Lacey SF, Badros AZ, Garfall A, Weiss B, Finklestein J, et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat Med. 2015;21:914–921. doi: 10.1038/nm.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]