Abstract
In recent years, Candida albicans infections treatment has become a growing problem because, among others, pathogenic strains are capable to develop resistance to the administered drugs. The elaboration of rapid and accurate method of resistance assessment is an important goal of many studies. They aim to avoid inappropriate dosage or drug choice, which may be life threatening in case of severe candidiasis. Here we propose a new protocol to predict C. albicans infections. The resistance prediction is based on high-resolution melt (HRM) analysis of ERG11 gene, especially, at the particularly unstable regions. Two statistically significant nucleotide polymorphisms were detected among twenty-seven strains isolated from saliva, one of which was silent mutation (Glu266Asp, Leu480Leu). We propose also HRM analysis as a convenient, simple and inexpensive method of preliminary selection of C. albicans DNA samples that vary in ERG11 nucleotide sequence within presumed region. Taken together, our study provides firm basis for the development of fast, simple and reliable methodology for diagnosis of C. albicans infections.
Introduction
Candida spp are opportunistic yeasts that normally inhabit human skin, genitourinary and gastrointestinal tracts. Under some conditions, Candida may overgrow and colonize mucosal and skin surfaces [5], which usually leads to easily treated infections. However, in patients with impaired immune system, severe candidiasis and life-threatening systemic infections can develop. Among Candida species, Candida albicans is the most frequent cause of human infections.
Several different mechanisms of drug resistance have been already described [4, 40]. Resistant Candida strains may overexpress membrane transporters which are responsible for drug removal from the cell [25, 28]. Efflux pumps can also be more active in drug-resistant strains than in susceptible ones [38]. Most commonly administered antifungal drugs are azole compounds and the main azole drugs target is lanosterol-14-α-demethylase, encoded by ERG11. Point mutations in ERG11, as well as overexpression of this gene, are usually involved in the development of resistance to azole-based drugs [6]. Mutations that lead to amino acid substitution may cause changes in the affinity of the enzyme for azole derivatives [6, 30, 41]. Thus, huge effort has been made to develop fast and reliable methods for detecting strains that bear mutations [1, 35, 39]. Most of these methods involve nucleic acid amplification and gel electrophoresis [35].
Among numerous ERG11 mutations, both homozygous and heterozygous nucleotide changes were reported to correlate with azole susceptibility [6, 29, 41, 42]. A huge progress in ERG11 mutations identification has been made in recent years and it is well known now, which of them are the most frequent and which are most probably involved in azole resistance [24]. However, detailed drug-resistant mutation pattern of ERG11 gene is still desired. We suggest that to achieve this aim, it is also important to consider mutations that do not directly change amino acid composition of the enzyme, but may also correlate with resistant phenotype, and thus may serve as convenient drug resistance markers. Likewise, silent polymorphism in drug-resistance transporter MDR1 gene (C3435T) was established as a risk factor for non-clear cell renal cell carcinoma [34]. It has been also shown that (C3435T) polymorphism may lead to the synthesis of protein product with different structural and functional properties, in spite of unchanged amino acid sequence [12]. Proposed molecular mechanism of this phenomena is the existence of frequent and infrequent codons in mRNA which influence the translation kinetics and temporal separation of folding events during protein synthesis causes conformational and functional differences of final protein [13]. Silent mutations in ERG11 gene of C. albicans occur with threefold higher frequency then amino acid changing mutations [21].
HRM (High-Resolution Melt) analysis is a precise and sensitive method, which can be successfully used for identification of species [20]. This technique allows to discriminate amplicons that differ in single nucleotide (single-nucleotide polymorphisms) as they generate various shapes of melting curves when heated after amplification [15]. Therefore, we propose HRM analysis as a rapid and robust method of preliminary scanning amplicons which allows to minimize the number of samples to be sequenced. Thus, it may contribute essentially to save money, time and labor, especially in case of diagnostic laboratories. Here we applied a recent PCR-based technique, high-resolution melting (HRM) analysis, to determine differences in ERG11 sequence isolated from various C. albicans strains and searched for ERG11 polymorphisms that are statistically significant in drug-resistance prediction.
Materials and Methods
Control Strains and Clinical Isolates
Three strains used as control strains were as follows: Ca11r—C. albicans (ATCC 10231) and Ca12r—C. albicans (ATCC 60193) and Ca1r—clinical C. albicans strain identified by MALDI. Seven clinical isolates came from vaginal candidiasis patients (Ca1V–Ca7V) and have been kindly provided by Dr. Agata Karowicz-Bilińska (Medical University of Lodz). 28 strains (Ca1s–Ca82s) were isolated from the saliva of healthy volunteers (Bioethics Committee agreement no. 6./KBBN-UŁ/II/2014). Clinical strains were isolated on yeast extract–peptone–dextrose (YEPD) agar plates supplemented with chloramphenicol. Then, they were directly identified as C. albicans species using chromogenic culture media (chromID®Candida, bioMérieux), HRM and sequence analysis of ITS2 regions (data not shown). All strains were stored at −70 °C as stocks in 8 % DMSO. Strains were cultured for 24 h at 37 °C in YEPD-agar containing 2 % glucose, 2 % peptone, and 1 % yeast extract.
Antifungal Susceptibility Testing
The susceptibility of reference and vaginal strains to antifungal drugs was tested according to the modified NCCLS M27-A microdilution procedures. Briefly, 1:1 dilutions of fluconazole (FCA), ketoconazole (KET), miconazole (MIC), itraconazole (ITR), amphotericin B (AMB), and 5 flucytosine (5FC) were prepared in 96-well plates in RPMI medium, frozen and thawed when needed. 24-h yeast culture in YPD medium was washed in RPMI medium and 2.5 × 103 cells mL−1 were seeded on a 96-well plate. IC50 was determined spectrophotometrically after 48 h of incubation. To determine the saliva isolates, susceptibility ATB FUNGUS3 (BioMerieux) tests were used according to manufacturer’s instructions.
DNA Isolation
Genetic material was extracted with modified Harju yeast genomic DNA isolation method [8]. Briefly, liquid culture of yeast was pelleted. Cell pellets were resuspended in 200 μl of lysis buffer [2 % Triton X-100, 1 % SDS, 100 mM NaCl, 10 mM Tris–HCl, 1 mM EDTA (pH 8.0)]. Cell suspension was heat shocked on ice bath followed by 95 °C water bath twice. Samples were then vortexed with chloroform, centrifuged and the aqueous layer was transferred to the ice-cold 96 % ethanol. After 5 min incubation, the probes were centrifuged and the supernatant was removed. DNA was washed with 70 % ethanol, pelleted, air-dried, resuspended in 30 µl of sterile water and stored at −20 °C.
HRM Analysis
For HRM analysis, the whole ERG11 gene was divided to 9 regions, aprox. 300 bp each. Primers were designed so as to obtain all specific PCR products at the same annealing temperature. Sequences of primers used for amplification are given in Table 1.
Table 1.
List of primers used for the amplification in HRM analysis
| PCR product size (bp) | Sequence of forward primer | Sequence of reverse primer | |
|---|---|---|---|
| Scan 1 | 268 | 5′-AGACAAAGAAAGGGAATTCAATC-3′ | 5′-TGCCATACTAAGTTGTAAACAAATGG-3′ |
| Scan 2 | 263 | 5′-CCCTTAGTGTTACACAACAGATCA-3′ | 5′-AAATTCATGACCTTTTGGACCT-3′ |
| Scan 3 | 263 | 5′-TTAGGGAAAATTATGACGGTTTAT-3′ | 5′-CTTTCATCAGTAACAAAATAATTCAAA-3′ |
| Scan 4 | 310 | 5′-TGCTAAATTTGCTTTGACTACTGA-3′ | 5′-AGCATCACGTCTCCAATAATGA-3′ |
| Scan 5 | 264 | 5′-TCTGATTTAGATAAAGGTTTTACCCC-3′ | 5′-TTGACCACCCATAAGAATACCA-3′ |
| Scan 6 | 282 | 5′-GGTGTGAAAATGACTGATCAAGA-3′ | 5′-TTTTCTAAAAATAGAATGTAATGGCA-3′ |
| Scan 7 | 274 | 5′-CCATCAGTCAATAACACTATTAAGGAA-3′ | 5′-TCCCAAACCCATAATCAACTTC-3′ |
| Scan 8 | 274 | 5′-TGCCAAAGCTAATTCTGTTTCA-3′ | 5′-TTTTTCCCAAATGATTTCTGCT-3′ |
| Scan 9 | 323 | 5′-GCCTGACCCTGATTATAGTTCAA-3′ | 5′-AAATAACCAGTGGACAAAAACCAT-3′ |
PCR was performed with Go Taq Master Mix (Promega), LCGreen (BioFire Diagnostics) and aprox. 0.2 µg DNA template was filled up with water to reach 15 µl of reaction mixture. HRM reactions were performed using the Bio-Rad CFX96 real-time PCR system. The amplification program was one cycle at 95 °C (2 min) and 95 °C (30 s), 60 °C (30 s), 72 °C (15 s) × 55 cycles followed by: 95 °C for 1 min, and 40 °C for 1 min. Melting of amplicons was conducted by rising the temperature from 67 °C to 91 °C with an 0.2 °C increase step. Melting curve analysis was performed by BioRad Precision Melt Analysis Software and verified visually.
PCR and DNA Sequencing
Whole ERG11 gene was amplified as two overlapping parts, each amplified with a unique oligonucleotide primer pair. Gene fragments were then sequenced from both ends with the same primers. The first part primer pair used was as follows: forward primer (5′-AGACAAAGAAAGGGAATTCAATC-3′), reverse primer (5′-TTGACCACCCATAAGAATACCA-3′); the second part forward primer (5′-CGTGATGCTGCTCAAAAGAA-3′), reverse primer (5′-AAATAACCAGTGGACAAAAACCAT-3′). PCR was carried out with Go Taq Master Mix (Promega) and the amplification program was one cycle at 95 °C for 3 min, followed by 45 cycles at 95 °C for 30 s, 58.1 °C for 30 s, and 72 °C for 45 s. Amplicons had the length of approx. 250–300 bp, were verified by agarose gel electrophoresis and were purified with a Wizard SV Gel and PCR Clean-up System (Promega).
DNA reads (Eurofins MWG Operon, Germany; Institute of Medical Biology of PAS, Poland) were screened for the presence of ambiguous nucleotide signals.
Results
Candida albicans Strains Exhibit Various Resistance to Antifungal Drugs
The susceptibility of reference strains and strains isolated from vagina was determined with microdilution method. MIC was defined as the 50 % inhibition of growth for 5 flucytosine and azole drug tests, and total inhibition of growth for amphotericin B. From among nine C. albicans strains tested, one (Ca5V) was classified as resistant to all azole compounds tested (Table 2). Five strains (Ca1V, Ca4V, Ca7V, Ca2V, Ca12r) have shown increased resistance to azole drugs and 5-flucytosine as assessed by drug concentration necessary for complete growth inhibition (data not shown).
Table 2.
Antifungal drug susceptibility of reference strains and strains isolated from vagina determined with microdilution method
| Strain | Susceptibility of reference strains and vaginal isolates, IC50 (mg/l) | |||||
|---|---|---|---|---|---|---|
| MIC | KET | ITR | FCA | AMB | 5FC | |
| Ca1r | 0.0625 | 0.125 | 0.25 | 0.5 | 0.25 | 0.25 |
| Ca1V | 0.0625 | 0.125 | 0.25 | 0.25 | 0.5 | 1 |
| Ca2V | 0.0625 | 0.125 | 0.25 | 0.25 | 0.5 | 1 |
| Ca3V | 0.0625 | 0.0625 | 0.25 | 0.25 | 0.25 | 0.125 |
| Ca4V | 0.0625 | 0.25 | 0.5 | 0.5 | 0.5 | 1 |
| Ca5V | 0.5 | 2 | 16 | 64 | 0.25 | 1 |
| Ca7V | 0.0625 | 0.125 | 0.25 | 0.25 | 0.25 | 0.25 |
| Ca11r | 0.0625 | 0.125 | 0.5 | 0.5 | 0.25 | 0.125 |
| Ca12r | 0.0625 | 0.125 | 0.5 | 0.5 | 0.25 | 0.5 |
The growth inhibition of saliva strains was determined visually on ATB FUNGUS3 tests after 24 h incubation (Table 3). Visual readings for most strains did not allow to establish MIC values as they showed very similar growth in all azole drug concentrations. Fourteen strains were classified as resistant (slight decrease in turbidity), and eleven were susceptible (no growth), one strain (Ca21s) was hard to classify (hazy growth). Strain Ca68s showed sharp difference in growth ability as 16 mg/l of fluconazole completely inhibited yeast growth, what is defined as susceptible dose-dependent according to NCCLS interpretive guidelines. Amphotericin B and 5-flucytosine efficiently inhibited growth of all strains.
Table 3.
Antifungal drug susceptibility of saliva strains determined with ATB FUNGUS3 test
| Resistant | Intermediate | Susceptible |
|---|---|---|
| Saliva yeast strain number | ||
| Ca1s | Ca21s | Ca6s |
| Ca2s | Ca68s (fluconazole susceptible dose-dependent) | Ca10s |
| Ca3s | Ca11s | |
| Ca25s | Ca20s | |
| Ca26s | Ca22s | |
| Ca36s | Ca23s | |
| Ca38s | Ca61s | |
| Ca40s | Ca70s | |
| Ca41s | Ca74s | |
| Ca44s | Ca80s | |
| Ca52s | Ca82s | |
| Ca59s | ||
| Ca69s | ||
| Ca81s | ||
HRM Analysis Allowed to Distinguish Five Variants of ERG11 Gene Related to Drug Resistance
HRM analysis was conducted to verify the utility of this method in analysis of C. albicans ERG11 gene profiles that correlate with drug resistance. Whole sequence of C. albicans ERG11 gene was analyzed as nine fragments. PCR followed by HRM analysis of products was conducted, which allowed to distinguish between different genetic variants of reference strains and vaginal isolates (Table 4). Initially, one to four variants/clusters of each scan were determined among 9 DNA samples. Then, the same patterns of 9 scans variants/clusters were classified as gene variants and marked 1.–5.
Table 4.
Variants of scans and ERG11 gene among reference strains and vaginal isolates identified by HRM analysis
| Strain name | Scan number | Gene variants | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |||
| Ca1r | A | A | C | C | A | B | B | C | D | → | 3. |
| Ca1V | A | A | B | B | A | C | A | A | A | → | 1. |
| Ca2V | A | A | A | A | A | A | A | B | B | → | 2. |
| Ca3V | B | A | B′ | B | B | B | C | A | C | → | 4. |
| Ca4V | A | A | B | B | A | C | A | A | A | → | 1. |
| Ca5V | A | A | A | A | A | A | A | B | B | → | 2. |
| Ca7V | A | A | B | B | A | C | A | A | A | → | 1. |
| Ca11r | A | A | C | B | A | B | B | C | A | → | 5. |
| Ca12r | A | A | A | A | A | A | A | B | B | → | 2. |
A, B, C, D—variants of ERG11 gene fragments among each scan, B′—scan 3 variant not distinguishable by means of HRM, 1–5—whole ERG11 sequence variants on the basis of scan variants
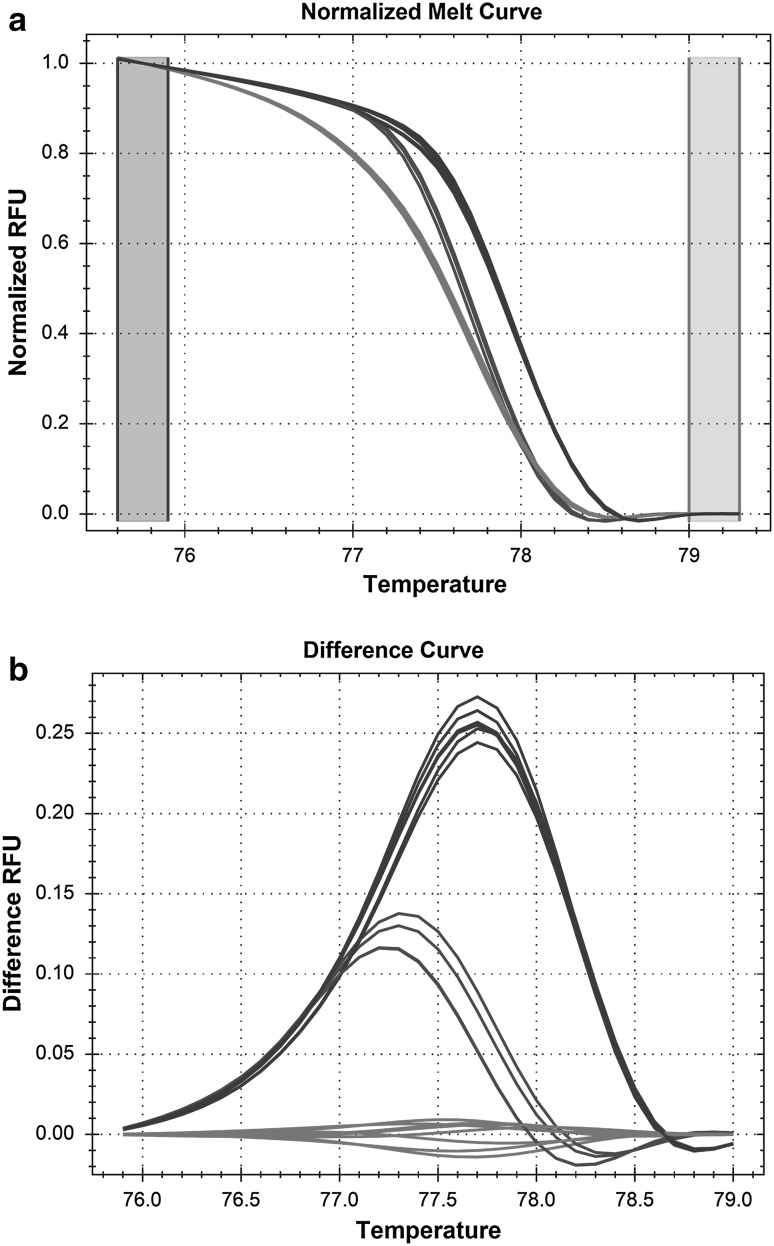
Isolates Ca1V, Ca4V, Ca7V all were classified as variant 1. as they have the same characteristic HRM profile in scan 3 (variant B) and in scan 6 (variant C), which was unique among 9 isolates tested. Figure 1 shows an example of HRM analysis. Scan 6 of ERG11 gene was amplified for 9 DNA samples and melting curves of PCR products were analyzed. Normalized melting curve analysis showed three variants of scan 6, which indicates differences in DNA sequence among tested samples (Fig. 1).
Fig. 1.
HRM analysis of ERG11 gene-scan 6 of Candida albicans reference strains and vaginal isolated. Both panels (a normalized melt curve, b difference curve) show three variants of the analyzed gene fragment among nine isolates tested indicating differences in DNA sequences. Red curves variant A, green curves variant B, blue curves variant C (Color figure online)
Isolates Ca2V, Ca5V, and Ca12r were classified as variant 2. of ERG11 gene as they represent unique pattern of nine scans variants (Table 4). Here, scan 8 and scan 9 are those, which differentiate the samples from all isolates as they are the only ones showing variant B in both ERG11 fragments.
Isolates Ca1r, Ca3V, and Ca11r were classified as variant 3., 4. and 5. of ERG11 gene, respectively, and their patterns of scans variants were unique among isolates tested.
To confirm differences in DNA sequence identified by HRM analysis, all DNA samples were amplified and sequenced. Indeed, isolates Ca1V, Ca4V, Ca7V (variant 1) had the same nucleotide sequence through whole ERG11 gene. Separate ERG11 variant was also found for Ca2V, Ca5V, Ca12r isolates (variant 2). Isolates Ca1r, Ca3V and Ca11r represent unique DNA sequence of ERG11. Sequence of fragment 3 of Ca3V sample (B′) occurred to be different from this part of Ca1V, Ca4V, Ca7V isolates, but this difference could not be detected by means of HRM. This difference in HRM and sequencing analysis is due to the principle of HRM analysis. The fluorescence signal comes from melting strands depending on their nucleotide content but not on the order of nucleotides. Thus, only by means of sequencing were we able to differentiate variant B (thymine in position 315 to cytosine and cytosine in position 411 to thymine—in strains Ca1V, Ca4V, Ca7V) from variant B′ (cytosine in position 315 to thymine and thymine in position 411 to cytosine—in strain Ca3V).
Regarding the yeast susceptibility to azoles, two gene variants (1 and 2) are related to increased drug resistance, except for strain Ca7V which is susceptible to all tested drugs. Strains numbered Ca1r, Ca3V, and Ca11r, having unique ERG11 patterns, are the most susceptible ones.
Sequencing of ERG11 Gene Revealed Two Point Mutations Associated with C. albicans Drug Resistance
Sequencing was the method of choice for analysis of ERG11 changes in C. albicans saliva strains. For ERG11 sequences of saliva strains the great majority of gene length was successfully sequenced but some fragments of gene were unreadable. The analysis of mutations in C. albicans strains of all origins was performed with gPLINK and Haploview software. Drug resistance and susceptibility of strains tested were referred to point mutations in ERG11 using allelic model. Two intermediate saliva strains (Ca21s, Ca68s) were classified as susceptible for allelic frequency tests. Among 21 point mutations found within ORFs, two were significantly associated with drug resistance (Table 5). Odds ratio based on allelic frequency was calculated in these two cases. Namely, nucleotide substitution at 798 position (Glu266Asp) corresponds to increase of drug resistance. Strains that possess mutated allele variant are 4.7-fold (odds ratio = 4.7143; CI 95 % 1.2368–17.9700) more probable to be drug resistant. In turn, mutation at 1440 position (Leu480Leu) significantly decreases the chance of a strain to be drug resistant (odds ratio = 0.11; CI 95 % 0.0127–0.9504).
Table 5.
Allele frequencies and Chi-square test results for C. albicans resistant (AFF) and susceptible (UNAFF) strains
| SNP | A1 | A2 | Model | AFF | UNAFF | CHISQ test | P |
|---|---|---|---|---|---|---|---|
| UTR_up | 2 | 4 | ALLELIC | 0/2 | 2/12 | 0.3265 | 0.5677 |
| Phe105Phe | 4 | 2 | ALLELIC | 14/16 | 19/21 | 0.004778 | 0.9449 |
| Asp116Glu | 1 | 4 | ALLELIC | 8/22 | 8/32 | 0.4321 | 0.511 |
| Lys119Lys | 3 | 1 | ALLELIC | 8/22 | 8/32 | 0.4321 | 0.511 |
| Lys128Thr | 2 | 1 | ALLELIC | 8/22 | 6/34 | 1.458 | 0.2272 |
| Ser137Ser | 2 | 4 | ALLELIC | 15/15 | 17/23 | 0.3886 | 0.5331 |
| Val159Ile | 1 | 3 | ALLELIC | 7/23 | 4/36 | 2.301 | 0.1293 |
| His183His | 2 | 4 | ALLELIC | 9/21 | 17/23 | 1.147 | 0.2841 |
| Le220Leu | 2 | 4 | ALLELIC | 6/24 | 11/29 | 0.5244 | 0.469 |
| Glu266Asp | 2 | 1 | ALLELIC | 9/7 | 6/22 | 5.495 | 0.01908* |
| Val332Val | 4 | 2 | ALLELIC | 12/12 | 17/19 | 0.04449 | 0.8329 |
| Leu340Leu | 3 | 1 | ALLELIC | 0/24 | 2/34 | 1.379 | 0.2402 |
| Lys342Lys | 3 | 1 | ALLELIC | 10/14 | 11/25 | 0.7814 | 0.3767 |
| Ser361Ser | 3 | 1 | ALLELIC | 0/2 | 4/10 | 0.7619 | 0.3827 |
| Leu370Leu | 2 | 4 | ALLELIC | 11/13 | 13/23 | 0.5671 | 0.4514 |
| Phe380Phe | 2 | 4 | ALLELIC | 2/22 | 4/32 | 0.1235 | 0.7253 |
| Tyr401Tyr | 2 | 4 | ALLELIC | 0/2 | 6/8 | 1.371 | 0.2416 |
| Asp428Asp | 2 | 4 | ALLELIC | 0/2 | 4/10 | 0.7619 | 0.3827 |
| Ala432Ala | 4 | 2 | ALLELIC | 14/12 | 12/24 | 2.609 | 0.1063 |
| Ala434Ala | 2 | 4 | ALLELIC | 14/12 | 14/22 | 1.364 | 0.2429 |
| Leu480Leu | 3 | 1 | ALLELIC | 1/25 | 8/22 | 5.378 | 0.0204* |
| Asn490Asn | 2 | 4 | ALLELIC | 1/25 | 6/24 | 3.323 | 0.06831 |
| UTR_down | 2 | 4 | ALLELIC | 1/1 | 6/8 | 0.03628 | 0.8489 |
One missense mutation (Glu266Asp) and one silent mutation (Leu480Leu) statistically significantly correlate with drug resistance (P value <0.05, marked *)
SNP single-nucleotide polymorphism, A1 allele 1, A2 allele2; 1 adenine, 2 cytosine, 3 guanine, 4 thymine
Pairwise linkage disequilibrium (LD) and r2 plots were generated on the basis of variants found in ERG11 gene (Fig. 2a, b). Two blocks of mutations that are inherited together with high frequency were identified. One block of strongly associated mutations comprised Asp116Glu, Lys119Lys, Lys128Thr, Ser137Ser, and Val159Ile. Second block contained Glu266Asp and Lys342Lys.
Fig. 2.
Pairwise linkage disequilibrium (LD) and r 2 plot. a Values on LD plot are D′ × 100 and lack of value indicates D′ = 1 × 100. Darker background indicates greater linkage. b Values on r 2 plot are r 2 × 100, lack of value indicates r 2 = 1 × 100. Denser color indicates greater linkage (Color figure online)
Sequencing of ERG11 gene revealed many heterozygous and homozygous mutations but only four of them changed amino acid sequence: D116E, K128T, V159I, E266D (Table 6).
Table 6.
Amino acid substitutions in clinical and reference strains
| Nucleotide position | Amino acid substitutions in ERG11 | Genotype | Strain name |
|---|---|---|---|
| 348 (scan 3) | D116E | TT | Ca1r, Ca1V, Ca3V, Ca4V, Ca11r, Ca2s, Ca3s, Ca6s, Ca10s, Ca20s, Ca21s, Ca23s, Ca26s, Ca38s, Ca40s, Ca41s, Ca44s, Ca68s, Ca74s, Ca80s, Ca81s, Ca82s |
| TA | Ca2V, Ca5V, Ca6V, Ca12r, Ca1s, Ca22s, Ca25s, Ca36s, Ca52s, Ca59s, Ca61s | ||
| AA | Ca11s, Ca69s, Ca70s | ||
| 383 (scan 3) | K128T | AA | Ca1r, Ca1V, Ca3V, Ca4V, Ca11s, Ca2s, Ca3s, Ca6s, Ca10s, Ca20s, Ca21s, Ca23s, Ca26s, Ca38s, Ca40s, Ca41s, Ca44s, Ca68s, Ca70s, Ca74s, Ca80s, Ca81s, Ca82s |
| AC | Ca2V, Ca5V, Ca6V, Ca12r, Ca1s, Ca22s, Ca25s, Ca36s, Ca52s, Ca59s, Ca61s | ||
| CC | Ca11r, Ca69s | ||
| 475 (scan 3) | V159I | GG | Ca1r, Ca1V, Ca2V, Ca3V, Ca4V, Ca5V, Ca11s, Ca12r, Ca2s, Ca3s, Ca6s, Ca10s, Ca20s, Ca21s, Ca23s, Ca26s, Ca38s, Ca40s, Ca41s, Ca44s, Ca68s, Ca70s, Ca74s, Ca80s, Ca81s, Ca82s |
| AA | Ca11r, Ca69s, Ca70s | ||
| AG | Ca1s, Ca22s, Ca25s, Ca36s, Ca52s, Ca59s, Ca61s | ||
| 798 (scan 5) | E266D | AA | Ca1r, Ca1V, Ca2V, Ca4V, Ca5V, Ca11r, Ca12r, Ca10s, Ca11s, Ca22s, Ca52s, Ca61s, Ca69s |
| CC | Ca3V, Ca3s, Ca23s, Ca40s, Ca41s, Ca44s | ||
| AC | Ca74s, Ca80s, Ca81s |
Discussion
Various mechanisms of drug resistance in C. albicans have been studied in recent years. Various authors agree that many different phenomena occurring simultaneously in a yeast cell are responsible for gaining drug resistance by C. albicans strains [7, 33, 40]. Mechanisms based on overexpression or hyperactivity of CDR1, CDR2, and MDR1 efflux pumps have been widely investigated [2, 19, 27]. Promising, but not yet very common are other approaches to the problem concerning mutations in CDR1 and CDR2 transcription factors [3], specific chromosome alterations [37], and mutations in various ergosterol synthesis pathway genes, like ERG5 or ERG3 [22]. Moreover, many genetic studies indicate that mutations in ERG11 or overexpression of these azole drugs target gene cause increased resistance to commonly administered antifungal drugs [14, 31, 43].
In this study, we present the application of HRM method to discriminate different variants of ERG11 gene in C. albicans strains on the basis of single-nucleotide polymorphisms. Among nine strains tested, three isolates Ca1V, Ca4V, Ca7V had the same variant of ERG11 gene sequence. Characteristic pattern is composed by mutations present in scan 3 and scan 6. Another distinctive sequence was determined by HRM analysis for Ca2V, Ca5V, and Ca12r isolates which represent variant 2 and their ERG11 gene sequence differs from other isolates in scan 8 and 9. Three other variants of ERG11 gene sequence were unique among strains tested. Sequence differences between variants were confirmed by sequencing reaction for all isolates included in HRM analysis.
These results indicate that HRM analysis could be a useful tool for classifying C. albicans isolates to distinctive variants of ERG11 gene. Similar approach has been described by Strzelczyk who used MSSCP method for preliminary selection of isolates presenting differences in ERG11 gene, prior to sequence analysis [36]. Rapid detection of ERG11 mutations was also performed by Loeffler et. [16] who used fluorescence resonance energy transfer and probed melting curves for this purpose. Another approach to determine ERG11 mutations consisted in the use of padlock probe and rolling circle amplification (RCA)-based method [39]. As the alternative to usual DNA sequencing, bioluminometric pyrosequencing method was employed to detect ERG11 mutations [10].
Considering low complexity of HRM reaction preparation and high resolution of analysis results, we propose HRM analysis as an alternative method of sample selection for subsequent sequencing. Nota bene, multi-well plate format is used for HRM reaction, which gives the possibility to predict mutations in tested sample using a panel of ERG11 gene variants for HRM analysis prepared in advance. Thus, HRM analysis should be seriously considered in attempts to accelerate diagnosis of Candida infection.
Drug susceptibility tests revealed that one out of nine strains was highly resistant to antifungal drugs (Ca5V) and was also resistant to itraconazole and fluconazole according to NCCLS breakpoints [26]. Five other strains presented increased resistance to at least one drug (Ca1V, Ca4V, Ca2V, Ca11r, and Ca12r) and only three of them were resistant to an azole drug. Namely, they were resistant to itraconazole with MIC = 0.5 mg mL−1, which is classified as dose-dependent susceptibility (S-DD) according to NCCLS.
In general, resistant strains represented variant 1. or variant 2. of ERG11 gene sequence with the exception of strain Ca11r, which was classified as distinct variant by HRM and by sequencing. It suggests the existence of another drug-resistance mechanism which causes slightly increased resistance to itraconazole of Ca11r strain. It is well documented that C. albicans strains develop many different mechanisms of resistance to azole drugs simultaneously [7, 30]. Some authors also suggest that due to the complexity of resistance in C. albicans it is necessary to analyze groups of resistant and susceptible strains that are matched, in order to reliably identify the mechanism responsible for azole resistance [40]. Moreover, heterogeneous mechanisms of resistance may develop within clonal origin subpopulations under azole exposure [23]. On the other hand, several studies indicate that some ERG11 point mutations may occur in azole susceptible, as well as in resistant strains [21, 30, 40]. This is in line with our observation in isolate Ca1V, Ca2V, and Ca7V that do not show increased resistance to any azole drug, although being members of variants 1 or 2. of ERG11, which we initially considered characteristic for drug-resistant strains.
Thus, in order to correlate C. albicans drug resistance with ERG11 gene variants using HRM method, it seems to be important to primarily recognize single-nucleotide polymorphism, which is related to increased resistance. Then, it could be possible to design more exact HRM tests with short fragments of ERG11 gene embracing statistically significant point mutations in ERG11 sequence. For this purpose, a numerous group of C. albicans isolates are needed. Here, we present ERG11 sequencing results for the collection of 27 C. albicans strains isolated from saliva of healthy volunteers. The statistical analysis was performed for strains of all origins. Among them, 21 nucleotide substitutions were recognized, and two of them were associated with changes in drug resistance with statistical significance (Chi square test, P < 0.05). Strains that carry substitution at position 798 (Glu266Asp) for at least one allele are 4.7-fold more probable to be azole resistant. This is in agreement with the results of Loffler who identified Glu266Asp substitution in 5 out of 19 resistant strains [17]. None of 19 susceptible strains showed this mutation. However, many authors suggest that substitution is not associated with resistance as it occurs in resistant, as well as in susceptible strains [24, 40]. Glu266Asp amino acid substitution was also shown not to be related to decreased affinity of fluconazole to ERG11 gene product, cytochrome P-450 [18]. Another substitution, which had statistical significance, was silent mutation at position 1440 (Leu480Leu) which statistically decreases probability of resistance in C. albicans strains. Nucleotide substitution at this site was also found by others [22, 23]; however, to the best of our knowledge no one have ever estimated its significance in drug resistance prediction. The impact of silent polymorphisms has been considered as a risk factor in disease development of higher eukaryotic organisms [34]. Silent mutations may cause structural and functional changes in synthesized protein [12]. Many mechanisms of this phenomena were proposed, one of which is change in translation kinetics and altered protein folding [13]. Other explanation may be that silent mutations cause native splicing donor site inactivation [11]. This change would shorten mRNA chain length of ERG11 and the usage of RT-PCR could confirm this hypothesis in our study. It is also possible that alternate splicing caused changed protein or small RNA binding or synonymous SNPs changed mRNA stability what consequently led to lower lanosterol demethylase expression and decrease of yeast drug resistance [32]. Also a polar effect to other gene can be considered, although it has only been shown for Saccharomyces cerevisiae mutants [9]. Taking the above into account, we suggest that silent mutations, frequently occurring in ERG11 gene, should also be considered as drug-resistance markers when establishing new genetic prediction methods.
On the basis of the presented data, HRM analysis seems to be a useful screening method that allows to select DNA samples for subsequent sequencing of ERG11 gene, thus cheapening and shortening the process of Candida infection diagnosis. However, it is not a reliable method of classification of C. albicans resistant and susceptible strains, firstly because other drug-resistance mechanisms may occur simultaneously, and secondly because our knowledge of changes in ERG11 responsible for azole resistance is not yet sufficient to design proper and trustworthy test based on ERG11 mutation sites. Nevertheless, the number of current studies on C. albicans drug resistance increases, and they lead to better understanding of the significance of ERG11 mutations, which makes scanning methods like HRM analysis a promising tool for the future diagnosis of C. albicans infections.
Acknowledgments
This work was supported by the Polish POIG (Grant Number 01.01.02-10-005/08) TESTOPLEK, supported by the EU through the European Regional Development Fund.
Author’s Contribution
Conceived and designed the experiments: AG, DS. Performed the experiments: MC, DS. Performed and analyzed the sequencing data: JD, MKM. Analyzed and interpreted the data: MC, AG, DS. Wrote the manuscript: MC, AG, DS. Revised the manuscript critically: JD.
Compliance with Ethical Standards
Conflict of interest
The authors declare that there are no conflicts of interest.
Human and Animal Rights Statements
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Chau AS, Mendrick CA, Sabatelli FJ, Loebenberg D, McNicholas PM. Application of real-time quantitative PCR to molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob Agents Chemother. 2004;48:2124–2131. doi: 10.1128/AAC.48.6.2124-2131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen LM, Xu YH, Zhou CL, Zhao J, Li CY, Wang R. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations. J Int Med Res. 2010;38:536–545. doi: 10.1177/147323001003800216. [DOI] [PubMed] [Google Scholar]
- 3.Coste A, Turner V, Ischer F, Morschhäuser J, Forche A, Selmecki A, Berman J, Bille J, Sanglard D. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics. 2006;172:2139–2156. doi: 10.1534/genetics.105.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowen LE. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat Rev Microbiol. 2008;6:187–198. doi: 10.1038/nrmicro1835. [DOI] [PubMed] [Google Scholar]
- 5.Eggimann P, Garbino J, Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect Dis. 2003;3:685–702. doi: 10.1016/S1473-3099(03)00801-6. [DOI] [PubMed] [Google Scholar]
- 6.Ge S-H, Wan Z, Li J, Xu J, Li R-Y, Bai F-Y. Correlation between azole susceptibilities, genotypes, and ERG11 mutations in Candida albicans isolates associated with vulvovaginal candidiasis in China. Antimicrob Agents Chemother. 2010;54:3126–3131. doi: 10.1128/AAC.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman GH, da Silva Ferreira ME, dos Reis Marques E, Savoldi M, Perlin D, Park S, Martinez PCG, Goldman MHS, Colombo AL. Evaluation of fluconazole resistance mechanisms in Candida albicans clinical isolates from HIV-infected patients in Brazil. Diagn Microbiol Infect Dis. 2004;50:25–32. doi: 10.1016/j.diagmicrobio.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Harju S, Fedosyuk H, Peterson KR. Rapid isolation of yeast genomic DNA: Bust n’ Grab. BMC Biotechnol. 2004;4:8. doi: 10.1186/1472-6750-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan CD, Jin H, Zhang IL, Belyanin A. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim T-H, Lee M-K. Evaluation of the V404I and V509 M amino acid substitutions of ERG11 gene in Candida albicans isolates by pyrosequencing. Folia Microbiol (Praha) 2010;55:301–304. doi: 10.1007/s12223-010-0045-2. [DOI] [PubMed] [Google Scholar]
- 11.Kimchi-Sarfaty C, Oh JM, Kim I-W, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 12.Komar AA. Silent SNPs: impact on gene function and phenotype. Pharmacogenomics. 2007;8:1075–1080. doi: 10.2217/14622416.8.8.1075. [DOI] [PubMed] [Google Scholar]
- 13.Komar AA. Genetics. SNPs, silent but not invisible. Science. 2007;315:466–467. doi: 10.1126/science.1138239. [DOI] [PubMed] [Google Scholar]
- 14.Lamb DC, Kelly DE, Schunck WH, Shyadehi AZ, Akhtar M, Lowe DJ, Baldwin BC, Kelly SL. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 15.Lin P-C, Liu T-C, Chang C-C, Chen Y-H, Chang J-G. High-resolution melting (HRM) analysis for the detection of single nucleotide polymorphisms in microRNA target sites. Clin Chim Acta. 2012;413:1092–1097. doi: 10.1016/j.cca.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Loeffler J, Hagmeyer L, Hebart H, Henke N, Schumacher U, Einsele H. Rapid detection of point mutations by fluorescence resonance energy transfer and probe melting curves in Candida species. Clin Chem. 2000;46:631–635. [PubMed] [Google Scholar]
- 17.Löffler J, Kelly SL, Hebart H, Schumacher U, Lass-Flörl C, Einsele H. Molecular analysis of cyp51 from fluconazole-resistant Candida albicans strains. FEMS Microbiol Lett. 1997;151:263–268. doi: 10.1016/S0378-1097(97)00172-9. [DOI] [PubMed] [Google Scholar]
- 18.Maebashi K, Kudoh M, Nishiyama Y, Makimura K, Kamai Y, Uchida K, Yamaguchi H. Proliferation of intracellular structure corresponding to reduced affinity of fluconazole for cytochrome P-450 in two low-susceptibility strains of Candida albicans isolated from a Japanese AIDS patient. Microbiol Immunol. 2003;47:117–124. doi: 10.1111/j.1348-0421.2003.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 19.Maebashi K, Niimi M, Kudoh M, Fischer FJ, Makimura K, Niimi K, Piper RJ, Uchida K, Arisawa M, Cannon RD, Yamaguchi H. Mechanisms of fluconazole resistance in Candida albicans isolates from Japanese AIDS patients. J Antimicrob Chemother. 2001;47:527–536. doi: 10.1093/jac/47.5.527. [DOI] [PubMed] [Google Scholar]
- 20.Mandviwala T, Shinde R, Kalra A, Sobel JD, Akins RA. High-throughput identification and quantification of Candida species using high resolution derivative melt analysis of panfungal amplicons. J Mol Diagn. 2010;12:91–101. doi: 10.2353/jmoldx.2010.090085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marichal P, Koymans L, Willemsens S, Bellens D, Verhasselt P, Luyten W, Borgers M, Ramaekers FC, Odds FC, Bossche HV. Contribution of mutations in the cytochrome P450 14α-demethylase (ERG11p, Cyp51p) to azole resistance in Candida albicans. Microbiol Read Engl. 1999;145(Pt 10):2701–2713. doi: 10.1099/00221287-145-10-2701. [DOI] [PubMed] [Google Scholar]
- 22.Martel CM, Parker JE, Bader O, Weig M, Gross U, Warrilow AGS, Kelly DE, Kelly SL. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14α-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother. 2010;54:3578–3583. doi: 10.1128/AAC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez M. Heterogeneous mechanisms of azole resistance in Candida albicans clinical isolates from an HIV-infected patient on continuous fluconazole therapy for oropharyngeal candidosis. J Antimicrob Chemother. 2002;49:515–524. doi: 10.1093/jac/49.3.515. [DOI] [PubMed] [Google Scholar]
- 24.Morio F, Loge C, Besse B, Hennequin C, Le Pape P. Screening for amino acid substitutions in the Candida albicansERG11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn Microbiol Infect Dis. 2010;66:373–384. doi: 10.1016/j.diagmicrobio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun. 2003;71:4333–4340. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards (1997) Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa. Approv. Stand. Ed. M27-A2
- 27.Niimi K, Maki K, Ikeda F, Holmes AR, Lamping E, Niimi M, Monk BC, Cannon RD. Overexpression of Candida albicans CDR1, CDR2, or MDR1 does not produce significant changes in echinocandin susceptibility. Antimicrob Agents Chemother. 2006;50:1148–1155. doi: 10.1128/AAC.50.4.1148-1155.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niimi M. Regulated overexpression of CDR1 in Candida albicans confers multidrug resistance. J Antimicrob Chemother. 2004;54:999–1006. doi: 10.1093/jac/dkh456. [DOI] [PubMed] [Google Scholar]
- 29.Carvalho VO, Okay TS, Melhem MSC, Szeszs MW, del Negro GMB. The new mutation L321F in Candida albicansERG11 gene may be associated with fluconazole resistance. Rev Iberoam Micol. 2013;30:209–212. doi: 10.1016/j.riam.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Perea S, Lopez-Ribot JL, Kirkpatrick WR, McAtee RK, Santillan RA, Martinez M, Calabrese D, Sanglard D, Patterson TF. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob Agents Chemother. 2001;45:2676–2684. doi: 10.1128/AAC.45.10.2676-2684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1093/jac/42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67:9609–9612. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- 33.Selmecki A, Forche A, Berman J. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot Cell. 2010;9:991–1008. doi: 10.1128/EC.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siegsmund M. Association of the P-glycoprotein transporter MDR1C3435T polymorphism with the susceptibility to renal epithelial tumors. J Am Soc Nephrol. 2002;13:1847–1854. doi: 10.1097/01.ASN.0000019412.87412.BC. [DOI] [PubMed] [Google Scholar]
- 35.Ślemp-Migiel A, Strzelczyk JK, Rother M, Wiczkowski A. MSSCP technology as a tool for the detection of genetic differences in ERG11 gene in Candida albicans sensitive and resistant to azoles. Med Mycol. 2009;16:5–9. [Google Scholar]
- 36.Strzelczyk JK, Slemp-Migiel A, Rother M, Gołąbek K, Wiczkowski A. Nucleotide substitutions in the Candida albicansERG11 gene of azole-susceptible and azole-resistant clinical isolates. Acta Biochim Pol. 2013;60:547–552. [PubMed] [Google Scholar]
- 37.Perepnikhatka V, Fischer FJ, Niimi M, Baker RA, Cannon RD, Wang Y-K, Sherman F, Rustchenko E. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J Bacteriol. 1999;181:4041–4049. doi: 10.1128/jb.181.13.4041-4049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandeputte P, Larcher G, Berges T, Renier G, Chabasse D, Bouchara J-P. Mechanisms of azole resistance in a clinical isolate of Candida tropicalis. Antimicrob Agents Chemother. 2005;49:4608–4615. doi: 10.1128/AAC.49.11.4608-4615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Kong F, Sorrell TC, Wang B, McNicholas P, Pantarat N, Ellis D, Xiao M, Widmer F, Chen SC. Rapid detection of ERG11 gene mutations in clinical Candida albicans isolates with reduced susceptibility to fluconazole by rolling circle amplification and DNA sequencing. BMC Microbiol. 2009;9:167. doi: 10.1186/1471-2180-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White TC, Holleman S, Dy F, Mirels LF, Stevens DA. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob Agents Chemother. 2002;46:1704–1713. doi: 10.1128/AAC.46.6.1704-1713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang M-J, Liu J-Y, Ni P-H, Wang S, Shi C, Wei B, Ni Y-X, Ge H-L. ERG11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013;13:386–393. doi: 10.1111/1567-1364.12042. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, Chen L, Li C. Susceptibility of clinical isolates of Candida species to fluconazole and detection of Candida albicansERG11 mutations. J Antimicrob Chemother. 2008;61:798–804. doi: 10.1093/jac/dkn015. [DOI] [PubMed] [Google Scholar]
- 43.Zhao J, Xu Y, Li C. Association of T916C (Y257H) mutation in Candida albicansERG11 with fluconazole resistance: T916C mutation in Candida albicansERG11 and fluconazole resistance. Mycoses. 2013;56:315–320. doi: 10.1111/myc.12027. [DOI] [PubMed] [Google Scholar]