Smooth muscle cells (SMCs) from distinct embryonic origins differ in their ability to support human umbilical vein endothelial cell (HUVEC) network formation. Lateral mesoderm-derived SMCs best supported endothelial cell network complexity and survival in vitro, in part through increased expression of midkine. A lineage-specific approach might be beneficial for vascular tissue engineering and therapeutic revascularization.
Keywords: Embryological origin, Lineage specific, Smooth muscle cells, Endothelial cells, Network formation
Abstract
Vascular smooth muscle cells (SMCs) from distinct anatomic locations derive from different embryonic origins. Here we investigated the respective potential of different embryonic origin-specific SMCs derived from human embryonic stem cells (hESCs) to support endothelial network formation in vitro. SMCs of three distinct embryological origins were derived from an mStrawberry-expressing hESC line and were cocultured with green fluorescent protein-expressing human umbilical vein endothelial cells (HUVECs) to investigate the effects of distinct SMC subtypes on endothelial network formation. Quantitative analysis demonstrated that lateral mesoderm (LM)-derived SMCs best supported HUVEC network complexity and survival in three-dimensional coculture in Matrigel. The effects of the LM-derived SMCs on HUVECs were at least in part paracrine in nature. A TaqMan array was performed to identify the possible mediators responsible for the differential effects of the SMC lineages, and a microarray was used to determine lineage-specific angiogenesis gene signatures. Midkine (MDK) was identified as one important mediator for the enhanced vasculogenic potency of LM-derived SMCs. The functional effects of MDK on endothelial network formation were then determined by small interfering RNA-mediated knockdown in SMCs, which resulted in impaired network complexity and survival of LM-derived SMC cocultures. The present study is the first to show that SMCs from distinct embryonic origins differ in their ability to support HUVEC network formation. LM-derived SMCs best supported endothelial cell network complexity and survival in vitro, in part through increased expression of MDK. A lineage-specific approach might be beneficial for vascular tissue engineering and therapeutic revascularization.
Significance
Mural cells are essential for the stabilization and maturation of new endothelial cell networks. However, relatively little is known of the effect of the developmental origins of mural cells on their signaling to endothelial cells and how this affects vessel development. The present study demonstrated that human smooth muscle cells (SMCs) from distinct embryonic origins differ in their ability to support endothelial network formation. Lateral mesoderm-derived SMCs best support endothelial cell network complexity and survival in vitro, in part through increased expression of midkine. A lineage-specific approach might be beneficial for vascular tissue engineering and therapeutic revascularization.
Introduction
Mural cells are essential for the stabilization and maturation of new endothelial cell networks [1–3]. In human embryonic development, nascent vascular networks emerge through vasculogenesis and angiogenesis. Subsequent stabilization and maturation of this network is achieved through recruitment of mural cells, involving pathways such as the platelet-derived growth factor-BB (PDGF-BB) and transforming growth factor-β (TGF-β) [4]. The critical role of mural cells has been demonstrated in PDGF-BB-deficient mice, which developed capillary micro-aneurysms in the absence of mural cells [3]. Furthermore, abnormal vascular morphogenesis, including changes in microvessel architecture, endothelial cell quantity and morphology, and transendothelial permeability, has been described in PDGF-BB- and PDGF-Rβ-deficient mice [5].
Both cell-cell and paracrine mechanisms are thought to regulate vessel maturation but remain poorly characterized [6, 7]. In particular, relatively little is known about the effect of the developmental origins of mural cells on their signaling to endothelial cells and how this affects vessel development. Mural cells have a variety of embryonic origins, which might affect their functional abilities [8, 9]. In chick embryos, isolated smooth muscle cells (SMCs) from the neural crest and the mesoderm exhibit different growth and transcriptional responses to TGF-β [9]. Historical in vivo data from chick embryos showed that SMC responses are not environment- but lineage-specific. These experiments demonstrated that SMCs derived from the nodose placode had superior ability to replace ablated cardiac neural crest SMCs compared with those of mesodermal origin, corroborating the presence of lineage-specific functionality in different SMC populations [10].
Regenerative cardiovascular medicine is making rapid progress in the treatment of ischemic or dysfunctional tissues through direct cell injection and tissue-engineered constructs. Adequate vascularization of cellular grafts after transplantation is critical for long-term perpetuation of homeostasis and functionality for which mural cells are paramount. However, to date, this topic remains poorly addressed, and translational approaches would benefit substantially from a better understanding of mural cell functionality. Furthermore, it remains unknown how the embryonic origin of human embryonic stem cell (hESC)-derived SMCs influences vascular network development and the signaling events involved in this process. We have previously generated a model of lineage-specific SMC development from hESCs or human induced pluripotent stem cells (iPSCs), allowing for derivation of mural cells from the lateral mesoderm (LM), paraxial mesoderm (PM), and neuroectoderm (NE) [11].
We report for the first time that endothelial network formation is substantially determined by the embryonic origin of smooth muscle cells. Gene expression profile and microarray data exhibited a distinct upregulation of angiogenesis- or vasculogenesis-related genes in LM-derived SMCs. We elucidated the functional role of midkine (MDK) as one mediator of this effect and demonstrated that a small interfering RNA (siRNA)-mediated knockdown results in substantially impaired vascular network formation in LM-derived SMC cocultures.
Materials and Methods
hESC Culture and Differentiation
hESCs (H9; WiCell Research Institute, Madison, WI, http://www.wicell.org) were maintained in a chemically defined medium (CDM-bovine serum albumin [BSA]) containing activin-A (10 ng/ml; R&D Systems, Minneapolis, MN, http://www.rndsystems.com) and fibroblast growth factor 2 (FGF2; 12 ng/ml; R&D Systems), as previously described [12]. Chemically defined medium consisted of Iscove’s modified Dulbecco’s medium (250 ml; Thermo Fisher Scientific Life Sciences, Waltham, MA, http://www.thermofisher.com), Ham’s F12 (250 ml; Thermo Fisher Scientific Life Sciences), Pen/Strep (5 ml; Thermo Fisher Scientific Life Sciences), insulin (350 μl; Roche Holding AG, Basel, Switzerland, http://www.roche.com), transferrin (250 μl; Roche Holding AG), chemically defined 100× lipid concentrate (5 ml; Thermo Fisher Scientific Life Sciences), and monothioglycerol (20 μl; Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). Differentiation to intermediate lineages and smooth muscle cells was performed, as previously described, in CDM-polyvinyl alcohol (PVA; 1 mg/ml; Sigma-Aldrich) [13]. In brief, early mesoderm differentiation was started with a combination of CDM-PVA, FGF2 (20 ng/ml), LY294002 (10 μM; Sigma-Aldrich), and bone morphogenetic protein 4 (BMP4; 10 ng/ml; R&D Systems) for 1.5 days. Consequently, either lateral mesoderm differentiation was started in CDM-PVA, FGF2 (20 ng/ml), and BMP4 (50 ng/ml) for 3.5 days or paraxial mesoderm differentiation was started in CDM-PVA, FGF2 (20 ng/ml), and LY294002 (10 μM; Sigma-Aldrich) for 3.5 days. To induce neuroectoderm differentiation, the cells were cultured in CDM-PVA, FGF (12 ng/ml), and SB431542 (10 μM; Tocris Bioscience, Bristol, United Kingdom, http://www.tocris.com) for 7 days. For smooth muscle cell differentiation, LM, PM, and NE cells were resuspended as single cells in CDM-PVA, PDGF-BB (10 ng/ml; PeproTech, Rocky Hill, NJ, http://www.peprotech.com), and TGF-β1 (2 ng/ml; PeproTech) for 6 days.
Generation of mStrawberry-Expressing hESCs
A lentiviral-based vector was used to allow the transduction of two reporter genes, encoding luciferase and the fluorochrome mStrawberry, driven by the constitutively active CAGGS promoter. To produce a transduced hESC line, P64 H9 hESCs were passaged 24 hours before lentiviral transduction and plated onto gelatin (0.1%)-coated six-well plates. Lentiviral transduction was performed by the addition of polycation protamine sulfate (10 μg/ml) and lentivirus at a multiplicity of infection (MOI) of 5, or polycation protamine sulfate alone as a negative control, to approximately 2 × 105 cells maintained in CDM with activin-A (10 ng/ml) and FGF2 (12 ng/ml). The cells were maintained in culture media for 48 hours, and the media were changed daily. At 24 hours before dissociating the transduced cells, the culture medium was supplemented with 10 μM rho kinase inhibitor. At 72 hours after transduction, the cells were incubated in TrypLE Express (Thermo Fisher Scientific Life Sciences) for 5 minutes at 37°C and then dissociated by gentle pipetting. The cells were plated onto a 10-cm plate that had been coated with 0.1% porcine gelatin and contained 1 × 105 irradiated mouse embryonic fibroblasts as feeder cells (feeder). The cells were maintained in Dulbecco’s modified Eagle’s medium /F12 medium, containing 20% knockout serum replacement and FGF2 (4 ng/ml) supplemented with rho kinase inhibitor (10 μM). The colonies that appeared were allowed to expand over a 7-day time period. Successfully transduced colonies were identified by fluorescence microscopy, picked, and expanded until a stable lentiviral luciferase mStrawberry reporter hESC line could be maintained in a feeder-free culture system using the chemically defined medium, as described previously.
Generation of Green Fluorescent Protein-Expressing HUVECs
A lentiviral-based vector was used to allow the transduction of the reporter gene, EGFP, driven by the constitutively active EF-1α promoter. To produce green fluorescent protein (GFP)-expressing HUVECs, P6 HUVECs were passaged 24 hours before lentiviral transduction and plated onto a T75 flask that had been coated with 0.1% porcine gelatin. Lentiviral transduction was performed by the addition of polycation protamine sulfate (10 μg/ml) and lentivirus at an MOI of 10, or polycation protamine sulfate alone as a negative control, to approximately 5 × 105 cells maintained in human large vessel endothelial cell growth medium (Cellworks, Buckingham, U.K., http://www.cellworks.co.uk). Cells were maintained in culture media for 48 hours after transduction, and the media were changed daily. This protocol produced a HUVEC line with 99.5% GFP+ cells, as determined by flow cytometry.
Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA extraction was performed using the Gen Elute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich), and 250 ng of RNA was converted to complementary DNA (cDNA) by reverse transcription using the Maxima First Strand cDNA Synthesis Kit as per the manufacturer’s instructions. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed with the Applied Biosystems 7900HT Fast Real-Time PCR System using SYBR Green PCR Master Mix (Thermo Fisher Scientific Life Sciences) and primers of the target genes. The obtained values were normalized to the housekeeping genes GAPDH and PBGD in the same run. Primer sequences are depicted in supplemental online Table 1.
Immunocytochemistry
The cells were fixed in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100/phosphate-buffered saline (PBS). This was followed by blocking in 3% BSA/PBS for 45 minutes at room temperature. Primary antibody incubations were performed at 4°C overnight. After incubation with primary antibody, the cells were washed and incubated with Alexa Fluor conjugated secondary antibodies for 45 minutes at room temperature. Finally, the cells were stained with 4′,6-diamidino-2-phenylindole for 10 minutes to visualize the nuclei. Images were acquired on a Zeiss LSM700 using ZEN software (Carl Zeiss AG, Jena, Germany, http://www.zeiss.com). A detailed description of the antibodies and dilutions used is provided in supplemental online Table 2.
Enzyme-Linked Immunosorbent Assay
For enzyme-linked immunosorbent assay (ELISA) of D6 PT smooth muscle cell supernatant, the Human Midkine ELISA Development Kit (PeproTech) was used as per the manufacturer’s instructions. Plate readings were obtained using a 2030 Multilabel Reader (VICTOR ×3; Perkin Elmer/Caliper Life Sciences, Hopkinton, MA, http://www.perkinelmer.com).
Three-Dimensional Cocultures
A schematic of three-dimensional (3D) coculture is presented in supplemental online Figure 1A. GFP-HUVECs and D6 PT SMCs derived from mStrawberry H9s were dissociated, centrifuged, and resuspended in warm CDM-PVA. The cells were counted, and cell combinations were prepared in 15-ml Falcon tubes, containing either 1.6 × 105 HUVECs alone or a combination of 1.6 × 105 HUVECs and 3.2 × 104 SMCs (ratio 5:1). Cell mixtures were then centrifuged and resuspended in 18 µl of ice-cold HUVEC medium (catalog number M-2953; TCS Cellworks, Buckingham, U.K., http://www.cellworks.co.uk). The tubes were spun down and the supernatant aspirated and resuspended in 18 μl of ice-cold HUVEC medium. Subsequently, 20 μl of ice-cold Matrigel was added and mixed with the cell suspension. Finally, 10 μl of each sample was added to each well of a prechilled Ibidi Angiogenesis µ-Slide (catalog number 81506; Ibidi, Martinsried, Germany, http://www.ibidi.com) in technical triplicates. The slides were incubated for 45 minutes at 37°C before adding (volume) warm HUVEC medium to each well. The medium was refreshed every second day throughout the experiment.
3D Paracrine Assay
A schematic of the 3D paracrine assay is presented in supplemental online Figure 1B. For paracrine 3D cocultures, the eight outer wells of each nine-well section of two Ibidi 2 × 9-well µ-slides were coated with Matrigel and incubated at 37°C for 30 minutes. GFP-HUVECs and D6 PT SMCs derived from mStrawberry H9s were dissociated, centrifuged, and resuspended in CDM-PVA. The cells were counted (6.4 × 105 HUVECs or 1.35 × 105 SMCs) and transferred to separate 15-ml Falcon tubes and centrifuged. HUVECs were resuspended in 78 μl of ice-cold HUVEC medium and mixed with 80 μl of ice-cold Matrigel. Next, 32 μl of HUVEC suspension was added to the middle wells of the 2 × 9-well Ibidi µ-slide. Each SMC sample was resuspended in 540 μl of warm HUVEC medium, and 60 μl was added to each of the eight outer wells of the µ-slide. For the HUVEC-alone sample, medium containing no cells was added to the eight outer wells. The slide was incubated for 1 hour at 37°C to allow the cells to attach and the Matrigel to solidify. Subsequently, the medium was aspirated from all the SMC wells of the μ-slide, and the chamber was filled with 600 μl of warm endothelial medium. The medium was refreshed every second day throughout the experiment.
Confocal Microscopy
For cocultures and paracrine assays, images were acquired in technical triplicates per each biological replicate. For the siRNA-mediated knockdown of MDK, quantification was further optimized. Three images were taken per each technical replicate, corresponding to nine images per biological replicate per condition. The images were obtained using a Zeiss LSM700 using ZEN software.
Image Quantification and Analysis
For quantification, three technical replicates, consisting of one image each, were analyzed per biological replicate. The LSM700 files were analyzed in a blinded fashion using ImageJ software (NIH, Bethesda, MD, http://www.imagej.nih.gov/ij/). For the total network area, the images were converted to binary images, and the threshold was set until exclusive visualization of the network area was obtained. The values obtained reflect micrometers squared. For quantification of the total cord length, lines were manually drawn on the network, their length measured, and the values converted to micrometers. Branch points were counted manually after transformation of all files to binary images. The data presented as bar charts consist of three independent biological replicates, each of which is the average of three analyzed images (technical replicates).
TaqMan Array
cDNA was synthesized from total RNA samples as described for qRT-PCR. For PCR amplification, the TaqMan master mix was combined with the cDNA, loaded on the TaqMan plate, and the array run, using the Applied Biosystems 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific Life Sciences).
Microarray Analysis
The genes analyzed in the TaqMan Human Angiogenesis Array were extracted from our previously performed microarray data set on smooth muscle cells derived from the lateral-plate mesoderm, paraxial mesoderm, and neuroectoderm after 12 days of PDGF-BB and TGF-β1 treatment [11]. Heat maps of relative gene expression were generated with Perseus (MaxQuant, version 1.4.1.3; Max Planck Institute of Biochemistry, Planegg, Germany, http://www.biochem.mpg.de/en). Array data sets are available from the ArrayExpress microarray data repository under accession number E-MTAB-781.
siRNA-Mediated Knockdown of MDK
MDK silencer select siRNA was synthesized by Ambion (s8625; Thermo Fisher Scientific Life Sciences). As a negative control, scrambled siRNA was used (silencer select negative control siRNA; Ambion; Thermo Fisher Scientific Life Sciences). SMCs were transfected with 10 nM siRNA twice, once on day 11 and once on day 12 (supplemental online data).
Statistical Analysis
The results are expressed as the mean ± SEM of at least three biological replicates of independent experiments. Statistical comparisons were performed using either Student’s t test for two groups of samples or one-way analysis of variance, with Bonferroni’s post hoc test in the case of multiple group comparisons, using GraphPad Prism software. Measuring two-sided significance, p < .05 was considered statistically significant.
Results
Generation of Embryonic Origin-Specific Intermediates and SMCs for 3D Coculture With HUVECs
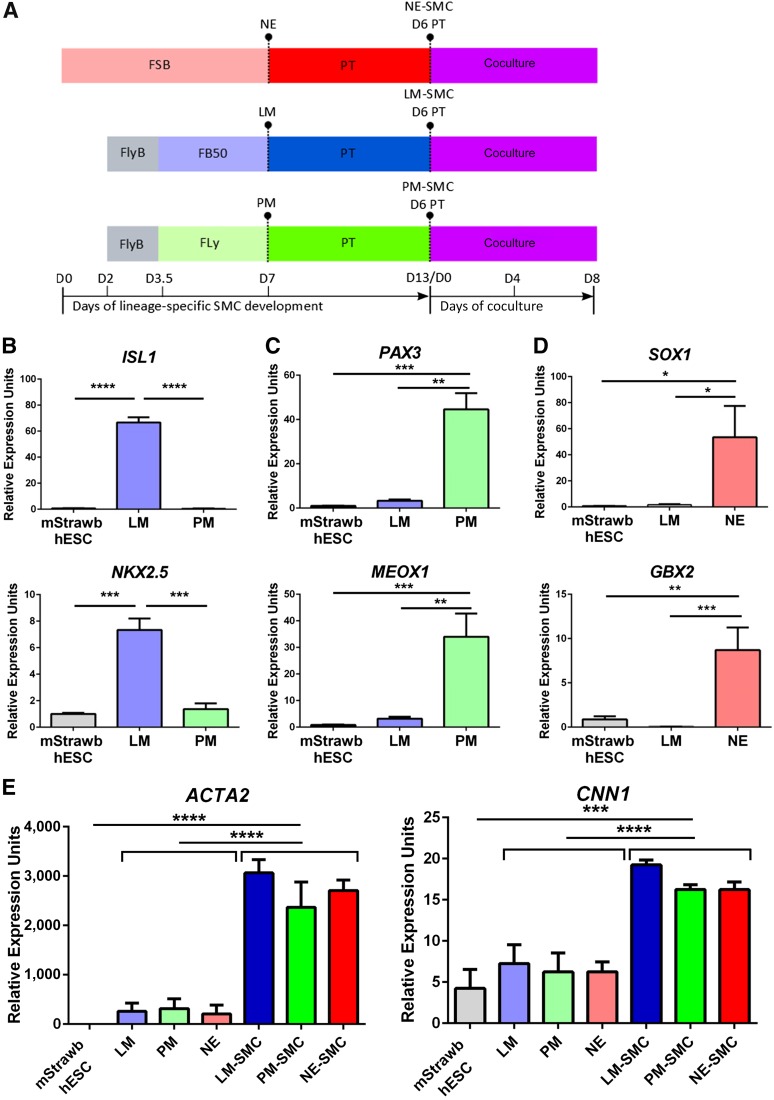
We first generated mStrawberry-expressing hESCs and GFP-expressing HUVECs (supplemental online Fig. 1A, 1B). mStrawberry-expressing hESCs were differentiated to LM, PM, and NE and subsequently treated with PDGF-BB and TGF-β1 (designated as PT) to induce SMC differentiation. On day 13 of the protocol, 3D Matrigel cocultures and paracrine assays were set up, in which HUVECs were used to model endothelial network formation. The respective potential of lineage-specific mural cells to support vasculogenesis was tested using embryonic origin-specific SMCs obtained after 6 days of differentiation in PT (Fig. 1A). To validate that lineage-specific SMCs were generated and used for the Matrigel vasculogenesis assays, lineage-specific marker expression was examined at the mRNA and protein levels. High expression of the LM markers ISL1 and NKX 2.5 was observed specifically in the LM population, as was the specific expression of the PM markers PAX3 and MEOX1 in PM (Fig. 1B, 1C). Expression of NE markers SOX1 and GBX2 was seen preferentially in NE, as shown previously (Fig. 1D). In accordance with the mRNA data, LM, PM, and NE, respectively, and specifically expressed ISL1, PAX3, and SOX1 at the protein level, as shown by immunocytochemistry (supplemental online Fig. 2A–2C). To show that SMCs were generated from the intermediate lineages, the expression of SMC markers CNN1 and ACTA2 was demonstrated at the mRNA and protein level before the cocultures were started (Fig. 1E; supplemental online Fig. 3A–3C). In conclusion, embryonic origin-specific SMCs were generated as shown by stage- and lineage-specific marker expression.
Figure 1.
Generation of embryonic origin-specific SMC populations for a three-dimensional coculture model. (A): Schematic representation of the in vitro differentiation of embryonic origin-specific SMCs. NE was differentiated from hESCs using FSB treatment for 7 days. hESCs were also differentiated to early mesoderm in FlyB for 36 hours and subsequently to LM in FB50 or PM in Fly for 3.5 days. For further differentiation into vascular SMCs, each of the three intermediate lineages was subjected to PT treatment for 6 days and then cocultured with HUVECs. (B–D): Validation of the identity of the intermediate lineages, LM, PM, and NE, using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis of lineage-specific markers (n = 3). (E): SMC marker expression in D6 PT-treated SMCs by qRT-PCR (∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; ∗∗∗∗, p < .0001; n = 3 independent biological replicates). Abbreviations: D, day; FB50, fibroblast growth factor 2 plus bone morphogenetic protein 4; FLy, fibroblast growth factor 2 plus LY294002; FlyB, fibroblast growth factor 2 plus LY294002 plus bone morphogenetic protein 4; FSB, fibroblast growth factor 2 plus SB431542; hESC, human embryonic stem cell; HUVECs, human umbilical vein endothelial cells; LM, lateral mesoderm; mStrawb, mStrawberry; NE, neuroectoderm; PM, paraxial mesoderm; PT, platelet-derived growth factor-BB plus transforming growth factor-β; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; SMC, smooth muscle cell.
LM-Derived SMCs Best Support HUVEC Network Formation in a 3D Coculture Model
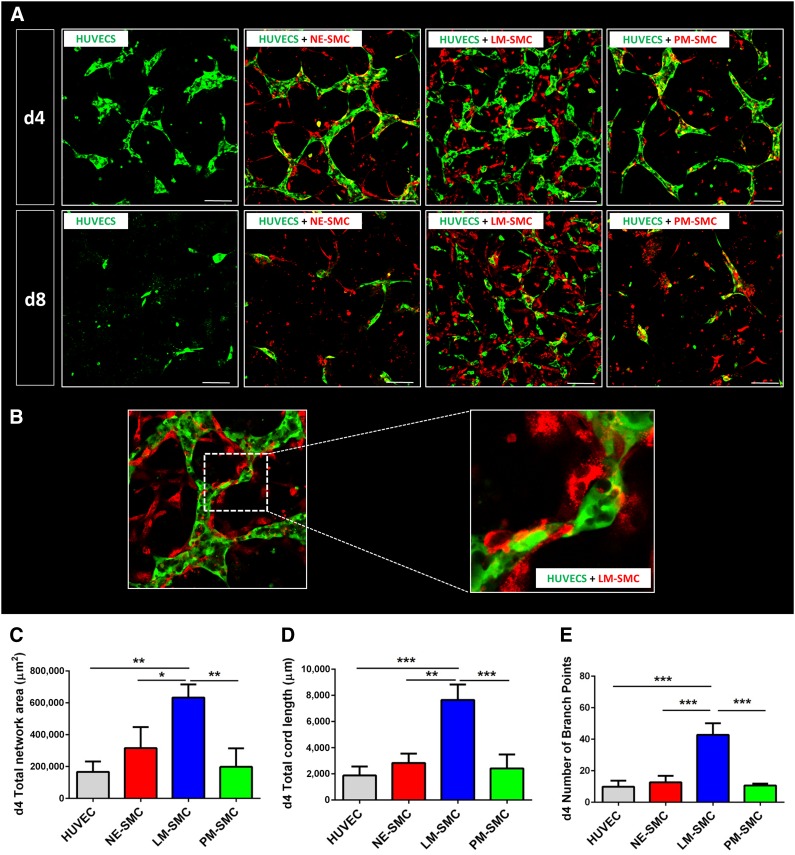
To investigate the effects of each SMC lineage on endothelial network formation, 3D cocultures were performed (supplemental online Fig. 4A). To show that mural cells improve endothelial network stability and complexity, a HUVEC-only assay was performed and compared with the other SMC groups. On day 4 of the coculture protocol, LM-derived SMCs accounted for the best endothelial network formation as reflected by the larger network area, higher total cord length, and greater number of branch points compared with PM- and NE-derived SMCs and HUVECs alone (Fig. 2A). The effect of the differential ability of SMCs to support endothelial network complexity first became apparent on day 2, peaked on day 4, and could be observed throughout all the time points until day 8. The absolute potential of SMCs to provide endothelial network stability became most evident on day 8, when the networks consisting of HUVECs alone had disintegrated entirely. As the difference in vasculogenic potency among the three lineages was best reflected on day 4, this time point was chosen for further endothelial network analysis and quantification. Self-assembly of SMCs and ECs was particularly robust in the LM-derived SMC group, with the SMCs providing physical support through wrapping around the endothelial tubes (Fig. 2B). Quantification of this effect demonstrated that LM-derived SMCs cocultures had the highest total endothelial network area on days 4 and 8 compared with the two other SMC lineages and to HUVECs alone (Fig. 2C). Furthermore, endothelial networks supported by SMCs of the LM origin accounted for a higher total cord length and a higher number of branch points than networks cocultured with the other two SMC lineages, at both time points (Fig. 2D, 2E, respectively).
Figure 2.
LM-derived SMCs best support HUVEC network formation in a three-dimensional coculture model. (A): Confocal images of HUVEC (green) cocultures with the three respective SMC types (red) on days 4 and 8 of the coculture protocol. (B): SMCs provide physical support to developing networks, wrapping around the endothelial network. (C): Quantification of total network area. (D): Quantification of total cord length. (E): Quantification of number of branch points. Quantitative data are shown for day 4 of the coculture (∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; n = 3 independent biological triplicates; scale bars = 100 μm). Abbreviations: d, day; HUVEC, human umbilical vein endothelial cell; LM, lateral mesoderm; NE, neuroectoderm; PM, paraxial mesoderm; SMC, smooth muscle cell.
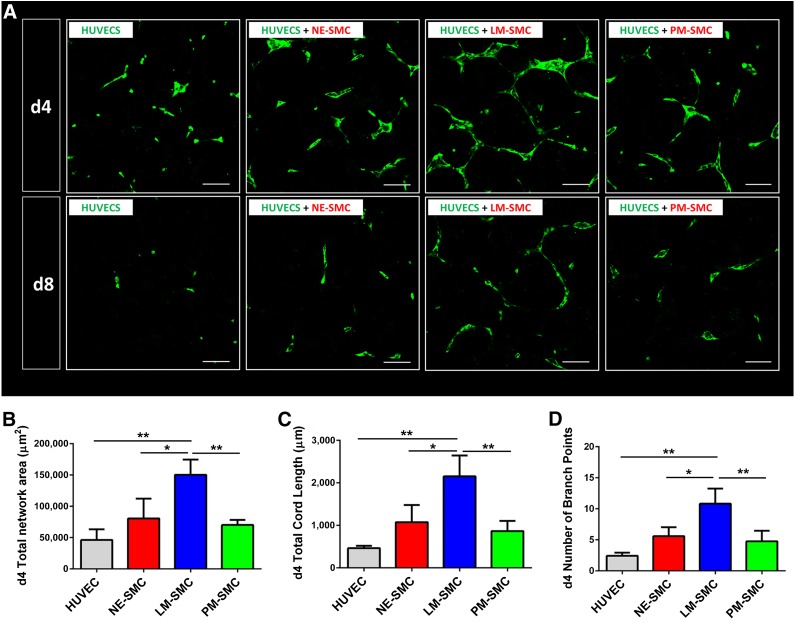
To elucidate whether this effect was paracrine in nature, 3D paracrine assays were performed. HUVECs were plated in the central well of a nine-well chamber and SMCs in the adjacent eight wells, with walls separating the wells only half way to the top. This setup allowed for SMC-conditioned media but not for SMCs to be in physical contact with the endothelial cells. Hence, any effect in the HUVEC network formation observed must be a paracrine, not a cell-cell-dependent, effect (supplemental online Fig. 4B). At days 4 and 8 of the coculture timeline, LM-derived SMCs presented as the best supportive cell type for endothelial network formation (Fig. 3A). Quantification of the day 4 paracrine assay data showed that HUVECs cocultured with LM-derived SMCs accounted for a greater total endothelial network area, with a higher total cord length and more branch points than HUVECs cocultured with SMCs of the two other lineages or HUVECs alone (Fig. 3B–3D). These results suggest that LM-derived SMCs best support endothelial network formation and that this effect is at least partly paracrine in nature.
Figure 3.
Paracrine effects of lineage-specific SMCs on HUVEC networks. HUVECs were exposed to supernatant conditioned by lineage-specific SMCs (supplemental online Fig. 4B). (A): Confocal images of paracrine cocultures taken on days 4 and 8 of the protocol. (B): Quantification of total network area. (C): Quantification of total cord length. (D): Quantification of number of branch points. Quantitative data are shown for day 4 of the coculture (∗, p < .05; ∗∗, p < .01; n = 3 independent biological replicates; scale bars = 100 μm). Abbreviations: d, day; HUVEC, human umbilical vein endothelial cell; LM, lateral mesoderm; NE, neuroectoderm; PM, paraxial mesoderm; SMC, smooth muscle cell.
Midkine Is One Mediator of Superior Vascular Network Formation in LM-Derived SMC Cocultures and Is Part of a Unique Angiogenic Expression Pattern
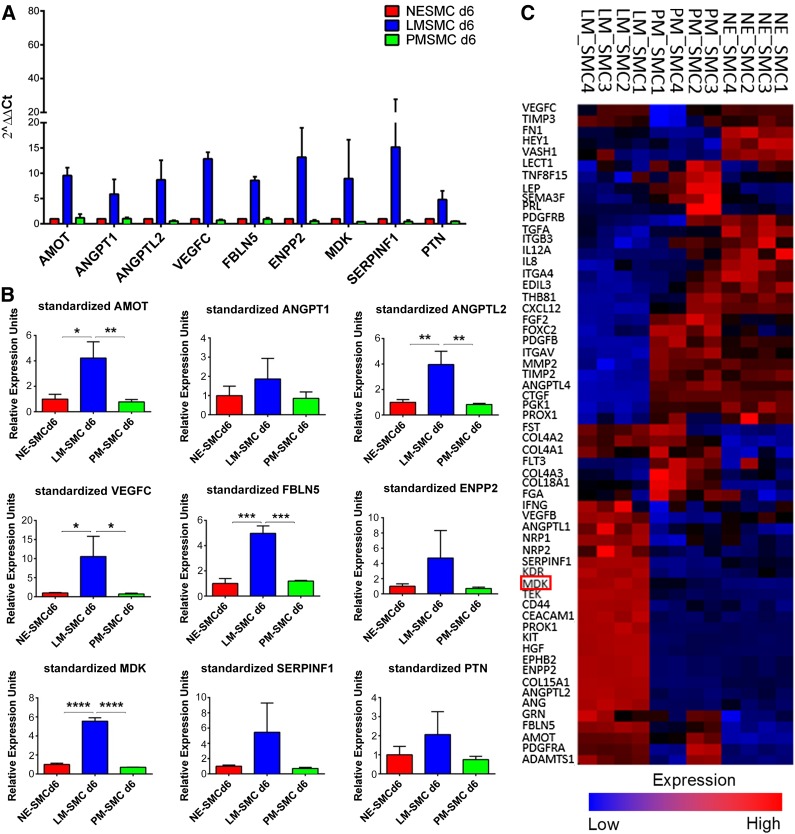
To identify the possible mediators of the LM-derived SMCs’ differential ability to support vascular network formation, a TaqMan Array Human Angiogenesis Panel (Thermo Fisher Scientific Life Sciences) was performed on D6 PT SMCs derived from LM, PM, and NE. This assay allows the screening of 94 genes involved in human angiogenesis and lymphangiogenesis. After evaluation of all the genes, the panel was narrowed to nine candidates that exhibited selective upregulation in LM-derived SMCs but not in SMCs of PM or NE origin. These included AMOT, ANGPT1, ANGPTL2, VEGFC, FBLN5, ENPP2, MDK, SERPINF1, and PTN (Fig. 4A). Validation of the Human Angiogenesis Panel was performed by qRT-PCR and showed a statistically significant upregulation of AMOT, ANGPTL2, VEGFC, FBLN, and MDK selectively in LM-derived SMCs. Compared with the other candidate genes, MDK showed the highest statistical significance, when comparing its expression in LM-derived SMCs against the expression levels in PM- and NE-derived SMCs (Fig. 4B). To further corroborate these data, we analyzed a microarray of D12 PT lineage-specific SMCs. The respective heat map is depicted in Figure 4C and includes all genes that accounted for an overall statistically significant difference among the three SMC lineages (p < .05). The expression pattern visualized demonstrated that LM-derived SMCs upregulate genes that are not expressed in PM- and NE-derived SMCs, thus constituting a list of genes uniquely confined to the embryonic origin of the LM lineage (Fig. 4C). Analysis of the microarray, including all 94 angiogenesis genes from the TaqMan array, was also performed (supplemental online Fig. 5). In summary, MDK was identified as one of nine candidate genes that were selectively expressed in the LM-derived SMC lineage, which exhibited a lineage specific angiogenic expression pattern.
Figure 4.
Putative mediators of network formation identified by TaqMan array and microarray. (A): Candidate genes differentially upregulated in LM-derived SMCs as per TaqMan Array Human Angiogenesis Panel. Transfection of scrambled small interfering RNA was used as a negative control. (B): Validation of the TaqMan Array by quantitative reverse transcription-polymerase chain reaction. (C): Lineage-specific gene signatures of the three embryonic origin-specific SMC lines per microarray. MDK is highlighted in red. Red (upregulation) and blue (downregulation) reflect expression from the mean across all samples (∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; ∗∗∗∗, p < .0001; n = 3 independent biological replicates). Abbreviations: AMOT, angiomotin; ANGPT1, angiopoietin 1; ANGPTL2, angiopoietin-like protein 2; d, day; ENPP2, ectonucleotide pyrophosphatase/phosphodiesterase family member 2; FBLN5, fibulin 5; LM, lateral mesoderm; MDK, midkine; NE, neuroectoderm; PM, paraxial mesoderm; PTN, pleiotrophin; SERPINF1, serpin peptidase inhibitor clade F; SMC, smooth muscle cell; VEGFC, vascular endothelial growth factor C.
SMC-Derived MDK Regulates Vascular Network Formation in LM-Derived SMC and HUVEC Cocultures
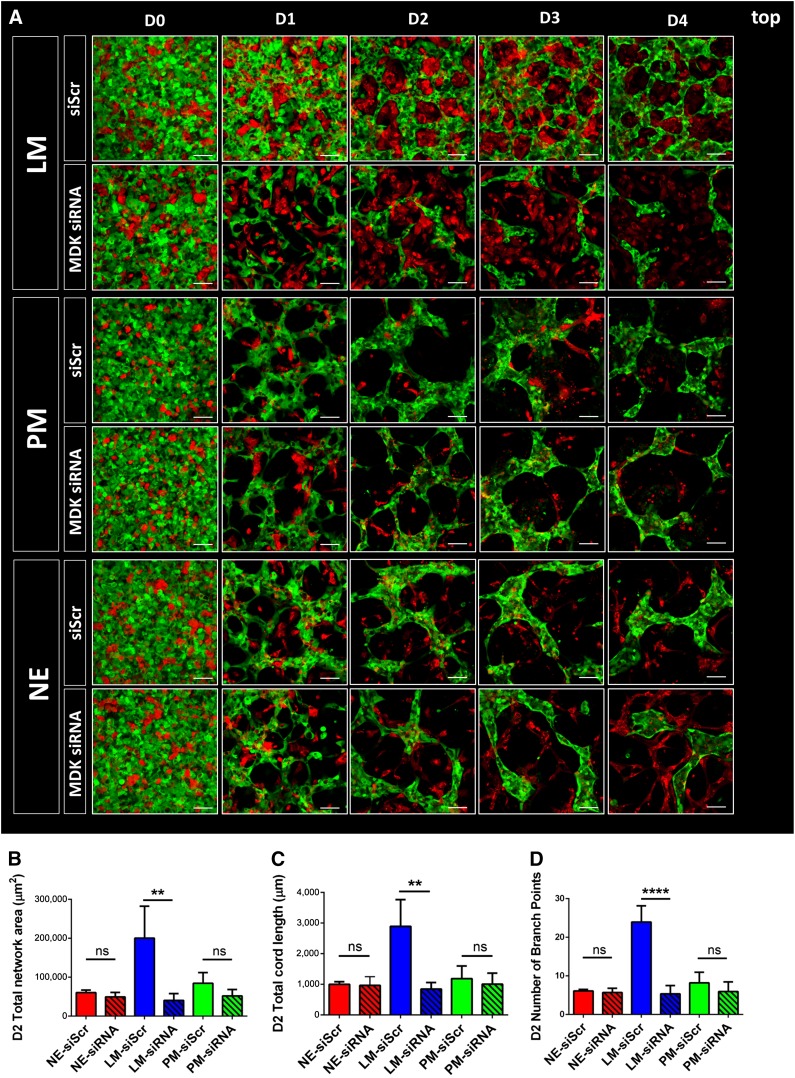
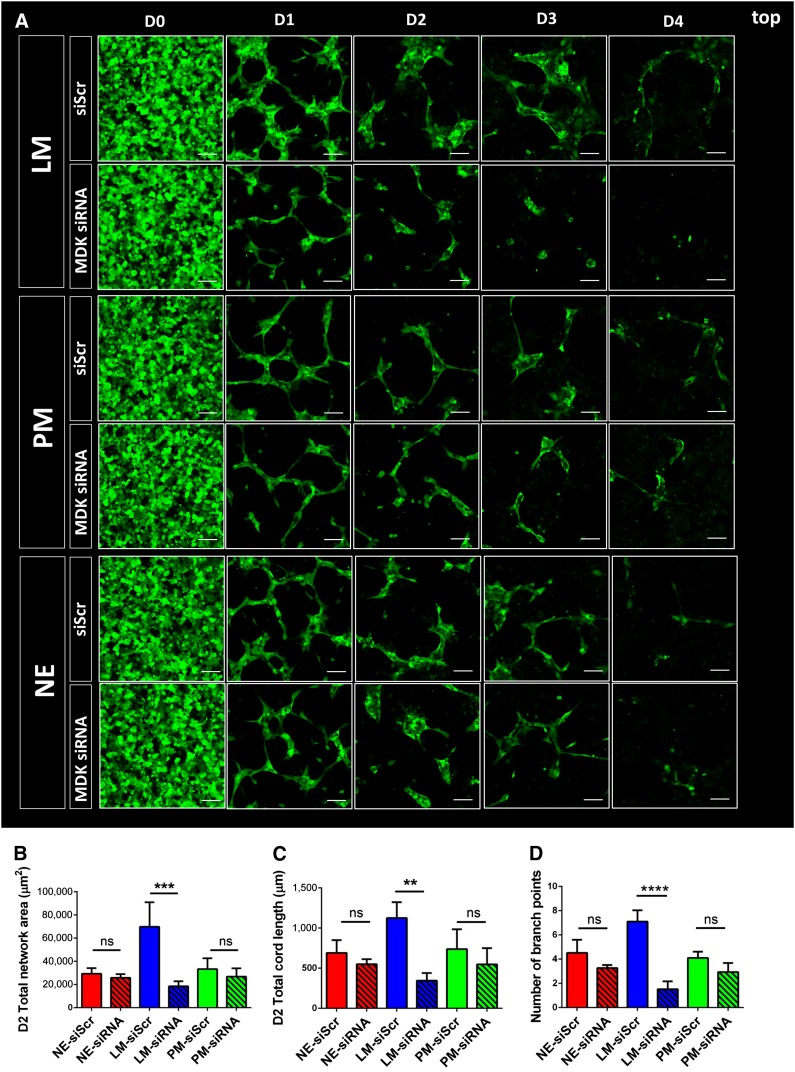
To examine the role of MDK in vascular network formation, an siRNA-mediated knockdown was performed. MDK siRNA was added to the SMC cultures on days 11 and 12 of the differentiation protocol, before starting the cocultures with the knocked-down SMCs on day 13 (supplemental online Fig. 6A). Confirmation of efficient knockdown of MDK was demonstrated by qRT-PCR and ELISA. In line with the results from the TaqMan array, ELISA confirmed that MDK was also selectively expressed in LM-derived SMCs at the protein level (supplemental online Fig. 6B, 6C). Cocultures of MDK-siRNA-treated SMCs with endothelial cells revealed substantial phenotypic changes in the LM-derived SMC cocultures. Impaired MDK expression resulted in a significant decrease in the total endothelial network area of LM-derived SMC cocultures over a continuous timeline of 4 days (Fig. 5A). Extensive quantification of this effect revealed a significant decrease in the total endothelial network area of LM-derived SMC cocultures over a 4-day period. As anticipated, no substantial differences in endothelial network formation were seen in the cocultures with SMCs derived from the PM or NE (Fig. 5B). Knockdown of MDK also resulted in a significant decrease in endothelial network complexity, as demonstrated by a significant decrease in total cord length and the number of branch points (Fig. 5C, 5D). To confirm our initial finding that LM-derived SMCs promote angiogenesis partly in a paracrine fashion, for which one mediator is MDK, we also performed paracrine cocultures with the MDK siRNA-treated SMCs. Over a course of 4 days, MDK knockdown selectively impaired endothelial network formation in LM-derived SMC paracrine assays. Again, no effect of MDK knockdown was seen in the PM- and NE-derived SMC cocultures (Fig. 6A). Quantification of this effect on day 2 of the paracrine assays showed a significant decrease in total endothelial network area and total cord length in the LM-derived SMC group (Fig. 6B, 6C). Similar to the effect observed in 3D cocultures, endothelial network complexity in the paracrine assay was also decreased, as demonstrated by the decreased number of branch points (Fig. 6D).
Figure 5.
siRNA-mediated knockdown of MDK in three-dimensional (3D) cocultures. (A): Effect of MDK knockdown in SMCs on HUVEC network formation in 3D coculture as per confocal microscopy over a 4-day timeline. Transfection of siScrwas used as a negative control. (B): Quantification of the effect of MDK siRNA-mediated knockdown on total network area in cocultures of the three SMC lineages on day 2. (C): Quantification of total cord length in cocultures containing MDK-siRNA treated SMCs on day 2. (D): Quantification of the effect of MDK siRNA-mediated knockdown on the number of branch points in cocultures of the three SMC lineages on day 2 (∗∗, p < .01; ∗∗∗∗, p < .0001; n = 3 independent biological replicates; scale bars = 100 μm). Abbreviations: D, day; HUVEC, human umbilical vein endothelial cell; LM, lateral mesoderm; MDK, midkine; NE, neuroectoderm; ns, not significant; PM, paraxial mesoderm; siRNA, small interfering RNA; siScr, scrambled siRNA; SMC, smooth muscle cell.
Figure 6.
siRNA-mediated knockdown of MDK in three-dimensional (3D) paracrine assays. (A): Effect of MDK knockdown in SMCs on human umbilical vein endothelial cell (HUVEC) network formation in 3D paracrine coculture as per confocal microscopy over a 4-day timeline. (B): Quantification of the effect of MDK siRNA-mediated knockdown on total HUVEC network area in cocultures with the three SMC lineages on day 2. (C): Quantification of total cord length in cocultures containing MDK-siRNA-treated SMCs on day 2. (D): Quantification of the effect of MDK siRNA-mediated knockdown on the number of branch points in cocultures of the three SMC lineages on day 2 (∗∗, p < .01; ∗∗∗, p < .001; ∗∗∗∗, p < .0001; n = 3 independent biological replicates; scale bars = 100 μm). Abbreviations: D, day; LM, lateral mesoderm; MDK, midkine NE, neuroectoderm; ns, not significant; PM, paraxial mesoderm; siRNA, small interfering RNA; siScr, scrambled siRNA; SMC, smooth muscle cell.
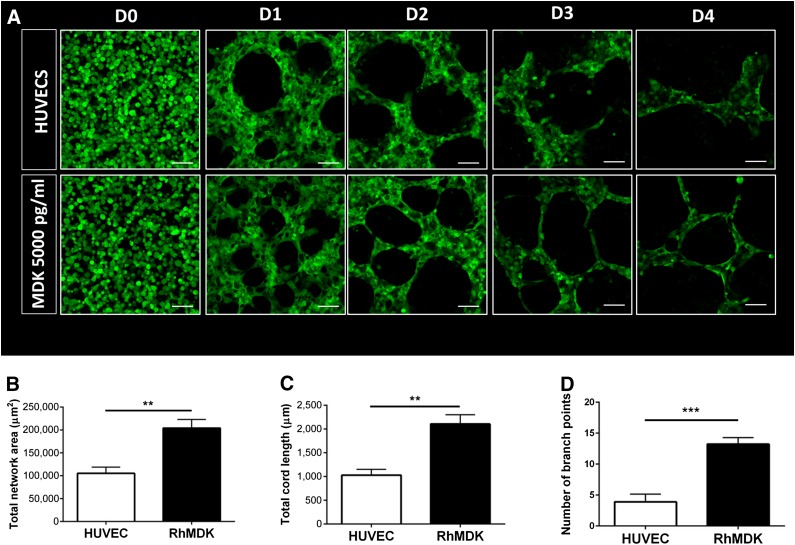
Finally, to examine whether MDK alone, in isolation from other angiogenic factors produced by SMCs, was able to promote human vasculogenesis, 3D Matrigel cultures with HUVECs and recombinant MDK were performed (Fig. 7A). Endothelial networks supplemented with recombinant MDK (5,000 pg/ml) accounted for larger and more complex networks, including a longer total cord length and higher number of branch points than with HUVECs alone. This finding corroborates the importance of this angiogenic factor in vascular network formation (Fig. 7B–7D). Collectively, these data show that MDK is an important factor in the selective ability of LM-derived SMCs to support human vasculogenesis, promoting not only network size but also complexity.
Figure 7.
Addition of recombinant MDK in three-dimensional (3D) HUVEC monocultures. (A): Effect of RhMDK on HUVEC network formation in 3D HUVEC monocultures as per confocal microscopy over a 4-day timeline. Quantification of the effect of RhMDK on total network area in HUVEC monocultures on day 2 (B); total cord length on day 2 (C); and number of branch points on day 2 (D) (∗∗, p < .01; ∗∗∗, p < .001; n = 3 independent biological replicates; scale bars = 100 μm). Abbreviations: D, day; HUVEC, human umbilical vein endothelial cell; MDK, midkine; RhMDK, recombinant human midkine.
Discussion
Microvascular network formation has not been devoted major scientific attention in regenerative cardiovascular research. However, recent attempts to regenerate ischemic tissue have rendered this field highly relevant for the generation of functional tissue grafts in vitro and in vivo. We have shown for the first time that the embryonic origin of mural cells has a functional effect on developing microvascular networks, with implications for regenerative cardiovascular medicine. Specifically, we have demonstrated that LM-derived SMCs provide superior support to endothelial network formation compared with SMCs originating from the PM or NE. Importantly, microarray and gene expression data identified MDK as one mediator of this effect, which was confirmed by loss- and gain-of-function studies.
Regenerative medicine has made remarkable progress in its endeavors to regenerate ischemic and dysfunctional body tissues. In the cardiovascular system, it has been demonstrated that transplantation of hESC-derived cardiomyocytes leads to remuscularization, electrical coupling, and partial restoration of cardiac function up to 4 weeks after transplantation in rodents [14, 15]. More recently, heroic efforts in the Murry Laboratory (University of Washington, Seattle, WA, http://pathology.washington.edu/research/labs/Murry/) have demonstrated that transplantation of 1 billion cardiomyocytes results in robust muscular grafts of infarcted nonhuman primate hearts [16]. However, for establishment of physiological homeostasis and long-term maintenance of graft function, efficient vascularization is likely to be necessary, particularly given that oxygenation by diffusion is only sufficient for grafts that are four to seven cell layers (∼100-µm) thick [17]. Similar reservations apply for tissue-engineered cardiac constructs, which have been shown to promote cardiac function after epicardial transplantation to rat hearts in the short term [18]. Several studies have shown that vascularization promotes cardiac graft development and maturation [19, 20]. However, the potential for vascularization to support regenerative medicine applications will only be realized through a deeper understanding of the ability of different cell types to contribute to vasculogenesis, angiogenesis, and, ultimately, arteriogenesis.
Relatively little is known to date about the embryonic origin of mural cells in microvascular development. Pericytes comprising the microvasculature of lung, liver, and gut have been tracked back to mesenchymal origin [21–23], and coronary pericytes originate from the epicardium [24, 25], which is a lateral-plate mesoderm derivative. In contrast, those comprising the microcerebrovasculature and thymus derive from the neural crest [26, 27]. A number of investigators have shown lineage-dependent functional differences for SMCs comprising large vessels. For NE-derived SMCs, a functional lineage-specific response was shown after angiotensin II treatment [28], and differentiation of this lineage revealed myocardin-related transcription factor B (MKL2) as sine qua non [9]. However, it remains unknown whether lineage-specific effects of mural cells exist in microvascular development. Owing to the high prevalence and increasing incidence of diseases involving pathologic features in the microvasculature, it is of great interest to elucidate the nature and mediators of lineage-specific effects in endothelial network formation. Furthermore, this information might aid attempts to revascularize ischemic body tissues and to bioengineer tissue constructs for regenerative purposes.
Searching for possible mediators of the ability of LM-derived SMCs to better support vascular network formation, we used a TaqMan Array Human Angiogenesis Panel (Thermo Fisher Scientific Life Sciences). Validation of this assay by qRT-PCR revealed that MDK was one of five genes that showed statistically significant upregulation in LM-derived SMCs. MDK is a well-studied target in oncology, with a direct correlation of its expression levels and microvessel density in salivary gland tumors and oral squamous cell carcinomas [29, 30]. An MDK-transfected breast carcinoma cell line was reported to exhibit greater vascular density and higher tumor and endothelial cell proliferative index compared with a mock-transfected control cell line [31]. In line with these findings, it has been shown that high expression levels in patients with invasive bladder cancer correlate with poor survival [32].
In our human in vitro system, knockdown of MDK resulted in significantly impaired vascular network formation. Suggesting a role in angiogenesis, MDK-deficient mice have been shown to exhibit fewer lung metastases of Lewis lung carcinoma cells and reduced tumorigenesis of neuroblastomas [33, 34]. In the cardiovascular field, MDK injection, as well as MDK gene transfer, into postinfarct rat hearts has been demonstrated to decrease cardiac remodeling through antiapoptotic and provasculogenic effects [35, 36]. However, these studies investigated the formation of endothelial cell networks without accounting for the effect of mural cells. In addition, it has not yet been demonstrated that SMCs and their embryonic origins play key roles in the production of this cytokine. Our model is the first to provide an important insight into the functional role of MDK during embryonic vascular development in a human system.
We have also shown that support through LM-derived SMCs results in a more developed endothelial network, reflected in the higher number of branch points. Knockdown of MDK resulted in fewer vascular branch points and supplementation of recombinant MDK supported network formation in HUVEC monocultures similar to those seen in LM-derived SMC cocultures. These results indicate that MDK is involved in generating a complex, branched endothelial network. At the same time, HUVECs alone appeared to form larger and less-complex endothelial cords. In the context of vascular network formation, the role of vascular endothelial growth factor (VEGF) and Notch in tip and stalk cell specification and vascular patterning has been extensively described; however, the role of MDK remains unknown [37–42].
In the present study, we have provided microarray data of lineage-specific D12 PT SMCs and shown that the three SMC lineages account for individual angiogenic gene expression signatures. Although our 3D endothelial network formation assay focused on early vascular development by using D6 PT SMCs, we were able to coherently show that more mature SMCs express the same genes as do D6 of SMC differentiation with PT and additionally upregulated others by D12. This TaqMan array and microarray provide a valuable library of targets to study in the context of vascular network formation. Thus, other putative mediators of the beneficial effects of LM-derived SMCs on endothelial network formation include angiomotin (AMOT), angiopoietin-like protein 2 (ANGPTL2), fibulin 5 (FBLN5), and VEGF-C.
In the present study, we chose Matrigel to study the effects of mural cells on endothelial network formation. We also wanted to elucidate the key signals responsible for the differential ability of embryonic origin-specific SMC lineages to allow for superior survival and growth of more complex endothelial networks. Matrigel assays are a widely used tool to study endothelial cell (EC) network formation and EC-mural cell crosstalk and are used as such by leading vascular laboratories worldwide [43–47]. In this context, Matrigel has been well established as a cord-forming assay. Although limitations exist regarding lumen formation, the main objective of the present study was to investigate branching and survival in a 3D context, which was well served by the system used. We have demonstrated that LM-derived SMCs also allow for the growth of more complex endothelial networks, with a higher number of branch points. This was not a mere consequence of superior survival but a reflection of the angiogenic effect of MDK, which was confirmed by loss- and gain-of-function studies.
That LM-derived SMCs exhibited the best supportive function was not surprising, because they emerge from a unique embryonic origin and express different sets of genes that are implicated in vasculogenesis. Whether these observations have an effect in vivo is a question that has to date not been addressed. Thus, it would be of interest to evaluate the vasculogenic potential of lineage-specific SMCs in a wound healing assay in vivo.
Although the present study is the first to demonstrate that the lineage specificity of mural cells has a functional impact on developing endothelial networks, it remains to be seen how the endothelium from different vascular beds will interact with smooth muscle cells of different types. Recent studies have shown that endothelial cells from different vascular territories display unique properties, and, indeed, these different endothelial cell populations have been demonstrated to have unique, tissue-specific signatures [48]. However, it is unclear how these signatures are obtained, whether through different intermediate developmental steps taken by the angioblasts or as a direct effect of the tissue-specific microenvironment. Further studies should investigate how to generate tissue-specific endothelial cells and whether these show distinct responses to the lineage-specific SMCs we have established. One future endeavor of tissue engineering and regenerative medicine will be the generation of embryonically and physiologically relevant body tissues in which lineage-specific vascular cells will play a key role.
Conclusion
Taken collectively, we have demonstrated that the embryonic origin of mural cells has a functional effect in endothelial network development, complexity, and survival, for which MDK is one important mediator. Fully exploiting the lineage-specific functionality of mural cells could prove critical for vascular tissue engineering and therapeutic revascularization.
Supplementary Material
Acknowledgments
This work was supported by the British Heart Foundation (BHF; Grants NH/11/1/28922 and G1000847), the U.K. Medical Research Council (MRC), and the Cambridge Hospitals National Institute for Health Research Biomedical Research Centre funding (S.S.). J.B. was supported by a Cambridge National Institute for Health Research Biomedical Research Centre Cardiovascular Clinical Research Fellowship and, subsequently, by a BHF Studentship (Grant FS/13/65/30441). L.L. was supported by an MRC award and a BHF grant (G1000847). C.C. is supported by the Independent Fellowship from the Institute of Molecular and Cell Biology, Singapore. W.G.B. is on a BHF Studentship (Grant FS/11/77/29327). D.I. received a University of Cambridge Commonwealth Scholarship. L.G. is supported by BHF Award RM/l3/3/30159. S.S. is supported by BHF Award FS/13/29/30024. We are grateful to Dr. Helle Jorgenson (University of Cambridge) for discussions and Donna Leaford (Wellcome Trust MRC Cambridge Stem Cell Institute) for her expertise and advice on the TaqMan Array. J.B. thanks Dr. Rachel G. Hoffman (King’s College, University of Cambridge) for unwavering support and inspiring discussions.
Author Contributions
J.B.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; L.L.: conception and design, collection and/or assembly of data, data analysis and interpretation; C.C.: conception and design, provision of study material or patients; W.G.B. and L.G.: data analysis and interpretation; D.I. and M.R.B.: administrative support, data analysis and interpretation; S.S.: conception and design, financial support, administrative support, data analysis and interpretation, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Kumar AH, Caplice NM. Clinical potential of adult vascular progenitor cells. Arterioscler Thromb Vasc Biol. 2010;30:1080–1087. doi: 10.1161/ATVBAHA.109.198895. [DOI] [PubMed] [Google Scholar]
- 2.Gaengel K, Genové G, Armulik A, et al. Endothelial-mural cell signaling in vascular development and angiogenesis. Arterioscler Thromb Vasc Biol. 2009;29:630–638. doi: 10.1161/ATVBAHA.107.161521. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl P, Johansson BR, Levéen P, et al. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277:242–245. doi: 10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- 4.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hellström M, Gerhardt H, Kalén M, et al. Lack of pericytes leads to endothelial hyperplasia and abnormal vascular morphogenesis. J Cell Biol. 2001;153:543–553. doi: 10.1083/jcb.153.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jain RK. Molecular regulation of vessel maturation. Nat Med. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 7.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 8.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27:1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 9.Topouzis S, Majesky MW. Smooth muscle lineage diversity in the chick embryo:. Two types of aortic smooth muscle cell differ in growth and receptor-mediated transcriptional responses to transforming growth factor-beta. Dev Biol. 1996;178:430–445. doi: 10.1006/dbio.1996.0229. [DOI] [PubMed] [Google Scholar]
- 10.Rosenquist TH, Kirby ML, van Mierop LH. Solitary aortic arch artery: A result of surgical ablation of cardiac neural crest and nodose placode in the avian embryo. Circulation. 1989;80:1469–1475. doi: 10.1161/01.cir.80.5.1469. [DOI] [PubMed] [Google Scholar]
- 11.Cheung C, Bernardo AS, Trotter MW, et al. Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat Biotechnol. 2012;30:165–173. doi: 10.1038/nbt.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 13.Cheung C, Bernardo AS, Pedersen RA, et al. Directed differentiation of embryonic origin-specific vascular smooth muscle subtypes from human pluripotent stem cells. Nat Protoc. 2014;9:929–938. doi: 10.1038/nprot.2014.059. [DOI] [PubMed] [Google Scholar]
- 14.Laflamme MA, Chen KY, Naumova AV, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 15.Shiba Y, Fernandes S, Zhu WZ, et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong JJ, Yang X, Don CW, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folkman J, Hochberg M. Self-regulation of growth in three dimensions. J Exp Med. 1973;138:745–753. doi: 10.1084/jem.138.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann WH, Melnychenko I, Wasmeier G, et al. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med. 2006;12:452–458. doi: 10.1038/nm1394. [DOI] [PubMed] [Google Scholar]
- 19.Dvir T, Kedem A, Ruvinov E, et al. Prevascularization of cardiac patch on the omentum improves its therapeutic outcome. Proc Natl Acad Sci USA. 2009;106:14990–14995. doi: 10.1073/pnas.0812242106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekine H, Shimizu T, Sakaguchi K, et al. In vitro fabrication of functional three-dimensional tissues with perfusable blood vessels. Nat Commun. 2013;4:1399. doi: 10.1038/ncomms2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Que J, Wilm B, Hasegawa H, et al. Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci USA. 2008;105:16626–16630. doi: 10.1073/pnas.0808649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asahina K, Zhou B, Pu WT, et al. Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology. 2011;53:983–995. doi: 10.1002/hep.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilm B, Ipenberg A, Hastie ND, et al. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- 24.Dettman RW, Denetclaw W, Jr, Ordahl CP, et al. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193:169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 25.Trembley MA, Velasquez LS, de Mesy Bentley KL, et al. Myocardin-related transcription factors control the motility of epicardium-derived cells and the maturation of coronary vessels. Development. 2015;142:21–30. doi: 10.1242/dev.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etchevers HC, Vincent C, Le Douarin NM, et al. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- 27.Foster K, Sheridan J, Veiga-Fernandes H, et al. Contribution of neural crest-derived cells in the embryonic and adult thymus. J Immunol. 2008;180:3183–3189. doi: 10.4049/jimmunol.180.5.3183. [DOI] [PubMed] [Google Scholar]
- 28.Owens AP, III, Subramanian V, Moorleghen JJ, et al. Angiotensin II induces a region-specific hyperplasia of the ascending aorta through regulation of inhibitor of differentiation 3. Circ Res. 2010;106:611–619. doi: 10.1161/CIRCRESAHA.109.212837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ota T, Ota K, Jono H, et al. Midkine expression in malignant salivary gland tumors and its role in tumor angiogenesis. Oral Oncol. 2010;46:657–661. doi: 10.1016/j.oraloncology.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Ruan M, Ji T, Wu Z, et al. Evaluation of expression of midkine in oral squamous cell carcinoma and its correlation with tumour angiogenesis. Int J Oral Maxillofac Surg. 2007;36:159–164. doi: 10.1016/j.ijom.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Choudhuri R, Zhang HT, Donnini S, et al. An angiogenic role for the neurokines midkine and pleiotrophin in tumorigenesis. Cancer Res. 1997;57:1814–1819. [PubMed] [Google Scholar]
- 32.O’Brien T, Cranston D, Fuggle S, et al. The angiogenic factor midkine is expressed in bladder cancer, and overexpression correlates with a poor outcome in patients with invasive cancers. Cancer Res. 1996;56:2515–2518. [PubMed] [Google Scholar]
- 33.Salama RH, Muramatsu H, Zou P, et al. Midkine, a heparin-binding growth factor, produced by the host enhances metastasis of Lewis lung carcinoma cells. Cancer Lett. 2006;233:16–20. doi: 10.1016/j.canlet.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 34.Kishida S, Mu P, Miyakawa S, et al. Midkine promotes neuroblastoma through Notch2 signaling. Cancer Res. 2013;73:1318–1327. doi: 10.1158/0008-5472.CAN-12-3070. [DOI] [PubMed] [Google Scholar]
- 35.Fukui S, Kitagawa-Sakakida S, Kawamata S, et al. Therapeutic effect of midkine on cardiac remodeling in infarcted rat hearts. Ann Thorac Surg. 2008;85:562–570. doi: 10.1016/j.athoracsur.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Sumida A, Horiba M, Ishiguro H, et al. Midkine gene transfer after myocardial infarction in rats prevents remodelling and ameliorates cardiac dysfunction. Cardiovasc Res. 2010;86:113–121. doi: 10.1093/cvr/cvp386. [DOI] [PubMed] [Google Scholar]
- 37.Gerhardt H, Golding M, Fruttiger M, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tammela T, Zarkada G, Wallgard E, et al. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature. 2008;454:656–660. doi: 10.1038/nature07083. [DOI] [PubMed] [Google Scholar]
- 39.Noguera-Troise I, Daly C, Papadopoulos NJ, et al. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 40.Ridgway J, Zhang G, Wu Y, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 41.Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–784. doi: 10.1038/nature05577. [DOI] [PubMed] [Google Scholar]
- 42.Leslie JD, Ariza-McNaughton L, Bermange AL, et al. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- 43.Patsch C, Challet-Meylan L, Thoma EC, et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orlova VV, Drabsch Y, Freund C, et al. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler Thromb Vasc Biol. 2014;34:177–186. doi: 10.1161/ATVBAHA.113.302598. [DOI] [PubMed] [Google Scholar]
- 45.Potus F, Ruffenach G, Dahou A, et al. Downregulation of microRNA-126 contributes to the failing right ventricle in pulmonary arterial hypertension. Circulation. 2015;132:932–943. doi: 10.1161/CIRCULATIONAHA.115.016382. [DOI] [PubMed] [Google Scholar]
- 46.Esser JS, Rahner S, Deckler M, et al. Fibroblast growth factor signaling pathway in endothelial cells is activated by BMPER to promote angiogenesis. Arterioscler Thromb Vasc Biol. 2015;35:358–367. doi: 10.1161/ATVBAHA.114.304345. [DOI] [PubMed] [Google Scholar]
- 47.Hassel D, Cheng P, White MP, et al. MicroRNA-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ Res. 2012;111:1421–1433. doi: 10.1161/CIRCRESAHA.112.279711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nolan DJ, Ginsberg M, Israely E, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.