Abstract
Over the last decade, research to improve success rates in reproductive medicine has focused predominantly on the understanding and optimization of embryo quality. However, the emergence of personalized medicine in ovulation induction and embryology has shifted the focus to assessing the individual status of the endometrium. The endometrium is considered receptive during an individually defined period, the window of implantation (WOI), when the mother permits a blastocyst to attach and implant. This individual receptivity status can now be objectively diagnosed using the endometrial receptivity array (ERA) developed in 2011. The ERA, together with a computational algorithm, detects the unique transcriptomic signature of endometrial receptivity by analyzing 238 differentially expressed genes and reliably predicting the WOI. We and others have illustrated the utility of this personalized diagnostic approach to discriminate between individual physiological variation in endometrial receptivity and unknown endometrial pathology, deemed as causal in recurrent implantation failure (RIF). An international randomized controlled trial (“The ERA as a diagnostic guide for personalized embryo transfer.” ClinicalTrials.gov Identifier: NCT01954758) is underway to determine the clinical value of this endometrial diagnostic intervention in the work-up for reproductive care. In this review, we analyse the current clinical practice in the diagnosis of the endometrial factor together with new avenues of research.
Key words: endometrial receptivity array (ERA), microRNA, window of implantation, implantation failure, assisted reproduction
Abstract
Zusammenfassung
Die Verbesserung der Erfolgsraten der assistierten Reproduktion hat sich bisher vor allem auf das Verständnis sowie die Optimierung der Embryoqualität konzentriert. Erst durch die Einführung der personalisierten Medizin im Rahmen der Ovulationsinduktion und in der Embryologie hat sich der Fokus auch auf den individuellen Status des Endometriums erweitert. Die endometriale Rezeptivität definiert den persönlichen Zeitraum, in dem sich der im Blastozystenstadium befindliche Embryo an das hormonell regulierte Endometrium einer Frau anheften und einnisten kann. Durch den 2011 patentierten Endometrial Receptivity Array (ERA) steht erstmals ein objektiver diagnostischer Test zur Bestimmung der rezeptiven Phase des Endometriums zur Verfügung. ERA identifiziert das transkriptomische Profil des Endometriums anhand der Signatur von 238 unterschiedlich exprimierten Genen in Verbindung mit einem computerbasierten Prädiktor und klassifiziert darauf beruhend den rezeptiven Status der Patientin. Die Bedeutung dieser personalisierten Untersuchung des Endometriums, die zwischen individueller physiologischer Variabilität der endometrialen Rezeptivität und unbekannter Pathologie des Endometriums unterscheiden kann, konnte sowohl durch uns als auch durch andere Gruppen bisher bei Patientinnen mit rezidivierendem Implantationsversagen (RIF) gezeigt werden. Auf diesen Ergebnissen aufbauend, wird der Nutzen von ERA in der Routinediagnostik bei unerfülltem Kinderwunsch aktuell in einer internationalen, randomisierten, kontrollierten Studie („The ERA as a diagnostic guide for personalized embryo transfer.“ ClinicalTrials.gov Identifier: NCT01954758) untersucht.
Schlüsselwörter: endometriale Rezeptivität, Implantationsfenster (WOI), assistierte Reproduktionstechniken (ART), micro-Ribonucleinsäure (RNA)
Introduction
Successful implantation of the embryo in the maternal endometrium is the result of a perfect synchrony between a viable blastocyst, the receptive endometrium, and appropriate communication between them 1. The most investigated element in the implantation triad is the embryo, which seeks to adhere to the endometrial epithelium and invade the decidualized stroma, initiating trophoblast invasion and placentation. Indeed, the understanding of human pre-implantation development is critical (for review see 2), as are the soluble ligands produced and received by their receptors to mediate this fundamental process (for review see 3). However, research to develop an understanding of the endometrial component of implantation has been largely neglected.
The maternal endometrium is receptive to an embryo only during the specific period of time in the menstrual cycle known as the window of implantation (WOI). Classically, this period is considered as occurring 8 to 10 days after ovulation and lasting 2 or 3 days, during which time a functional and transient ovarian steroid-dependent status is acquired to enable the blastocyst to implant. This classical definition was established on the grounds of a relevant clinical study 4 but without basic research supporting it. In this important contribution, published by Wilcox et al. in 1999 4, the day of ovulation was defined on the basis of changes in urinary excretion of the estradiol metabolite estrone 3-glucuronide and the progesterone metabolite pregnanediol 3-glucuronide, which were measured by radioimmunoassay. The authors developed an algorithm to identify the day of ovulation based on the ratio of these urinary hormone metabolites, and claimed the test was similar to measurement of the luteinizing hormone (LH) peak 4. However, 26 years later, the method proposed by these authors to identify ovulation has not been clinically adopted. Further, we now recognize limitations of the use of LH measurements in urine or even in blood to predict ovulation 5. Nevertheless, the clinical community has since assumed that the endometrium in all patients becomes receptive during the indicated time frame (8 to 10 days after ovulation), regardless of individual characteristics or hormonal treatments received (i.e., natural cycles or controlled ovarian stimulation).
Human Endometrial Receptivity
To date no single molecular or histological biomarker has been identified to objectively and reliably diagnose endometrial receptivity. In the absence of such a diagnosis, the endometrium has been supported by progesterone or human chorionic gonadotropin (hCG) as the only “endometrial treatment” in patients undergoing assisted reproductive techniques (ART). Accordingly, embryo transfer (ET) has been guided only by the quality and developmental stage of the embryo and the thickness of the endometrial layer. However, we have demonstrated that in 25 % of cases repeated implantation failure is attributable to endometrial origin 6, which is consistent with the clinical relevance of endometrial receptivity in successful pregnancy 7.
In 1950 Noyes et al. histologically defined the endometrial dating criteria for evaluating the endometrium 8. However, multiple randomized 9, 10 and prospective studies 11, 12, 13, 14, 15, 16, 17 questioned the accuracy and reproducibility of the Noyes method to diagnose endometrial receptivity or fertility status.
Subsequent research has focused on discovering biochemical markers to assess endometrial status. Although myriad molecular mediators, including growth factors, cytokines, chemokines, lipids, and adhesion molecules, have been identified in the endometrium 1, 7, so far, none of these molecules has been established as an endometrial biomarker in clinical practice 18.
Developments in molecular biology techniques, along with global transcriptomic analyses, have enabled the investigation of the genomics of human endometrial development 19. Transcriptomic analyses identify actively expressed genes at the mRNA level at any given time 20. Human endometrial transcriptomic analyses reveal that differential gene expression patterns exist during different phases of the menstrual cycle 21, 22, including during the receptive phase 19, 23. Further, differential transcriptomic profiles have been uncovered in patients with repetitive implantation failure 24, 25, 26 as well as endometrial pathologies such as endometriosis or endometrial cancer 27, 28, and gene expression patterns have been defined during controlled ovarian stimulation (COS) and hormonal replacement therapy (HRT) cycles 29, 30. These efforts enabled the discovery of the unique genomic signature of endometrial receptivity that became the basis of the endometrial receptivity array (ERA) 31. This assay diagnoses the molecular status of the receptive endometrium according to its transcriptomic signature, regardless of its histological appearance 31.
Endometrial Receptivity Array
The ERA is a novel diagnostic method clinically available worldwide that classifies the endometrium as receptive, pre-receptive, or post-receptive 6. The test requires a small biopsy of endometrial tissue taken during scheduled treatment at either 7 days after the luteinizing hormone peak (LH + 7) in a natural cycle, or at the end of 5 days of progesterone administration after estrogen priming in a hormonal replacement therapy cycle (P + 5). RNA extracted from the tissue is applied to a microarray to determine the transcriptomic profile of 238 genes. This transcriptomic profile, when coupled to a computational predictor, objectively identifies whether this endometrium is receptive, pre-receptive or post-receptive by clustering analysis against sample training sets 6, 31. The 238 genes analyzed by ERA were chosen according to the expression data of 14 previous papers by our group searching for the transcriptomic signature of endometrial receptivity in natural cycles, COS, HRT and even in patients with intrauterine device (IUD) (for review see 19). Although these genes were selected by t-test with an absolute fold change > 3 and a false discovery rate < 0.05, the clinical validation was done with a training set in real patients 31. Importantly, the result obtained by ERA is independent of the histological appearance of the endometrium, and has been demonstrated to be more accurate than histological dating 32 and completely reproducible even with up to 40 months between samples 32. This finding is consistent with the idea that the receptivity status remains the same within an individual woman throughout her lifetime, but that different hormonal treatments and states such as pregnancy may change the endometrium since it is a hormonally regulated organ.
Analysis of over 6000 ERA results, performed by our group, indicates that, in approximately 30 % of patients, the endometrial biopsy is classified as non-receptive. In these instances, the predictor describes whether the tissue is pre-receptive (85.0 %) or post-receptive (12.6 %). Based on these findings, the algorithm then recommends the timing of progesterone treatment for the individual patient to find her personalized WOI, thereby obtaining an optimal chance of successful implantation through personalized embryo transfer (pET) (Fig. 1).
Fig. 1.
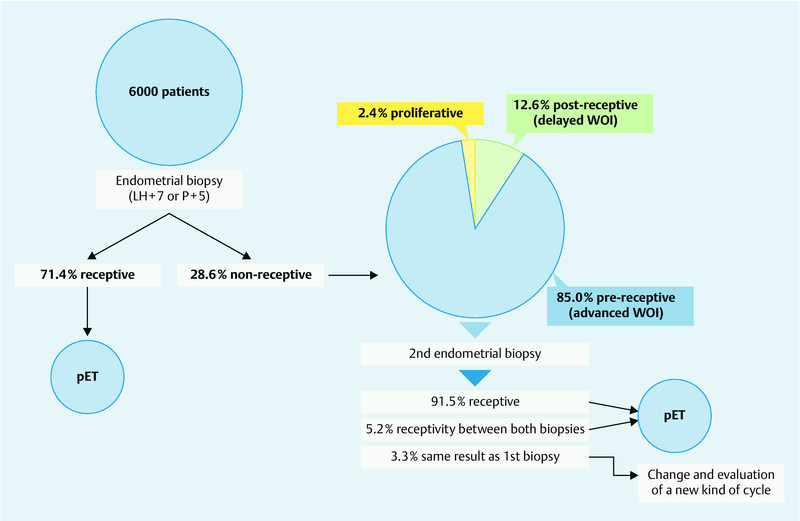
Clinical algorithm for personalized embryo transfer (pET), including the percentage probability (unpublished data provided by C. Simon).
Personalized Embryo Transfer
The clinical application of the ERA has been studied in a prospective, interventional, multicenter, clinical trial in 85 patients with recurrent implantation failure (RIF) versus 25 controls undergoing IVF for the first time 6. The endometrial biopsy was classified as receptive in 74.1 % of patients with RIF; when embryo transfer was performed according to the timing indicated by ERA diagnosis, patients achieved a 33.9 % implantation rate and a 51.7 % pregnancy rate. However, displacement of the WOI was observed in one out of four patients with RIF as diagnosed by ERA 6. In these 26.3 % of patients, when embryo transfer was performed according to ERA-diagnosed timing of the WOI, pregnancy and implantation rates rose to the level of normally receptive controls, in this initial study 7 patients underwent pET.
A clinical case of a successful personalized embryo transfer in a patient having experienced four IVF and three oocyte donation failures has been reported 34. This patient was diagnosed with a displacement of the WOI using the ERA. Therefore, personalized embryo transfer of two blastocysts was performed after 7 days of progesterone (P + 7) in an HRT cycle, resulting in a successful twin pregnancy after 7 previous repeated implantation failures. Similarly, in a pilot study of 17 patients undergoing oocyte donation who had experienced failed implantations with routine embryo transfer, the implantation rate was increased from 12.9 to 34.5 % and the pregnancy rate from 23.5 to 52.9 % when pET was performed following ERA diagnosis 33. All 17 patients were initially diagnosed with a displaced WOI, whereby the endometrium biopsy was classified pre-receptive in 16 patients and post-receptive in one patient 33.
The value of the diagnosis of endometrial receptivity during the routine infertility work-up of patients undergoing assisted reproductive technology is currently being explored in an international, multicenter, prospective, randomized, interventional and controlled study – “The ERA as a diagnostic guide for personalized embryo transfer” – comparing fresh embryo transfer versus elective delayed embryo transfer or pET (Clinical trails.gov, Identifier: NCT01954758).
MicroRNAs: New Molecules Advancing Our Reproductive Knowledge
Despite the wealth of information uncovered in recent years, technologies continue evolving to discover all transcripts across the transcriptome. In 2014, Hu et al. reported the first global gene expression profile of the human endometrium using next-generation, high-throughput RNA sequencing (RNA-seq) 34. This RNA-seq-based transcriptome comparison of pre-receptive and receptive human endometrium revealed a total of 2372 differentially expressed genes, including metallothionein family members, HAP1, ZCCHC12, MRAP2, OVGP1, regulatory factors (GLI2, CDC25A, TLR9, MT1G and SLC5A1), and transcription factors (AP2 and SP1) that have not previously been linked to endometrial receptivity. In addition, the discovery of microRNAs (miRNAs) as potential post-transcriptional regulators of gene expression 35 represents a breakthrough in biology during the last ten years and has become an extremely active research field 36, 37, 38, 39.
MiRNAs are small, non-coding RNA sequences of 18 to 25 nucleotides that regulate gene expression post-transcriptionally 40. These molecules do not encode proteins; instead, miRNAs target mRNAs through complementary base pairing to the 3′-untranslated region for degradation or repression, thereby functioning as gene silencers 41. Based on the degree of sequence homology, one miRNA can potentially target a broad range of genes and one gene can be regulated by several miRNAs 40. Initially, long precursors (pri-miRNAs) are transcribed and processed to shorter precursors (pre-miRNAs) in the nucleus 37; these precursors are exported into the cytoplasm and incorporated into the RNA-induced silencing complex (RISC) to bind an mRNA target 42, as shown in Fig. 2.
Fig. 2.
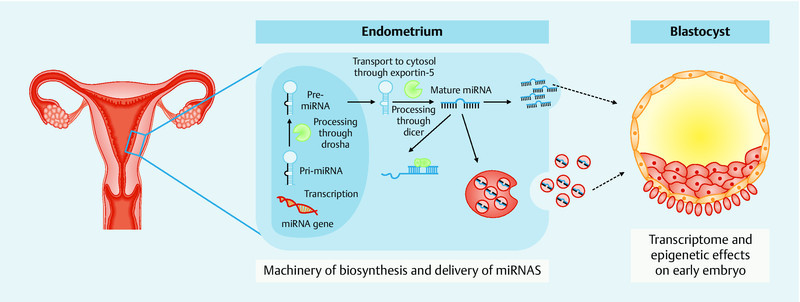
Diagram showing the process of miRNA synthesis and the potential role of miRNAs in the embryo-maternal dialogue.
As has been found for mRNAs, miRNAs are differentially expressed in the endometrium across the menstrual cycle 43. Further, the endometrial epithelium releases miRNAs that are secreted into the endometrial fluid 43. Profiling of miRNA and mRNA transcripts in human endometrium suggests that the hormonal regulation of miRNAs leads to a suppression of cell proliferation by down-regulating the expression of some cell cycle genes in the endometrial epithelium during the secretory phase 44. By isolating endometrial epithelial cells from endometrial biopsies of 14 fertile women in the late-proliferative and mid-secretory phases, Kuokkanen et al. identified miRNA-29B, miRNA-29C, miRNA-30B, miRNA-30D, miRNA-31, miRNA-193A-3P, miRNA-203, miRNA-204, miRNA-200C, miRNA-210, miRNA-582-5P, and miRNA-345 as significantly increased in the secretory endometrium. Indeed, a subset of miRNAs, namely hsa-miR-30b and hsa-miR-30d, are significantly upregulated, whereas hsa-miR-494 and hsa-miR-923 are downregulated, in receptive endometrium (LH + 7) versus pre-receptive endometrium (LH + 2) in fertile women 45. These findings support the previously reported upregulation of hsa-miR-30b and hsa-miR-30-d during the acquisition of endometrial receptivity 46. The involvement of miRNAs in failed embryo implantation has been suggested in patients with recurrent implantation failure 47. The existence of 13 differentially expressed miRNAs (miRNA-145, miRNA-23b, miRNA-99a, miRNA-27b, miRNA-652, miRNA-139-5p, miRNA-195, miRNA-342-3p, miRNA-150, miRNA-374b, miRNA-32, miRNA628-5b, miRNA-874) has been described in patients with recurrent implantation failure; these may regulate the expression of up to 3800 genes 47.
Recently, our group has demonstrated that hsa-miR-30d is secreted by the human endometrial epithelium into the endometrial fluid either free or in exosome-associated form, and can be incorporated into the pre-implantation embryo to potentially modify its transcriptome 43. The internalization of this miRNA results in an indirect overexpression of genes encoding for certain molecules involved in embryonic adhesion, such as ITGB3, ITGA7, and CDH5 43. Furthermore, it has been suggested that miRNAs can be secreted by the human embryo 48; hsa-miR-191, hsa-mi-372, and hsa-miR-645 are differentially expressed according to the fertilization method, chromosomal status, and pregnancy outcome. Together, these findings reinforce the concept of maternal-embryonic cross-talk that uses many different languages, with miRNAs as one of them.
Conclusion
The receptivity status of the endometrium can now be diagnosed reliably by the ERA test, an objective molecular tool based on the transcriptomic signature of human endometrial receptivity, to identify the WOI. The ERA can guide and improve our clinical practice by introducing and enabling a personalized diagnosis of the WOI and, accordingly, a personalized embryo transfer. In the near future, the challenge will be to identify biomarkers of endometrial receptivity that could be assessed by non-invasive methods. MiRNAs may be interesting candidate molecules to consider, particularly with the potential role of maternal endometrial miRNAs as transcriptomic modifiers of the preimplantation embryo.
Footnotes
Conflict of Interest CS is inventor of the ERA patent and holds shares in Igenomix, the company commercializing the ERA test. MR & FV are employees of Igenomix.
References
- 1.Cha J, Vilella F, Dey S K, Simón C. Washington, DC: Science/AAAS; 2013. Molecular Interplay in successful Implantation; pp. 44–48. [Google Scholar]
- 2.Niakan K K, Han J, Pedersen R A. et al. Human pre-implantation embryo development. Development. 2012;139:829–841. doi: 10.1242/dev.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thouas G A, Dominguez F, Green M P. et al. Soluble ligands and their receptors in human embryo development and implantation. Endocr Rev. 2015;36:92–130. doi: 10.1210/er.2014-1046. [DOI] [PubMed] [Google Scholar]
- 4.Wilcox A J, Baird D D, Weinberg C R. Time of implantation of the conceptus and loss of pregnancy. N Engl J Med. 1999;340:1796–1799. doi: 10.1056/NEJM199906103402304. [DOI] [PubMed] [Google Scholar]
- 5.Direito A, Bailly S, Mariani A. et al. Relationships between the luteinizing hormone surge and other characteristics of the menstrual cycle in normally ovulating women. Fertil Steril. 2013;99:279–285. doi: 10.1016/j.fertnstert.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz-Alonso M, Blesa D, Diaz-Gimeno P. et al. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertil Steril. 2013;100:818–824. doi: 10.1016/j.fertnstert.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Achache H, Revel A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update. 2006;12:731–746. doi: 10.1093/humupd/dml004. [DOI] [PubMed] [Google Scholar]
- 8.Noyes R W, Herting A T, Rock J. Dating the endometrial biopsy. Fertil Steril. 1950;1:3–25. doi: 10.1016/j.fertnstert.2019.08.079. [DOI] [PubMed] [Google Scholar]
- 9.Coutifaris C, Myers E R, Guzick D S. et al. Histological dating of timed endometrial biopsy tissue is not related to fertility status. Fertil Steril. 2004:1264–1272. doi: 10.1016/j.fertnstert.2004.03.069. [DOI] [PubMed] [Google Scholar]
- 10.Murray M J, Meyer W R, Zaino R J. et al. A critical analysis of the accuracy, reproducibility, and clinical utility of histologic endometrial dating in fertile women. Fertil Steril. 2004;81:1333–1343. doi: 10.1016/j.fertnstert.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 11.Balasch J, Fabregues F, Creus M. et al. The usefulness of endometrial biopsy for luteal phase evaluation in infertility. Hum Reprod. 1992;7:973–977. doi: 10.1093/oxfordjournals.humrep.a137782. [DOI] [PubMed] [Google Scholar]
- 12.Balasch J, Vanrell J A, Creus M. et al. The endometrial biopsy for diagnosis of luteal phase deficiency. Fertil Steril. 1985;44:699–701. doi: 10.1016/s0015-0282(16)48990-9. [DOI] [PubMed] [Google Scholar]
- 13.Scott R T, Snyder R R, Strickland D M. et al. The effect of interobserver variation in dating endometrial histology on the diagnosis of luteal phase defects. Fertil Steril. 1988;50:888–892. doi: 10.1016/s0015-0282(16)60367-9. [DOI] [PubMed] [Google Scholar]
- 14.Scott R T, Snyder R R, Bagnall J W. et al. Evaluation of the impact of intraobserver variability on endometrial dating and the diagnosis of luteal phase defects. Fertil Steril. 1993;60:652–657. doi: 10.1016/s0015-0282(16)56216-5. [DOI] [PubMed] [Google Scholar]
- 15.Gibson M, Badger G J, Byrn F. et al. Error in histologic dating of secretory endometrium: variance component analysis. Fertil Steril. 1991;56:242–247. doi: 10.1016/s0015-0282(16)54479-3. [DOI] [PubMed] [Google Scholar]
- 16.Landis J R, Koch G G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 17.Ordi J, Creus M, Quinto L. et al. Within-subject between-cycle variability of histological dating, alpha versus beta 3 integrin expression, and pinopod formation in the human endometrium. J Clin Endocrinol Metab. 2003;88:2119–2125. doi: 10.1210/jc.2002-021659. [DOI] [PubMed] [Google Scholar]
- 18.Aghajanova L, Simon C, Horcajadas J. Are favourite molecules of endometrial receptivity still in favour? Expert Rev Obstet Gynecol. 2008;3:487–501. [Google Scholar]
- 19.Ruiz-Alonso M, Blesa D, Simon C. The genomics of the human endometrium. Biochim Biophys Acta. 2012;1822:1931–1942. doi: 10.1016/j.bbadis.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Galliano D, Pellicer A. MicroRNA and implantation. Fertil Steril. 2014;101:1531–1544. doi: 10.1016/j.fertnstert.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 21.Borthwick J M, Charnock-Jones D S, Tom B D. et al. Determination of the transcript profile of human endometrium. Mol Hum Reprod. 2003;9:19–33. doi: 10.1093/molehr/gag004. [DOI] [PubMed] [Google Scholar]
- 22.Ponnampalam A P, Weston G C, Trajstman A C. et al. Molecular classification of human endometrial cycle stages by transcriptional profiling. Mol Hum Reprod. 2004;10:879–893. doi: 10.1093/molehr/gah121. [DOI] [PubMed] [Google Scholar]
- 23.Horcajadas J A, Pellicer A, Simon C. Wide genomic analysis of human endometrial receptivity: new times, new opportunities. Hum Reprod Update. 2007;13:77–86. doi: 10.1093/humupd/dml046. [DOI] [PubMed] [Google Scholar]
- 24.Koler M, Achache H, Tsafrir A. et al. Disrupted gene pattern in patients with repeated in vitro fertilization (IVF) failure. Hum Reprod. 2009;24:2541–2548. doi: 10.1093/humrep/dep193. [DOI] [PubMed] [Google Scholar]
- 25.Altmäe S, Martinez-Conejero J A, Salumets A. et al. Endometrial gene expression analysis at the time of embryo implantation in women with unexplained infertility. Mol Hum Reprod. 2010;16:178–187. doi: 10.1093/molehr/gap102. [DOI] [PubMed] [Google Scholar]
- 26.Tapia A, Vilos C, Marín J C. et al. Bioinformatic detection of E47, E2F1 and SREBP1 transcription factors as potential regulators of genes associated to acquisition of endometrial receptivity. Reprod Biol Endocrinol. 2011;27:9–14. doi: 10.1186/1477-7827-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuzaki S. DNA microarray analysis in endometriosis for development of more effective targeted therapies. Front Biosci (Elite Ed) 2011;3:1139–1153. doi: 10.2741/e317. [DOI] [PubMed] [Google Scholar]
- 28.Habermann J K, Bündgen N K, Gemoll T. et al. Genomic instability influences the transcriptome and proteome in endometrial cancer subtypes. Mol Cancer. 2011;10:132. doi: 10.1186/1476-4598-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon C, Oberyé J, Bellver J. et al. Similar endometrial development in oocyte donors treated with either high- or standard-dose GnRH antagonist compared to treatment with a GnRH agonist or in natural cycles. Human Reprod. 2005;20:3318–3327. doi: 10.1093/humrep/dei243. [DOI] [PubMed] [Google Scholar]
- 30.Horcajadas J A, Riesewijk A, Polman J. et al. Effect of controlled ovarian hyperstimulation in IVF on endometrial gene expression profiles. Mol Hum Reprod. 2005;11:195–205. doi: 10.1093/molehr/gah150. [DOI] [PubMed] [Google Scholar]
- 31.Diaz-Gimeno P, Horcajadas J A, Martínez-Conejero J A. et al. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril. 2011;95:50–60. doi: 10.1016/j.fertnstert.2010.04.063. [DOI] [PubMed] [Google Scholar]
- 32.Diaz-Gimeno P, Ruiz-Alonso M, Blesa D. et al. The accuracy and reproducibility of the endometrial receptivity array is superior to histology as a diagnostic method for endometrial receptivity. Fertil Steril. 2013;99:508–517. doi: 10.1016/j.fertnstert.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Alonso M, Galindo N, Pellicer A. et al. What a difference two days make: “personalized” embryo transfer (pET) paradigm: a case report and pilot study. Hum Reprod. 2014;29:1244–1247. doi: 10.1093/humrep/deu070. [DOI] [PubMed] [Google Scholar]
- 34.Hu S, Yao G, Wang Y. et al. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-Seq. J Clin Endocrinol Metabol. 2014;99:E2744–E2753. doi: 10.1210/jc.2014-2155. [DOI] [PubMed] [Google Scholar]
- 35.Lee R C, Feinbaum R L, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 36.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 37.Bartel D P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Liang H, Zhang J. et al. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Rosenbluth E M, Shelton D N, Sparks A E. et al. MicroRNA expression in the human blastocyst. Fertil Steril. 2013;99:855–861. doi: 10.1016/j.fertnstert.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Lim L P, Lau N C, Garrett-Engele P. et al. Microarray analysis shows that some microRNAs downregulate large numbers of traget mRNAs. Nature. 2005;7027:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 41.Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 42.Lee J Y, Kim S, Hwang do W. et al. Development of a dual-luciferase reporter system for in vivo visualization of MicroRNA biogenesis and posttranscriptional regulation. J Nucl Med. 2008;49:285–294. doi: 10.2967/jnumed.107.042507. [DOI] [PubMed] [Google Scholar]
- 43.Vilella F, Moreno-Moya J M, Balaguer N. et al. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development. 2015;142:3210–3221. doi: 10.1242/dev.124289. [DOI] [PubMed] [Google Scholar]
- 44.Kuokkanen S, Chen B, Ojalvo L. et al. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol Reprod. 2010;82:791–801. doi: 10.1095/biolreprod.109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altmae S, Martinez-Conejero J A, Esteban F J. et al. MicroRNAs miR-30b, miR-30d, and miR-494 regulate human endometrial receptivity. Reprod Sci. 2013;20:308–317. doi: 10.1177/1933719112453507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sha A G, Liu J L, Jiang X M. et al. Genome-wide identification of micro-ribonucleic acids associated with human endometrial receptivity in natural and stimulated cycles by deep sequencing. Fertil Steril. 2011;96:150–155. doi: 10.1016/j.fertnstert.2011.04.072. [DOI] [PubMed] [Google Scholar]
- 47.Revel A, Achache H, Stevens J. et al. MicroRNAs are associated with human embryo implantation defects. Human Reprod. 2011;26:2830–2840. doi: 10.1093/humrep/der255. [DOI] [PubMed] [Google Scholar]
- 48.Rosenbluth E M, Shelton D N, Wells L M. et al. Human embryos secrete microRNAs into culture media-a potential biomarker for implantation. Fertil Steril. 2014;101:1493–1500. doi: 10.1016/j.fertnstert.2014.01.058. [DOI] [PubMed] [Google Scholar]


