This work describes a robust, simple method for generating cerebral organoids from human induced pluripotent stem cells by using a chemically defined hydrogel material and chemically defined culture medium. This method greatly facilitates access to cerebral organoid technology, enables scalable applications, and provides a potential pathway to translational applications where defined components are desirable.
Keywords: Organoids, Brain, Stem cells, Adrenoleukodystrophy, In vitro techniques
Abstract
Tissue organoids are a promising technology that may accelerate development of the societal and NIH mandate for precision medicine. Here we describe a robust and simple method for generating cerebral organoids (cOrgs) from human pluripotent stem cells by using a chemically defined hydrogel material and chemically defined culture medium. By using no additional neural induction components, cOrgs appeared on the hydrogel surface within 10–14 days, and under static culture conditions, they attained sizes up to 3 mm in greatest dimension by day 28. Histologically, the organoids showed neural rosette and neural tube-like structures and evidence of early corticogenesis. Immunostaining and quantitative reverse-transcription polymerase chain reaction demonstrated protein and gene expression representative of forebrain, midbrain, and hindbrain development. Physiologic studies showed responses to glutamate and depolarization in many cells, consistent with neural behavior. The method of cerebral organoid generation described here facilitates access to this technology, enables scalable applications, and provides a potential pathway to translational applications where defined components are desirable.
Significance
Tissue organoids are a promising technology with many potential applications, such as pharmaceutical screens and development of in vitro disease models, particularly for human polygenic conditions where animal models are insufficient. This work describes a robust and simple method for generating cerebral organoids from human induced pluripotent stem cells by using a chemically defined hydrogel material and chemically defined culture medium. This method, by virtue of its simplicity and use of defined materials, greatly facilitates access to cerebral organoid technology, enables scalable applications, and provides a potential pathway to translational applications where defined components are desirable.
Introduction
It has become increasingly apparent that three-dimensional (3D) culture of both pluripotent and differentiated cells results in important emergent biological features absent in traditional two-dimensional (2D) plate culture. Indeed, the traditional concept of cells having the same phenotype in 2D and 3D is erroneous, with widely divergent phenotypes developing in response to their niche microenvironment, polarity, and extracellular matrix interactions [1]. Of particular note is the ability of pluripotent stem cells (PSCs), such as induced pluripotent stem cells (iPSCs) or embryonic stem cells (ESCs), to self-organize under various conditions to form complex tissue structures that recapitulate important developmental features and adult structural and functional characteristics typical of each particular tissue [2–4]. The cellular and structural complexity of these organoids and their fidelity to the structure of corresponding tissues in vivo make them particularly intriguing as readily accessible in vitro models for a wide range of physiologic and metabolic studies, for pharmaceutical screens, and as models for human pathological conditions. In addition, if scale-up methods for organoid generation were readily available and compliant with current good manufacturing practices, cells from organoids could potentially be used in therapeutic applications [5, 6].
There is presently a particular need for the development of organoid-based in vitro disease models for human developmental neurological conditions that are difficult to reproduce in experimental animal models and for neurodegenerative disorders with complex genetics (e.g., Alzheimer’s disease, Parkinson’s disease, and adrenoleukodystrophy). In these conditions, genes, gene modifiers, and environmental factors all contribute to disease and thus cannot be easily reduced to linear experiments that would illuminate the mechanism of action. As such, cerebral organoids (cOrgs) derived from patient and disease-specific iPSCs may be a unique opportunity for the generation of new knowledge leading to clinically relevant interventions. In this regard, a means of generating relatively large and complex cOrgs was recently described [5]. In that report the cOrgs were shown to contain neurons and structural arrangements representing several different brain regions within individual organoids and, furthermore, demonstrated features of normal human brain development and structure. The utility of this approach was elegantly demonstrated by generation of iPSCs from patients with the neurodevelopmental disorder microcephaly and subsequent production of cOrgs that exhibited disease-specific features of this condition. As such, the study demonstrated the potential for the creation of accessible in vitro models of human neurological diseases by using the innate ability of pluripotent stem cells to differentiate into neural lineages and self-organize in a manner that recapitulates normal (or abnormal) development. Similarly, using ESCs, Kadoshima et al. also recently reported the generation of remarkably complex cOrgs that showed spontaneous development of intracortical polarity, curving morphology, and complex zone separations, which recapitulated normal human brain development [6]. Thereby, these reports substantiated the complexity achievable in cOrgs derived from PSCs and their applicability to in vitro disease-modeling applications.
We are focused on establishing an in vitro disease model for adrenoleukodystrophy (ALD), which in a subpopulation of affected boys results in a devastating, progressive, degenerative neurologic condition. ALD is an X-linked peroxisomal disease that affects the nervous system, adrenal cortex, and testis, resulting from mutations in the ABCD1 gene [7]. Several phenotypes can result from this mutation, including adrenomyeloneuropathy, a cerebral adult form, isolated Addison’s disease, and cerebral childhood adrenoleukodystrophy (ccALD)—the most severe form of ALD characterized by rapid neurologic decline from demyelination within the cerebral white matter [7]. More than 643 mutations in the ABCD1 gene have been associated with ALD; however, correlations between specific mutations and ALD phenotypes have remained elusive, thus implying that additional genetic, epigenetic, and/or environmental modifiers may be involved [8]. Currently, hematopoietic cell transplantation is the only treatment able to stabilize ccALD, with early treatment being critical for optimal long-term outcome [9]. Therefore, establishing early screening mechanisms to identify which patients with ALD mutations will present a ccALD phenotype is an enormous clinical need, which is not yet met with the currently explored methods, such as increased cerebral spinal fluid (CSF) cytokine levels [10], diffusion tensor brain imaging [11], and chitotriosidase activity in plasma and CSF [12]. ALD patient iPSC-derived cOrgs could serve as a powerful in vitro model by which to study gene expression, epigenetics, and effects of environmental factors, potentially illuminating mechanisms of action and leading to clinically relevant interventions as well as potential biomarkers that could be used in early ccALD screening.
Although the method used by Lancaster et al. [5] to generate cOrgs was highly effective, it has a high degree of complexity in execution, requires costly neural induction cell culture constituents, and involves the use of a xenobiotic extracellular matrix material. Here we report our development of a novel method for generation of cOrgs that addresses these issues. The method is robust, simple, does not require neural induction components beyond those included in the Essential 8 (E8) medium, and uses a chemically defined hydrogel, termed Cell-Mate3D. Histological, immunohistochemical, and gene expression analysis combined with calcium-signaling studies confirmed the cerebral organoid phenotype, including evidence for forebrain, midbrain, and hindbrain specification. Overall, this system may facilitate both basic research and translational applications where defined components are desirable.
Materials and Methods
Preparation of Cell-Mate3D Dry Blend
Sodium hyaluronan (HA-Na) (original molecular weight [MW] = 1,600–1,800 kDa; polydispersity index [PDI] < 4.0) and Chitosan (CT) protonated with formic acid (CTNH3+) (original MW = 400–600 kDa; PDI < 3.0) were used in this study. Hyaluronan (HA; Lifecore Biomedical, Chaska, MN, http://www.lifecore.com) was used as received. CT (NovaMatrix; FMC Health and Nutrition, Princeton, NJ, http://www.fmcbiopolymer.com) was received as a base at 85%–87.5% degree of deacetylation and was protonated with formic acid to 100% of available amine groups. Protonated chitosan-base was prepared as a 0.1% (wt/vol) solution, filter-sterilized (0.2 µm), and aseptically filled into 120-ml sterile vials. The CT solution was lyophilized, reduced to small leaflets, and mechanically blended with small particles of HA-Na at the mass ratio of HA-Na = 1.0: CT = 1.44; carrying a charge ratio of CT-n+ = 2.0: HA-Na-n− = 1.0.
Preparation of Cell-Mate3D Hydration Fluid
Hydration fluid was produced by preparing a solution containing 37.5% of 10% LMD dextran 40 in 5% dextrose injection solution USP grade (Pfizer, New York, NY, http://www.pfizer.com) and 51.75% of 0.9% sodium chloride injection solution USP grade (Pfizer). pH was adjusted to 6.5 with a solution of 0.6% glycerol phosphate disodium salt (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com) diluted in sterile water for injection USP grade (Pfizer) and remaining volume consisted of sterile water for injection USP grade (Pfizer).
Preparation of Cell-Mate3D Cocoon
The HA-CT dry blend was brought to room temperature and mixed by vigorous vortexing. The dry blend was hydrated by pipetting a cell suspension in hydration fluid under continuous agitation. A total of 500 µl of the cell suspension containing 19 million iPSCs (as whole colonies) was used to hydrate 35.5 mg of the dry blend. The cell-loaded matrix was transferred into a 1-ml syringe barrel by centrifuging at 1,000g through a funnel apparatus. This process produced a 400- to 500-µl cylinder of cell-embedded matrix, which was then extruded from the syringe barrel, cut to 100-µl sections, and placed into E8 culture medium (Invitrogen, Carlsbad, CA, http://www.invitrogen.com).
Derivation and Culture of iPSCs
Normal and ccALD iPSC lines described in Table 1 were used. Cells were derived on irradiated MEF cultures and transferred to Matrigel (Corning, Corning, NY, http://www.corning.com) and E8 Medium (Thermo Fisher Scientific Life Sciences, Waltham, MA, http://www.thermofisher.com) for additional feeder-free expansion. Cell lines were reprogrammed by using Cytotune IPS 1.0 (WT2, WT3, and WT4; Thermo Fisher Scientific Life Sciences) or Cytotune 2.0 (ALD3; Thermo Fisher Scientific Life Sciences) or were derived by using retroviral gene delivery using the reprogramming factors Oct4, Sox2, Klf4, and c-Myc (WT1, ALD1, and ALD2).
Table 1.
iPSC lines and derivation methods
Culture of Cells in Cell-Mate3D and Induction of Cerebral Organoids
IPSC-embedded matrices were cultured in E8 medium (Thermo Fisher Scientific Life Sciences), and medium was changed every 3–4 days. Organoids spontaneously occurred within the matrix and emerged out from the matrix between day 10 (D10) and D14.
RNA Isolation, Expression, and Analysis
Organoids were collected and lysed in RLT buffer (Qiagen, Venlo, The Netherlands, http://www.qiagen.com) and stored at −80°C until processed. RNA was isolated from cell lysates by using the RNA mini plus kit (Qiagen) according to manufacturer’s instructions. Off-column DNase treatment was performed by using Turbo DNase (Thermo Fisher Scientific Life Sciences) according to manufacturer’s instructions. cDNA was synthesized by using Superscript III reverse transcriptase (Thermo Fisher Scientific Life Sciences) per manufacturer’s instructions. Each polymerase chain reaction (PCR) consisted of 20 ng of cDNA, 6 µl of 1× SYBR Green Mix PCR reaction buffer (Thermo Fisher Scientific Life Sciences), and 100 nM primers (Integrated DNA Technologies, Coralville, IA, https://www.idtdna.com; supplemental online Table 1) plus RNase free water to equal 12 µl total. All reactions were run in triplicate on a Realplex mastercycler (Eppendorf, Hamburg, Germany, https://www.eppendorf.com) by using the following program: 50°C for 2 minutes, 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and 59°C for 60 seconds, followed by 95°C for 15 seconds, 59°C for 20 seconds, a 20-minute ramp to 95°C, and 95°C for 15 seconds. Results of gene expression were calculated by using expression relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), where ΔCt = (Ct gene of interest − Ct GAPDH). The Δ (ΔCt) was then calculated relative to D0 of appropriate IPS line RNA, where Δ (ΔCt) = ΔCt gene − ΔCt control. The fold change relative to the control for each gene was then calculated, where fold change = 2-Δ(ΔCt). Samples with a Ct value greater than 35 were considered to represent no gene expression.
Mechanical Properties Testing
The mechanical properties of the Cell-Mate3D cocoons, with and without cells, were measured by using an AR-G2 rheometer from TA Instruments (Schaumburg, IL, http://www.tainstruments.com). The cocoons were prepared from 35.5 mg of the dry blend hydrated with 500 µl of hydration fluid under continuous agitation. Cell-laden cocoons were prepared with 19 million WT1 iPSCs in the hydration fluid. After hydration, gels were shaped into cylindrical disks with a diameter of 16 mm and height of 3 mm and placed in 5 ml of E8 culture medium for 24 hours. Cocoons were transferred to the Peltier plate and trimmed to the 8-mm parallel plate with a gap of 2.2 mm. Strain sweeps were performed at a constant frequency to select a percent strain within the linear viscoelastic regimen of the matrix. Frequency sweeps were then performed from 100 to 0.1 rad/second with 1% strain at 25°C. The Young’s modulus, E, was calculated from the following formula: E = 3 × sqrt(G′2 + G′′2), where values of the G′, the storage modulus, and G′′, the loss modulus, were both included at 1 rad/second in the calculation. Data are given as average ± SE from six independent experiments performed on different days, each with a single replicate (n = 6). A Student’s t test statistical analysis was used to compare E for the gels with and without cells.
Histology and Immunohistochemistry
Samples were placed in 10% neutral buffered formalin solution and fixed at room temperature for 3.5 hours. After fixation, samples were transferred to 70% ethanol solution until they were processed for routine paraffin embedding. Samples were then sectioned 4-µm thick and stained with hematoxylin and eosin. For immunohistochemical staining, sections were cut at 4 µm, deparaffinized, and rehydrated, followed by incubation with 3% hydrogen peroxide to quench endogenous peroxidase activity and 15 minutes in serum-free protein block (DAKO, Glostrup, Denmark, http://www.dako.com). Sections were then subjected to appropriate antigen retrieval methods (if needed) and incubated with the primary antibody at room temperature for 60 minutes (supplemental online Table 2). Stained sections were examined by using an Olympus BH-2 microscope (Olympus America, Center Valley, PA, http://www.olympusamerica.com) and imaged with a SPOT Insight 4 megasample digital camera and SPOT Advanced software (Diagnostic Instruments Inc., Sterling Heights, MI, http://www.spotimaging.com).
Cryosectioning
Organoids were collected at D14 and D28, washed twice with phosphate-buffered saline (PBS; Sigma-Aldrich) and placed in 10% formalin at 4°C overnight. Samples were washed with PBS and placed in 30% sucrose overnight at 4°C. Organoids were then placed in a mold, and most of the sucrose was removed with blotting paper. Samples were blocked in optimal cutting temperature compound (Tissue-Tek; Sakura Finetek, AV Alphen aan Den Rijn, The Netherlands, http://www.sakura.eu) for 20 minutes in dry ice. Sections of 8-µm thickness were cut using a cryostat, model CM3050 S (Leica Biosystems, Wetzlar, Germany, http://www2.leicabiosystems.com). Sections were mounted on superfrost plus slides (Thermo Fisher Scientific, Waltham, MA, https://www.thermofisher.com) and stored at −20°C before processing for immunocytochemistry.
Immunocytochemistry
Slides were placed in PBS (Sigma-Aldrich) containing 0.1% Tween-20 (Sigma-Aldrich) (PBS-T) for 10 minutes. Sections were blocked with 1% bovine serum albumin (Sigma-Aldrich) and 0.1% Tween-20 in PBS for 30 minutes at room temperature. Sections were incubated with primary antibodies diluted in blocking buffer at 4°C overnight. Sections were washed three times with PBS-T for 10 minutes each. Sections were incubated with secondary antibody (Alexa Fluor 488 or 555) in blocking buffer for 30 minutes at room temperature. All antibodies and dilutions used are detailed in supplemental online Table 1. Samples were washed with PBS-T, and slides were mounted with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA, http://vectorlabs.com). Stained sections were examined with a Bio-Rad confocal microscope (Bio-Rad Laboratories, Hercules, CA, http://www.bio-rad.com).
Gross Sample Imaging
Gross images were obtained by using a SMZ1500 stereoscope (Nikon, Chiyoda, Tokyo, Japan, http://www.nikon.com) and an EVOLT E-300 camera (Olympus, Shinjuku, Tokyo, http://www.olympus-global.com).
Calcium Imaging
Organoids were incubated in 10 µM Fluo-3 AM with PowerLoad (Thermo Fisher Scientific Life Sciences) in culture medium at 37°C for 1 hour. Organoids were placed in a perfusion chamber, stabilized by using a polyester mesh, and superfused continuously with extracellular medium containing 146 mM NaCl, 3 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 1.25 mM NaH2PO4, 10 mM HEPES, 10 mM glucose, and 1 mM Na pyruvate (pH 7.4). Elevated potassium solutions contained 50 mM KCl and 99 mM NaCl, as well as the other constituents listed above. Frozen stock solutions of 100 mM glutamate were diluted 1,000-fold in culture medium on the day of the experiment to yield 100 μM final concentration. All chemicals were purchased from Sigma-Aldrich. Glutamate or elevated potassium solutions were both applied for 30–90 seconds via the superfusate by using a four-way valve to switch solutions.
The perfusion chamber was placed on the stage of an inverted microscope (Olympus model IX-70) equipped with a ×40 oil objective, a Λ DG-4 fluorescence lamp and wavelength switcher, and a Hamamatsu Orca E2 camera (Hamamatsu Photonics, Hamamatsu City, Japan, http://www.hamamatsu.com). MetaFluor (Molecular Devices, Sunnyvale, CA, http://www.moleculardevices.com) was used to acquire images at 1- to 2-second intervals, and Fluo-3 fluorescence intensity in regions of interest corresponding to cell bodies was plotted versus time using GraphPad Prism graphing software (GraphPad Software Inc., La Jolla, CA, http://www.graphpad.com).
The cOrg used for Calcium imaging was derived from cell line WT1 on D49. This cOrg was cultured in E8 medium for 33 days and in neural induction medium containing a 1:1 mixture of DMEM/F12 (Thermo Fisher Scientific Life Sciences) and Neurobasal, 1:200 N2 (Thermo Fisher Scientific Life Sciences), 1:100 B27 (Thermo Fisher Scientific Life Sciences), 3.5 μl 2-mercaptoethanol per liter (Thermo Fisher Scientific Life Sciences), 1:4,000 insulin (Sigma-Aldrich), 1:100 l-glutamine (Thermo Fisher Scientific Life Sciences), and 1:200 minimal essential medium nonessential amino acids (Thermo Fisher Scientific Life Sciences) for an additional 18 days.
Statistical Analysis
The outcome variable for the quantitative reverse-transcription polymerase chain reaction data was ΔΔCt multiplied by 1,000. Means and standard deviations were calculated and presented at each time point for each gene within each cell line (supplemental online Tables 3–7). Analysis of variance F-tests were performed to compare the means at D0 versus D14, D0 versus D28, and D14 versus D28. Because multiple comparisons were performed, Tukey-Kramer adjustments for the p values were made to account for multiple testing within trait. All analysis was performed by using Statistical Analysis Software (version 9.3; SAS Institute Inc., Cary, NC, http://www.sas.com). A two-sided p value <.05 was considered statistically significant.
Results
Formation of iPSC Embedded Cell-Mate3D Hydrogel
Cell-Mate3D hydrogels embedded with iPSCs were formed as previously described [13], except that here a chemically defined hydration fluid composed of buffered dextran was used. Briefly, as cells suspended in hydration fluid were added to the dry blend of hyaluronic acid and Chitosan under high agitation (vortexing), a gel formed instantly through polyelectrolytic complexation between HACOO- and CTNH3+. The gel was then formed into a cylinder by placing it into a syringe barrel-funnel apparatus followed by brief centrifugation. It was extruded from the syringe barrel, cut into several 100-ul sections, and cultured in E8 medium (Thermo Fisher Scientific Life Sciences). iPSCs were embedded as large, undisrupted colonies.
Mechanical Properties of Cell-Mate3D Hydrogels
Oscillatory rheology was used to characterize the mechanical properties of Cell-Mate3D with and without iPSCs. The Cell-Mate3D samples had a storage modulus, G′, greater than the loss modulus, G′′, indicating that gels had formed. The Young’s modulus of Cell-Mate3D with and without iPSCs was calculated to be 9,838 ± 694 Pa and 10,111 ± 1,673 Pa, respectively, and there was no statistically significant difference between these two measurements (p > .05).
Histologic Analysis
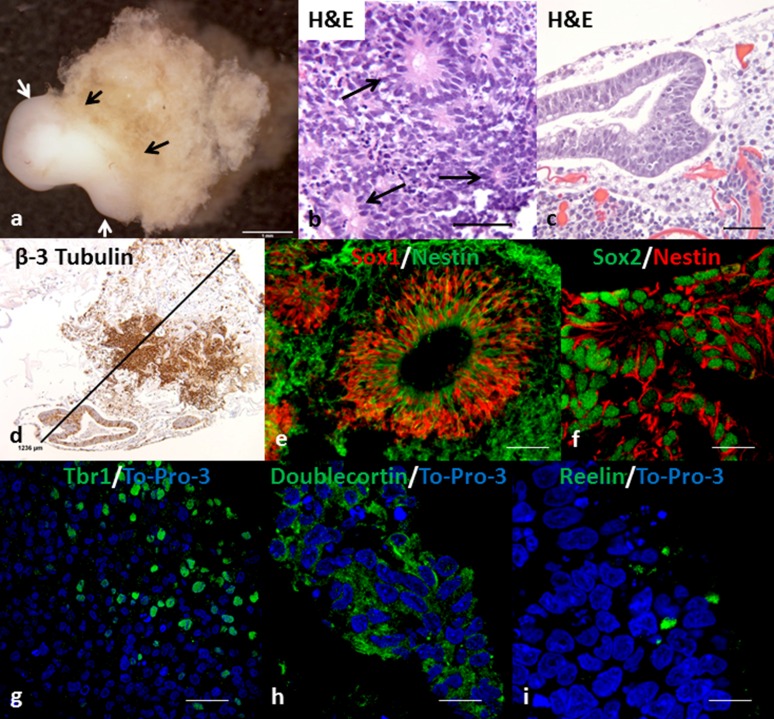
In our initial experiments using the WT4 human iPSC line cultured in E8 medium in Cell-Mate3D hydrogels, we noted the emergence of surface structures on the constructs between D10 and D14. At this stage, the structures were up to approximately 1 mm in greatest dimension and had an approximately spherical shape. Histologic examination of these structures revealed a neural morphology including the formation of neural rosettes and other neuroepithelial structures consistent with the development of cOrgs (Fig. 1). Using immunohistochemical stains, we further evaluated these structures at D14 and found that the majority of cells were strongly positive for β-3 tubulin, Sox2 (nuclear), and nestin (cytoplasmic), consistent with neural differentiation (Fig. 1). Noting the marked resemblance of these structures to the cOrgs recently reported by Lancaster et al. [5], we undertook further experiments to determine whether this was a generalized phenomenon among various iPSC lines.
Figure 1.
Gross appearance, histology, and immunofluorescence of cerebral organoids (cOrgs) derived from human induced pluripotent stem cells (hiPSCs) embedded in Cell-Mate3D matrix and cultured in E8 medium. (A): Gross photograph of day 28 cerebral organoid (arrows) attached to matrix. Scale bar = 1 mm. (B): Neural rosettes (arrows) in cOrg derived from WT3 induced pluripotent stem cell line at day 14 (D14). Hematoxylin and eosin stain. Scale bar = 50 µm. (C): Neural tube-like structure in D14 cOrg derived from ALD1 hiPSC line. Hematoxylin and eosin stain. Scale bar = 50 µm. (D): D14 cOrg derived from ALD1 hiPSC line and immunohistochemically stained for β-3 tubulin. Scale bar = 1,236 µm. (E): Immunofluorescent stain showing colocalization of Sox1 (red) and nestin (green) in a cell of a neural tube-like structure and surrounding area in a cOrg from WT1 hiPSC line at D14. Scale bar = 60 µm. (F): Immunofluorescent stain showing Sox2 (green) and nestin (red) colocalization in neural rosettes in a cOrg from ALD2 hiPSC line at D14. Scale bar = 18 µm. (G–I): Immunofluorescent staining of neurocortical-like regions in D28 cOrgs for Tbr1 (ALD3) (G), doublecortin (ALD1) (H), and reelin (WT3) (I). Scale bars = 30 µm (G) and 15 µm (H, I).
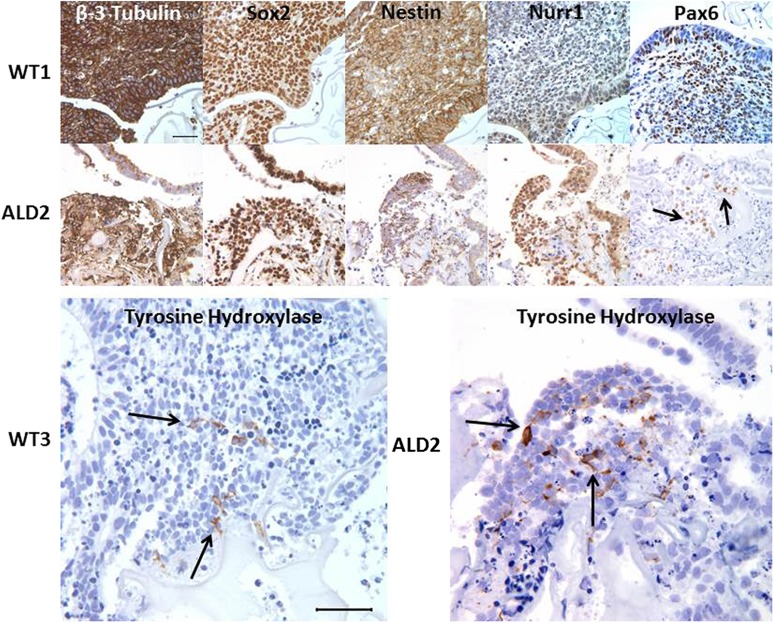
Six additional human iPSC lines (Table 1), including three derived from normal control subjects and three derived from patients with ccALD, were tested in this system. All demonstrated the formation of cOrgs, which were typically visible by D10. Histologic and immunohistochemical analysis of D14 cOrgs in all cases showed β-3 tubulin, Sox2, and nestin immunoreactivity (Fig. 2), as seen in the initial cOrgs derived from the WT4 cell line. In addition, the forebrain neural progenitor marker Pax6 was frequently found in loose clusters of cells (Fig. 2). Furthermore, frozen sections of cOrgs stained for both Sox1 and nestin, and paraffin sections stained for both Sox2 and nestin, showed frequent cells that were double-labeled, consistent with neural stem/progenitor cells (Fig. 1). Many cells in cOrgs derived from all iPSC lines were positive for the midbrain marker nuclear receptor related 1 (Nurr1). Moreover, small clusters of cells in many of the organoids from all iPSC lines were also positive for tyrosine hydroxylase and had dendritic morphology consistent with the formation of dopaminergic neurons (Fig. 2). More rarely, cells expressing olig2, a marker of oligodendrocyte and motor neuron progenitor cells, were found in some cOrgs from all cell lines (data not shown).
Figure 2.
Representative immunohistochemical staining panels for neural markers on day 14 cerebral organoids (cOrgs) derived from control (WT1 and WT3) or cerebral childhood adrenoleukodystrophy patient (ALD2) human induced pluripotent stem cells (hiPSCs). The majority of cells within all cOrgs derived from either control or adrenoleukodystrophy (ALD)-patient hiPSCs stained for β-3 tubulin, Sox2, and nestin. More localized expression of Nurr1 and Pax6 was seen in most cOrgs. Tyrosine hydroxylase positive cells, which often exhibited neuronal-like processes, were found in loose groupings in many of the cOrgs. Scale bars = 50 µm. Specific immunohistochemical stains (brown stain) with hematoxylin (blue) nuclear stain. Abbreviation: Nurr1, nuclear receptor related 1.
Additional experiments were carried out to D28 on all cell lines (n = 7). D28 cOrgs were histologically and immunohistochemically similar to those at D14, but tended to be larger, with some attaining 2.5–3.0 mm in greatest dimension (Fig. 1), and these often showed increased complexity with additional nodules budding from the initial organoid. Furthermore, compact regions exhibiting early stratification of cells near the surfaces of the cOrgs were also found that showed cells labeling for T-box brain 1 (Tbr1), doublecortin (Dcx), or reelin (Fig. 1), indicating early stages of corticogenesis. Some of the cOrgs also completely separated from the surface of the hydrogel and were free-floating in the cell culture plate. Concomitant with the increase in size of D28 cOrgs, there was increased cell death in central regions of many organoids, as indicated by nuclear pyknosis, karyorrhexis, and cell fragmentation.
Gene Expression in Cerebral Organoids
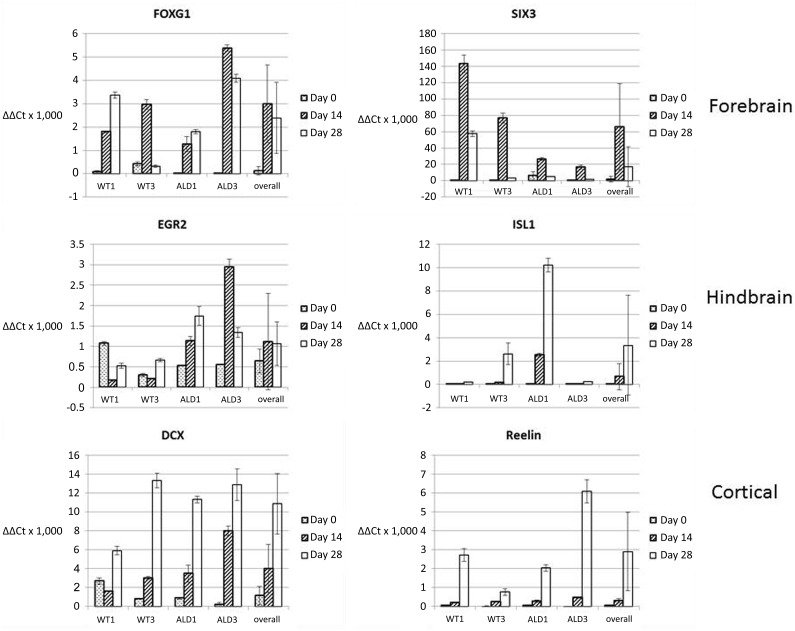
To further characterize cOrg development, we analyzed expression of forebrain (Forkhead box G1 [FoxG1] and sine oculis homeobox 3 [SIX3]), hindbrain (early growth response 2 [EGR2] and Islet 1 [ISL1]), and corticogenesis (DCX and Reelin) markers in undifferentiated iPSCs, D14 cOrgs, and D28 cOrgs derived from two control and two ALD patient hiPSC lines (Fig. 3; supplemental online Tables 3–7). Both forebrain markers were significantly upregulated in cOrgs above those in undifferentiated iPSCs in all four cell lines individually, and the combined analysis showed significantly increased expression at both time points for FOXG (p < .0004) or for SIX3 only D14 (p = .0001). Both hindbrain markers were also expressed in cOrgs derived from all cell lines and, with the exception of EGR2 in the WT1 cell line, were expressed at a level significantly (p < .05) above the undifferentiated iPSCs at one or both time points. However, because of the variation in expression of EGR2 among the cell lines, the differences in expression did not attain significance in the overall analysis (p > .30). Expression of ISL1 was significantly elevated above expression in undifferentiated iPSCs in the combined analysis on D28 (p < .02). Both corticogenesis markers were expressed and significantly upregulated in all cell lines above the undifferentiated iPSCs. DCX expression in the overall analysis was significantly elevated at both D14 (p < .02) and D28 (p < .0001), and Reelin expression in the overall analysis was significantly elevated at D28 (p < .0001).
Figure 3.
Quantitative reverse-transcription polymerase chain reaction analysis of gene expression for forebrain, hindbrain, and corticogenesis markers in undifferentiated human induced pluripotent stem cells, day 14 cerebral organoids (cOrgs), and day 28 cOrgs. Values indicated are mean ± SD. Data are given for gene expression for each individual cell line at D0 (undifferentiated induced pluripotent stem cell), D14, and D28, and the “overall” analysis included pooled data from all four cell lines (WT1, WT3, ALD1, and ALD3) at each time point. Abbreviations: DCX, doublecortin; EGR2, early growth response 2; FOXG1, Forkhead box G1; ISL2, islet 2; SIX3, sine oculis homeobox 3.
Physiologic Assessment of Cerebral Organoids
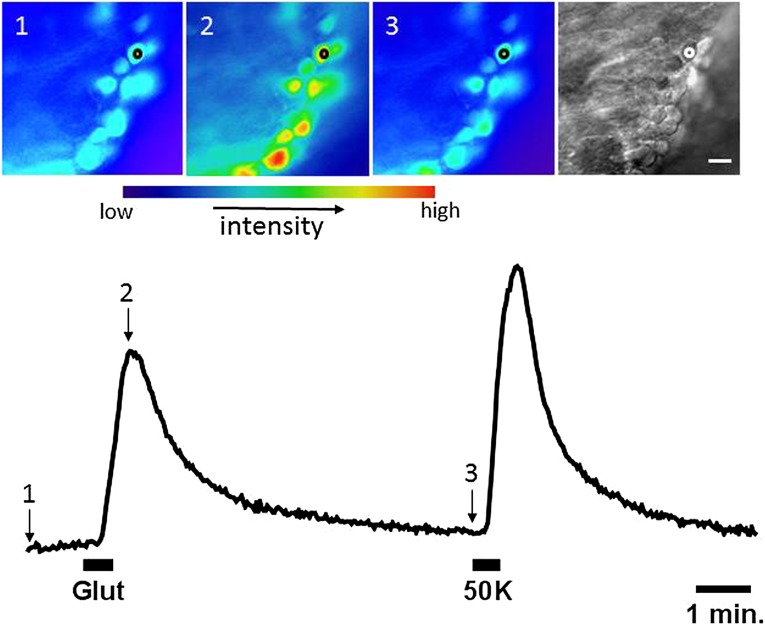
To assess the functional characteristics of putative neurons in the cOrgs, responses to depolarization (evoked by elevation of extracellular potassium) and to bath-application of the excitatory neurotransmitter, glutamate, were examined by using the calcium indicator Fluo-3. Increases in intracellular calcium concentration in response to superfusion with 50 mM K were observed in three different cOrgs derived from WT4. One cOrg that was cultured for 33 days in E8 medium and 18 days in neurobasal medium showed up to 10–20 cells responding per field (Fig. 4). In this same cOrg, all of the cells in three different fields also responded to 100 μM glutamate with an increase in intracellular calcium concentration (Fig. 4). These responses are consistent with those expected to be observed in functioning neurons.
Figure 4.
Physiologic response of cells within a cerebral organoid derived from control human induced pluripotent stem cell line WT1 to glutamate and potassium (K+). Intracellular calcium concentration increased upon bath application of glutamate (100 µM) and elevated potassium concentration (50 K; 50 mM). Upper: False color images showing Fluo-3 fluorescence intensity before (1), at the peak (2), and after recovery (3) from the response to glutamate. Image at far right is an IR transmitted light image of the same field. Scale bar = 10 µm. Lower: Plot of fluorescence intensity over time showing Fluo-3 fluorescence in the region indicated by the circle on the images above. Glutamate and elevated potassium were bath applied during the times indicated by the black bars. Abbreviation: Glut, glutamate.
Discussion
cOrgs derived from patient and disease-specific iPSCs could be an exceptionally valuable tool for in vitro modeling of a variety of human neurological disorders, for the study of neurodevelopment, for neurophysiologic studies, for pharmaceutical drug screening, and as a source of therapeutic cells. In the present study, we describe a novel method for generating cOrgs that, by its simplicity, rapidity, and use of only defined cell culture and matrix components, may serve to accelerate the use and further development of this technology.
Methods available to date have relied upon the use of a xenobiotic extracellular matrix component (Matrigel) that could impose limits on downstream applications of cOrg technology. Generation of cOrgs using our method eliminates the need for this component and thus enables translational applications such as the production of therapeutic cells and use of the cell embedded hydrogel for pharmaceutical or toxicity screening where consistent and reproducible manufacturing are of critical importance. The method may also serve to accelerate studies of the mechanisms involved in PSC differentiation. For example, a recent study suggests that spontaneous differentiation is a default of the primed iPSC and that this is regulated by concerted or opposing actions of Wnt and Fgf2 [14]. Our method of cOrg generation could serve as a unique tool for the study of such foundational signaling in health and disease, including ALD.
We also provide preliminary evidence for the use of this method for the in vitro modeling of ALD and potentially other neurodegenerative diseases. Previously, the potential for cOrgs to model human neurodevelopmental disorders in vitro was elegantly demonstrated by Lancaster et al. [5] in showing disease-specific alterations in cOrgs derived from patients with microencephaly. cOrgs produced in that study showed remarkable size and complexity, which included cortical epithelia that exhibited biologically relevant neuronal layers [4]. In addition, suspension culture has been extensively used to generate complex neuronal tissue-like structures from PSCs. This paradigm involved forming PSCs into embryoid bodies, followed by the addition of conditioned hepatocellular carcinoma medium [15, 16] or, more recently, employment of the “serum free embryoid body-like” (SFEB) method.
The SFEB method uses PSC colonies, or colonies that are dissociated and reaggregated and then cultured in defined medium combined with potent chemical inhibitors to encourage specific cell lineages, such as telencephalic precursors [16] or cortical neuroepithelia [17]. Although stratification of both dividing and nondividing cortical cells, and sequential layering of deep layer and upper layer neurons in SFEB culture has been shown, an in vivo-like neuroepithelium phenotype that exhibits an inside-out-like pattern has not been achieved by using this method [17–20]. Lancaster et al. [5] overcame this difficulty by employing their method, which included the formation of embryoid bodies (EBs), followed by placement of the EBs into Matrigel, and exposure to specific kinase inhibitors and a combination of neural induction media. While Although highly effective, this method was complex and relied on the use of Matrigel, which tends to limit its use for some applications. Another limitation noted by Lancaster et al. [5] was that cOrgs tended to develop central areas of cell death, which limited their growth, which was circumvented by the use of spinner flasks to create convection and improve nutrient and gas exchange. This tendency for central regions to begin to show cell death was also found in cOrgs generated by our method and will require the further development of methods to improve nutrient and gas exchange, such as the use of spinner flasks, gas-permeable culture dishes, and/or flow-through methods of cell culture.
Although the concept of what defines a tissue organoid is still evolving, a recently proposed definition states that they: (a) contain multiple organ-specific cell types, (b) are capable of recapitulating some specific function of the organ, and (c) cells and structures are grouped together and spatially organized similar to an organ [4]. In our constructs, virtually all cells expressed the general neural marker β-3 tubulin, and many also coexpressed the neural progenitor cell markers Sox1/nestin or Sox2/nestin. Further immunohistochemical evidence of regional specification was seen in the expression of the midbrain marker Nurr1 in many cells and expression of the forebrain marker Pax6 or cortical markers Tbr1, doublecortin, or reelin in local groups of cells. Regional specification was further supported by our gene expression analysis that showed evidence for expression of forebrain (FoxG1 and Six3) in all cell lines and hindbrain (Egr2 and Isl1) markers. Finally, cells within the putative cOrgs showed immunohistochemical evidence of distinct neural cell types, including tyrosine hydroxylase (dopaminergic neurons), reelin (Cajal-Retzius cells), and doublecortin (cortical neurons). Along with this, our gene expression analysis further indicated cortical differentiation with significantly increased expression in cOrgs of both doublecortin (D14 and D29) and reelin (D28). Together, these findings support our conclusion that our putative cOrgs exhibit a diversity of neural-specific cell and region-specific differentiation pathways that meet cOrg criterion 1 above. Next, our physiologic studies indicated that cell populations in our putative cOrgs responded to glutamate in a manner consistent with neurons, and thus met criterion 2 for appropriate cell functionality. Finally, the putative cOrgs in our studies showed multiple neuroepithelial structures such as rosettes and larger neural tube-like structures as well as sites of early corticogenesis consistent with developmental architecture of the central nervous system and, therefore, fulfill the requirements for having the appropriate structural organization of criterion 3 (above).
Although the mechanism underlying neural differentiation observed in our chemically defined system was not elucidated in this study, it is possible that cell-matrix interactions and/or the stiffness of the microenvironment could play a role in cOrg development described here. HA has been found to be present in the extracellular matrix in the neural stem cell niche and in the embryonic and adult central nervous system, and it plays multiple roles in proliferation and differentiation [21]. HA, a major component of the Cell-Mate3D matrix, is known to interact with several cell surface receptors, including CD44, the receptor for hyaluronic acid mediated motility (RHAMM) [22, 23], TLR2, and TLR4 [24, 25]; however, which, if any, of these might mediate or influence the neural differentiation in our system has yet to be determined. Others have demonstrated that matrix stiffness alone can influence cell fate, and that culturing cells on or in materials of varying stiffness can direct PSCs to a number of lineages, including neuronal lineages [26–28]. In our preliminary assessment, we found a Young’s modulus for the Cell-Mate3D hydrogel alone to be 10,111 ± 3,742 Pa and that of an iPSC-embedded Cell-Mate3D matrix to be 9,838 ± 1,552 Pa. There was no statistically significant difference between these two measurements, and therefore the entrapment of iPSCs in the Cell-Mate3D gels had no effect on the mechanical properties of the gels. A decrease in the Young’s modulus of a hydrogel after entrapment of cells has been previously demonstrated and was attributed to embedding cells that are softer than the gel and the disruption of interactions between the gel fibers [29]. Interestingly, Engler et al. showed that mesenchymal stem cells differentiate to a neural lineage on a substrate stiffness of 0.1–1 kPa [26], and Leipzig and Shoichet showed that neural stem/progenitor cells will differentiate to neurons, astrocytes, and oligodendrocytes when cultured on substrates of stiffness between 0.6 and 7 kPa, whereas on stiffer substrates (≥10 kPa), they will differentiate to oligodendrocytes [30]. Native brain tissue has been measured to range from 0.5–1 kPa [31–33]. Other groups have demonstrated similar results using materials containing HA [34–36]. Thus, using the oscillating plate rheometer methodology the Cell-Mate3D hydrogel had a higher elastic modulus than was previously reported to promote neural differentiation, suggesting that additional factors may be influencing this phenomenon in our system.
Conclusion
Here, we demonstrated that iPSCs derived from both ALD-affected individuals and patient controls consistently form cOrgs under the study conditions described here. Therefore, this method is suitable for the development of an in vitro model of ALD or other neurodevelopmental or neurodegenerative diseases. Although not intended to assess differences between ALD and control patient-derived cOrgs these initial studies did not reveal any unequivocal differences in frequency of cOrg formation, size, complexity, expression of differentiation markers, or gene expression. However, because the pathogenesis of ccALD likely involves more subtle alterations in metabolism of neurons, astrocytes, or oligodendrocytes we envision that further metabolomic, proteomic, and gene expression studies of much finer resolution than these initial studies will be needed to assess the cOrg model of ALD.
Supplementary Material
Acknowledgments
We thank Paula Overn for the production of excellent histologic preparations and the development of the immunohistochemical and immunofluorescent stains critical to this work. Gross images were obtained by using a Nikon SMZ1500 stereoscope and support by Grant Barthel at the University of Minnesota-University Imaging Centers.
Footnotes
Co-senior authors of this study.
Author Contributions
B.A.L.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; J.H.B.: financial support, provision of study material or patients; A.L.V., C.B.U., K.T.H., S.L.V., and C.M.S.: collection and/or assembly of data; S.S.: collection and/or assembly of data, data analysis and interpretation; C.R.E., P.J.O., and W.C.: provision of study material or patients; Q.W.: data analysis and interpretation; F.P. and S.A.K.: collection and/or assembly of data, manuscript writing; E.K.: data analysis and interpretation, final approval of manuscript; J.R.D. and J.T.: final approval of manuscript; T.D.O.: conception and design, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
J.H.B. is an employee of, has an inventor/leadership position at, and has compensated stock options in BRTI Life Sciences, Inc. B.A.L. is an employee of and has compensated stock options in Bioactive Regenerative Therapeutics, Inc., which provided the Cell-Mate3D materials for this study. T.D.O. holds equity in and is entitled to royalties from BRTI, the company that sells Cell-Mate3D. P.J.O. is a compensated consultant as the Chair of the Institutional Review Board of the National Marrow Donor Program, has compensated honoraria from Genzyme, and is also invited to speak at meetings. The other authors indicated no potential conflicts of interest.
References
- 1.Baker BM, Chen CS. Deconstructing the third dimension: How 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sasai Y, Eiraku M, Suga H. In vitro organogenesis in three dimensions: Self-organising stem cells. Development. 2012;139:4111–4121. doi: 10.1242/dev.079590. [DOI] [PubMed] [Google Scholar]
- 3.Karus M, Blaess S, Brüstle O. Self-organization of neural tissue architectures from pluripotent stem cells. J Comp Neurol. 2014;522:2831–2844. doi: 10.1002/cne.23608. [DOI] [PubMed] [Google Scholar]
- 4.Lancaster MA, Knoblich JA. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 5.Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadoshima T, Sakaguchi H, Nakano T, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex [published correction appears in Proc Natl Acad Sci USA 2014;111:7498] Proc Natl Acad Sci USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger J, Gärtner J. X-linked adrenoleukodystrophy: Clinical, biochemical and pathogenetic aspects. Biochim Biophys Acta. 2006;1763:1721–1732. doi: 10.1016/j.bbamcr.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Berger J, Forss-Petter S, Eichler FS. Pathophysiology of X-linked adrenoleukodystrophy. Biochimie. 2014;98:135–142. doi: 10.1016/j.biochi.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller WP, Rothman SM, Nascene D, et al. Outcomes after allogeneic hematopoietic cell transplantation for childhood cerebral adrenoleukodystrophy: The largest single-institution cohort report. Blood. 2011;118:1971–1978. doi: 10.1182/blood-2011-01-329235. [DOI] [PubMed] [Google Scholar]
- 10.Lund TC, Stadem PS, Panoskaltsis-Mortari A, et al. Elevated cerebral spinal fluid cytokine levels in boys with cerebral adrenoleukodystrophy correlates with MRI severity. PLoS One. 2012;7:e32218. doi: 10.1371/journal.pone.0032218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKinney AM, Nascene D, Miller WP, et al. Childhood cerebral X-linked adrenoleukodystrophy: diffusion tensor imaging measurements for prediction of clinical outcome after hematopoietic stem cell transplantation. AJNR Am J Neuroradiol. 2013;34:641–649. doi: 10.3174/ajnr.A3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orchard PJ, Lund T, Miller W, et al. Chitotriosidase as a biomarker of cerebral adrenoleukodystrophy. J Neuroinflammation. 2011;8:144. doi: 10.1186/1742-2094-8-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindborg B, Brekke JH, Scott CM, et al. A novel chitosan-hyaluronan based hydrocolloid supports in vitro culture and differentiation of human mesenchymal stem/stromal cells (MSC) Tissue Eng A. 2015;21:1952–1962. doi: 10.1089/ten.TEA.2014.0335. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Okamura D, Li M, et al. An alternative pluripotent state confers interspecies chimaeric competency. Nature. 2015;521:316–321. doi: 10.1038/nature14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathjen J, Lake JA, Bettess MD, et al. Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J Cell Sci. 1999;112:601–612. doi: 10.1242/jcs.112.5.601. [DOI] [PubMed] [Google Scholar]
- 16.Rathjen J, Haines BP, Hudson KM, et al. Directed differentiation of pluripotent cells to neural lineages: Homogeneous formation and differentiation of a neurectoderm population. Development. 2002;129:2649–2661. doi: 10.1242/dev.129.11.2649. [DOI] [PubMed] [Google Scholar]
- 17.Mariani J, Simonini MV, Palejev D, et al. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eiraku M, Sasai Y. Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat Protoc. 2011;7:69–79. doi: 10.1038/nprot.2011.429. [DOI] [PubMed] [Google Scholar]
- 19.Eiraku M, Watanabe K, Matsuo-Takasaki M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Nasu M, Takata N, Danjo T, et al. Robust formation and maintenance of continuous stratified cortical neuroepithelium by laminin-containing matrix in mouse ES cell culture. PLoS One. 2012;7:e53024. doi: 10.1371/journal.pone.0053024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brekke J, Thacker K. Hyaluronan as a biomaterial. In: Hollinger J, Guelcher S, editors. Introduction to Biomaterials. Boca Raton, FL: CRC Press; 2006. pp. 219–248. [Google Scholar]
- 22.Preston M, Sherman LS. Neural stem cell niches: Roles for the hyaluronan-based extracellular matrix. Front Biosci (Schol Ed) 2011;3:1165–1179. doi: 10.2741/218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhary M, Zhang X, Stojkovic P, et al. Putative role of hyaluronan and its related genes, HAS2 and RHAMM, in human early preimplantation embryogenesis and embryonic stem cell characterization. Stem Cells. 2007;25:3045–3057. doi: 10.1634/stemcells.2007-0296. [DOI] [PubMed] [Google Scholar]
- 24.Termeer C, Benedix F, Sleeman J, et al. Oligosaccharides of hyaluronan activate dendritic cells via toll-like receptor 4. J Exp Med. 2002;195:99–111. doi: 10.1084/jem.20001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang D, Liang J, Noble PW. Hyaluronan as an immune regulator in human diseases. Physiol Rev. 2011;91:221–264. doi: 10.1152/physrev.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engler AJ, Sen S, Sweeney HL, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Abdeen AA, Kilian KA. Rewiring mesenchymal stem cell lineage specification by switching the biophysical microenvironment. Sci Rep. 2014;4:5188. doi: 10.1038/srep05188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Yong KMA, Villa-Diaz LG, et al. Hippo/YAP-mediated rigidity-dependent motor neuron differentiation of human pluripotent stem cells. Nat Mater. 2014;13:599–604. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott CM, Forster CL, Kokkoli E. Three-dimensional cell entrapment as a function of the weight percent of peptide-amphiphile hydrogels. Langmuir. 2015;31:6122–6129. doi: 10.1021/acs.langmuir.5b00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leipzig ND, Shoichet MS. The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials. 2009;30:6867–6878. doi: 10.1016/j.biomaterials.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Pashuck ET, Cui H, Stupp SI. Tuning supramolecular rigidity of peptide fibers through molecular structure. J Am Chem Soc. 2010;132:6041–6046. doi: 10.1021/ja908560n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meijering E, Jacob M, Sarria J-CF, et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- 33.Paramonov SE, Jun HW, Hartgerink JD. Self-assembly of peptide-amphiphile nanofibers: The roles of hydrogen bonding and amphiphilic packing. J Am Chem Soc. 2006;128:7291–7298. doi: 10.1021/ja060573x. [DOI] [PubMed] [Google Scholar]
- 34.Seidlits SK, Khaing ZZ, Petersen RR, et al. The effects of hyaluronic acid hydrogels with tunable mechanical properties on neural progenitor cell differentiation. Biomaterials. 2010;31:3930–3940. doi: 10.1016/j.biomaterials.2010.01.125. [DOI] [PubMed] [Google Scholar]
- 35.Pan L, Ren Y, Cui F, et al. Viability and differentiation of neural precursors on hyaluronic acid hydrogel scaffold. J Neurosci Res. 2009;87:3207–3220. doi: 10.1002/jnr.22142. [DOI] [PubMed] [Google Scholar]
- 36.Brännvall K, Bergman K, Wallenquist U, et al. Enhanced neuronal differentiation in a three-dimensional collagen-hyaluronan matrix. J Neurosci Res. 2007;85:2138–2146. doi: 10.1002/jnr.21358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.