Abstract
A sub-group of class I Caseinolytic proteases (Clps) function as molecular chaperone and confer thermotolerance to plants. We identified class I Clp family consisting of five ClpB/HSP100, two ClpC, and two ClpD genes from bread wheat. Phylogenetic analysis showed that these genes were highly conserved across grass genomes. Subcellular localization prediction revealed that TaClpC and TaClpD subgroup proteins and TaClpB1 proteins are potentially targeted to chloroplast, while TaClpB5 to mitochondria, and TaClpB2, TaClpB3, and TaClpB4 to cytoplasm. Spatio-temporal expression pattern analysis revealed that four TaClpB and TaClpD2 genes are expressed in majority of all tissues and developmental stages of wheat. Real-time RT-PCR analysis of expression levels of Clp genes in seven wheat genotypes under different abiotic stresses revealed that genes coding for the cytosolic Clps namely TaClpB2 and TaClpB3 were upregulated under heat, salt and oxidative stress but were downregulated by cold stress in most genotypes. In contrast, genes coding for the chloroplastic Clps TaClpC1, TaClpC2, and TaClpD1 genes were significantly upregulated by mainly by cold stress in most genotypes, while TaClpD2 gene was upregulated >2 fold by salt stress in DBW16. The TaClpB5 gene coding for mitochondrial Clp was upregulated in all genotypes under heat, salt and oxidative stresses. In addition, we found that biotic stresses also upregulated TaClpB4 and TaClpD1. Among biotic stresses, Tilletia caries induced TaClpB2, TaClpB3, TaClpC1, and TaClpD1. Differential expression pattern under different abiotic and biotic stresses and predicted differential cellular localization of Clps suggest their non-redundant organelle and stress-specific roles. Our results also suggest the potential role of Clps in cold, salt and biotic stress responses in addition to the previously established role in thermotolerance of wheat.
Keywords: ClpATPases, ClpB/HSP100, ClpC, ClpD, Triticum aestivum, high temperature, salinity, oxidative stress
Introduction
Caseinolytic protease (Clp) was first identified in Escherichia coli as a heat shock inducible ATP-dependent protease complex that is capable of hydrolyzing casein (Hwang et al., 1987). Subsequent studies showed that ClpATPases fall within the AAA+ superfamily and it functions in assembly and disassembly of protein complexes (Neuwald et al., 1999; Frickey and Lupas, 2004). Members of this family form large hexameric ring structures in ATP dependent manner. These proteins have two AAA+ like domains, which consist of a RecA-like nucleotide-binding domain (NBD) and an α-helical domain (Lee et al., 2004). Under stress conditions, these proteases function as molecular chaperone by minimizing the protein aggregation and also target the damaged proteins for degradation (Snider et al., 2008). ClpATPases associate with ClpP, a proteolytic subunit for degradation of damaged proteins (Porankiewicz et al., 1999; Shikanai, 2001). This process maintains the quality of cellular proteins, thereby increases the tolerance of plants during stress conditions. ClpATPases are divided into two types namely; class I ClpATPases (ClpA, ClpB/Hsp100, ClpC, ClpD, and ClpE) and class II ClpATPases (ClpM, ClpN, ClpX, and ClpY). Class I ClpATPases have two ATP binding domains, whereas class II ClpATPases have one ATP binding domain (Singh et al., 2010). Class I ClpATPase (ClpB/HSP100, ClpC andClpD) gene families are well characterized in rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana) (Singh et al., 2010). Rice and Arabidopsis genomes encode 9 and 6 Class I ATPase family members, respectively (Singh et al., 2010). ClpB/HSP100 protein functions with the HSP70/DnaK chaperone system to remodel the denatured protein aggregates (Doyle and Wickner, 2009). In Arabidopsis and rice, the ClpB proteins have been divided into three groups based on sub-cellular localization namely, cytoplasmic ClpB (ClpB-cyt), chloroplastic ClpB (ClpB-p), and mitochondrial ClpB (ClpB-m) isoforms (Singh et al., 2010). Among three groups, the ClpB-Cyt/HSP100 is well studied in plants. It is involved in both basal and acquired thermotolerance, and also negatively influence primary root growth in maize (Zea mays) (Nieto-Sotelo et al., 2002). Arabidopsis ClpB-p is involved in chloroplast development (Myouga et al., 2006; Lee et al., 2007), while tomato (Solanum lycopersicum) ClpB-p (LeHSP100/ClpB) is necessary for acquired thermotolerance (Yang et al., 2006). AtClpB-m transcripts were upregulated during high temperature stress (Lee et al., 2007). In rice, OsClpB-cyt, OsClpB-m, and OsClpB-p expression were also upregulated by ABA treatment (Singh et al., 2010). Plant ClpC proteins function as stromal molecular chaperones involved in importing and protecting unfolded newly synthesized proteins, and play a vital role in chloroplast function, leaf development, and Fe homeostasis (Park and Rodermel, 2004; Sjögren et al., 2004; Adam et al., 2006; Wu et al., 2010). In Arabidopsis, AtClpC function as principle stromal protease responsible for maintaining homeostasis and also confers a novel protein quality control mechanism for chloroplast pre-protein import (Sjögren et al., 2014). In rice and Arabidopsis, ClpD expression was found to be induced by multiple abiotic stressessuch as dehydration, cold stress, high light, and during natural senescence (Kiyosue et al., 1993; Weaver, 1999; Zheng et al., 2002; Singh et al., 2010). Overexpression of Arabidopsis AtHSP101 conferred the basal thermotolerance in transgenic rice (Katiyar-Agarwal et al., 2003). Thus, class I Clp proteins play an important role in stress responses, growth and development of plants.
Wheat is an important staple food and its production needs to be increased upto 50% to meet the food demand of the growing population by the end of 2050 (Godfray et al., 2010). One of the limitations to achieve this target is abiotic stresses that drastically reduce the wheat grain production (Viswanathan and Khanna-Chopra, 2001). Among abiotic stresses, high temperature stress adversely impacts wheat grain production in tropical and sub-tropical areas of the world (Ortiz et al., 2008; Lobell et al., 2012). An increase in temperature by 1°C above optimum during reproductive stage may reduce the wheat yield by 4–10% (Aggarwal et al., 2000; Brisson et al., 2010). Despite its importance, the genetic basis of heat tolerance is poorly understood, and no major QTLs (Quantitative Trait Loci) have been identified in wheat. At cellular level, heat shock proteins, also known as molecular chaperons, are the most studied candidate genes for high temperature response. Since class I Clp proteins perform important role in stress responses in different plant species, we examined the potential role of this class of genes in different stresses including high temperature stress in wheat. This study was conducted to determine the structural aspects and expression pattern of class I ClpATPases in wheat. The results revealed differential expression pattern of wheat class I ClpATPases in different tissues and developmental stages and in response to different stresses, and thus suggest their potential roles in biotic and abiotic stress tolerance of wheat.
Materials and methods
Identification and classification of Class I ClpATPases genes in wheat
The protein sequences of Class I ClpATPase genes from Arabidopsis (3 ClpB proteins—At1g74310, At2g25140, At5g15450; 2 ClpC proteins—At3g48870, At5g50920; and 1 ClpD protein—At5g51070) (Zheng et al., 2002) and rice (3 ClpB proteins—Os05g44340, Os02g08490, Os03g31300; 4 ClpC proteins—Os04g32560, Os12g12580, Os11g16590, Os1g16770; and 2 ClpD proteins—Os02g32520, Os04g33210) (Singh et al., 2010) were used as query and blastp search was performed in TriFLDB (http://TriFLDB.psc.riken.jp/) (Mochida et al., 2009) and NCBI database to identify the wheat class I ClpATPase gene homologs. Clustal omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and MEGA 5 software (Tamura et al., 2011) were used for removal of redundant cDNA clones. The identified wheat Class I ClpATPase homologs were reconfirmed by tblastn search in rice genome database TIGR (Ouyang et al., 2007) and in Arabidopsis genome database TAIR (https://www.arabidopsis.org/). Isoelectric point (pI) and molecular weight (Mw) was computed through ExPASy tool (http://web.expasy.org/compute_pi/). Protein subcellular localization was predicted through WoLF PSORT tool (Horton et al., 2007) and TargetP 1.1 server (Emanuelsson et al., 2007).
Multiple sequence alignment, phylogenetic analysis, and domain prediction
Multiple sequence alignment was carried out using Clustal omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and Simple Modular Architecture Research Tool (SMART), (Letunic et al., 2012) was used to predict the conserved domains in class I ClpATPase protein sequences. The COILS Server (http://www.ch.embnet.org/software/COILS_form.html) was used to predict the coiled-coil region in TaClp proteins (Lupas et al., 1991). Rice, class I ClpATPase protein sequences were used as query and tblastn search was performed in phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html) against the available grass genomes such as maize (Z. mays), sorghum (Sorghum bicolor), brachypodium (Brachypodium distachyon) and setaria (Setaria italica) to identify the respective homologs. For phylogenetic analysis, the amino acid sequences of Clp proteins from Arabidopsis and grass genomes were aligned using MUSCLE program (Tamura et al., 2011). Phylogenetic trees were constructed using Neighbor Joining method with pairwise deletion and poisson correction in MEGA 5 software (http://megasoftware.net/) (Tamura et al., 2011). Bootstrap analysis was performed with 2000 replicates to evaluate the reliability of different phylogenetic groups. MEME Suite web server was used to identify the conserved motifs in class I ClpATPase protein sequences (Bailey et al., 2009). Exon/Intron organization of class I ClpATPase genes were generated using gene structure display server program (Hu et al., 2015).
Digital expression analysis
A blastn search was carried out using the identified TaClps genes against Triticum aestivum NCBI UniGene database to identify their UniGene IDs. The EST sequences of wheat Clp genes corresponding to their UniGene IDs were downloaded from NCBI UniGene database for digital expression analysis. These ESTs expression information were used to characterize the tissue specific expression of TaClp genes (Table 3). We also used wheat array expression data information to study the spatio-temporal expression pattern, abiotic and biotic stress inducible expression pattern of identified wheat Clp genes (except for TaClpC2, which doesn't have probe in affymetrix wheat genome array). Genevestigator v3 database was used for normalization and meta-analysis of the wheat array expression datasets (https://genevestigator.com/gv/doc/intro_plant.jsp) (Hruz et al., 2008). The following abiotic and biotic wheat array experiment's datasets were used in our study: NCBI GEO Accession GSE12936 (Chain et al., 2009), GSE21386 (Wang et al., 2010), GSE22080 (unpublished), GSE34445 (Zhu et al., 2012), GSE9915 (Bolton et al., 2008), GSE13660 (Desmond et al., 2008), GSE31762 (Krugman et al., 2010), and EBI Array Express IDs E-MEXP-1193 (Wan et al., 2008), E-MEXP-971 (Mott and Wang, 2007), E-MEXP-1488 (Ergen et al., 2009), and E-MEXP-1523 (Qin et al., 2008). The details of the wheat array experiments used in study were given in Supplementary Table S2.
Plant materials and growth conditions
Seven genotypes of wheat (T. aestivium) cultivars WR544, C306, DBW16, NIAW34, PBW343, HD2687, and HW2019 were used in this study. The cultivars WR544, C306, DBW16, and NIAW34 were thermotolerant, whereas PBW343, HD2687, and HW2019 were thermosensitive. Seeds of these genotypes were germinated on cotton bed in falcon tubes as described by Singh et al. (2010). Seedlings were grown at 22–25°C under 16 h/8 h light/dark regime, respectively, in the culture room. Plants were watered regularly at an interval of 2–3 days to retain optimum moisture for plant growth. Fifteen days old seedlings with uniform size were selected and used for stress treatments and molecular analysis.
Stress treatments
High temperature stress was imposed to the plants by placing the plants growing in the falcon tube in an incubator at 42 (±2)°C for 3 h. Cold stress was imposed by placing the plants in cold chamber at 2 (±2)°C for 3 h. Salt stress was imposed by saturating the cotton pad with 150 mM NaCl and sampling was done after 6 h. For oxidative stress, seedlings were treated with 10 mM H2O2 and kept in dark conditions for 6 h. Well watered seedlings grown at 22–25°C under 16 h/8 h light/dark regime in the culture room served as control. After the treatments, leaf tissues from 10 plants were pooled and immediately frozen in liquid nitrogen and stored at −80°C till further use.
qRT-PCR analysis
High quality RNA was extracted using RNeasy® plant mini kit (QIAGEN) by following the manufacturer's instructions. Further the extracted RNA was subjected to DNaseI (Fermentas) treatment to obtain DNA-free RNA. First strand cDNA was synthesized from 1 μg of total RNA by using Superscript–III reverse transcriptase (Invitrogen, USA). Three biological replicates were used for qRT-PCR (We isolated RNA from three different samples from the pooled leaf tissues of each treatment of each genotypes). For qRT-PCR analysis, the cDNA was diluted to 1:4 times and 1.5 μL of diluted cDNA was used as a template in a 10 μL reaction volume in 96 well plates by following the manufacturer's instruction. List of primers used in this study is given in Supplementary Table S3. Quantitative RT-PCR analysis was carried out by using the Realplex4 system (Eppendorf) using iQ™ SYBR® Green (Bio-Rad). Expression data was normalized using endogenous control ubiquitin [Unigene ID Ta50503] gene expression. The expression was represented in the form of relative fold change which is the relative change in expression of TaClp genes under stress conditions as compared with their expression under control conditions. Expression level of TaClp gene in control condition was considered as 1. Relative fold change was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The error bars represents the standard deviation of the expression of the three biological replicates. A two-tailed unpaired t-test was performed to analyze the significance of the difference of the gene expression between control and stress condition.
Results
Identification and classification of class I Clp genes in wheat
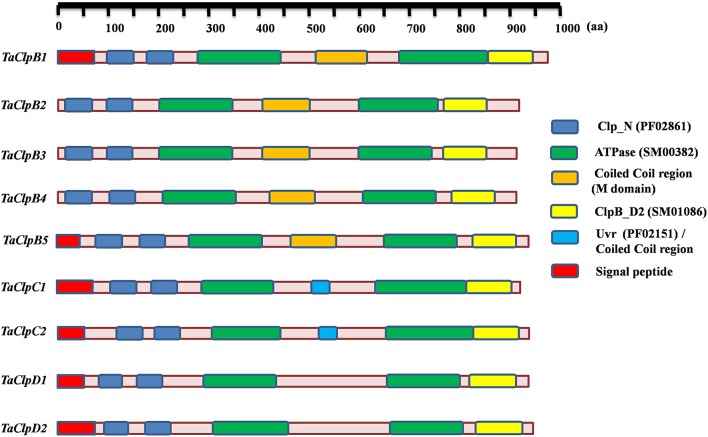
Arabidopsis genome encodes 6 Class I ClpATPases, while rice genome encodes 9 Class I ClpATPases (Zheng et al., 2002; Singh et al., 2010). The protein sequences of Class I ClpATPases from Arabidopsis and rice were used as query for blastp search in the wheat database, TriFLDB (Mochida et al., 2009) and NCBI database to identify the respective wheat homologs. Initially 16 wheat cDNA clones were identified. Seven cDNA sequences were redundant and hence eliminated based on multiple sequence alignment analysis. Wheat ClpATPase homologs were reconfirmed by performing tblastn search in rice genome annotation project (RGAP) database and Arabidopsis genome (TAIR) database. Finally, nine wheat Class I ClpATPase homologs were identified which include five ClpB genes, two ClpC genes, and two ClpD genes (Table 1). The subcellular localization of wheat Clp proteins were predicted through WoLF PSORT tool (Horton et al., 2007) and TargetP 1.1 server (Emanuelsson et al., 2007). TaClpB2, TaClpB3, and TaClpB4 localized to cytoplasm, TaClpB5 localized to mitochondria, while TaClpB1, TaClpC1, TaClpC2, TaClpD1, and TaClpD2 were localized to chloroplast (Table 1). Presence of a plastid transit peptide in the N-terminal of TaClpB1, TaClpC1, TaClpC2, TaClpD1, and TaClpD2 proteins, and a mitochondrial targeting sequence in the N-terminal of TaClpB5 conformed to the subcellular localization prediction. Simple Modular Architectural Related Tool (SMART) (Letunic et al., 2012) was used to predict and locate the conserved functional domains present in wheat Class I ClpATPases proteins (Table 2). The position of the conserved domains of the wheat Clp proteins were pictorially represented in Figure 1. All the wheat Clp proteins have two conserved Clp_N domains (PF02861) in the N-terminal and two ATPase domains which are essential for hexamerization and chaperone function (Krzewska et al., 2001; Weibezahn et al., 2004). A coiled coiled domain (M domain) located between the two nucleotide binding domains (NBD) is characteristic of ClpB proteins (Mishra and Grover, 2015). The M domain is variable in both sequence and length. It is absent in ClpA, while presence of a shorter M domain is predicted for ClpC and ClpD proteins in higher plants (Mishra and Grover, 2015). Our results revealed that M domain is present in ClpB and ClpC proteins while it is absent in ClpD proteins of wheat (Figure 1). The coiled coil region (M Domain) of ClpC1 (510–535) and ClpC2 (530–550) is located within the Uvr domain (PF02151), and is shorter in ClpC than that of ClpB (Figure 1 and Table 2). M domain can adapt different orientation in a single hexamer. The horizontal orientation represses the ClpB activity by preventing binding of Hsp70, while tilted orientation activates the ClpB activity (Carroni et al., 2014). The interaction of M domain of ClpB with Hsp70 chaperone system is essential for regulating the intrinsic disaggregase activity of ClpB hexamer (DeSantis and Shorter, 2012; Carroni et al., 2014). Similar to wheat ClpC, the M domain of bacterial ClpC is half the size of that in ClpB and it corresponds to the N terminal coiled coil (M domain) region of ClpB (Mishra and Grover, 2015; Nishimura and van Wijk, 2015). The N-domain and the NBD1-M domain of bacterial ClpC interacts with the C-terminal domain of MecA which facilitates hexamerization and activation of ClpC. This ClpC-MecA complex plays an important role in disaggregation and translocation of the unfolded proteins to ClpP for degradation (Schlothauer et al., 2003; Kirstein et al., 2006; Wang et al., 2011).
Table 1.
Wheat ClpB/Hsp100, ClpC, and ClpD genes and their predicted cellular localization.
| Gene | TriFLDBcDNA sequence id | NCBI GenBank ID | Size (aa) | Mw (K Da) | Pi | Cellular localization |
|---|---|---|---|---|---|---|
| TaClpB1 | tplb0010c03 | AK330787 | 975 | 108.73 | 6.66 | Chloroplast |
| TaClpB2 | RFL_Contig4216 | AK333710 | 918 | 101.06 | 5.84 | Cytoplasm |
| TaClpB3 | AF174433 | AF174433 | 913 | 100.90 | 5.95 | Cytoplasm |
| TaClpB4 | RFL_Contig5540 | AK335181 | 913 | 99.79 | 6.12 | Cytoplasm |
| TaClpB5 | JV864025.1 | 933 | 104.12 | 5.54 | Mitochondria | |
| TaClpC1 | RFL_Contig3528 | AK332945 | 920 | 101.80 | 6.55 | Chloroplast |
| TaClpC2 | RFL_Contig4439 | AK333957 | 938 | 102.74 | 6.26 | Chloroplast |
| TaClpD1 | RFL_Contig1508 | AK330701 | 937 | 101.78 | 6.39 | Chloroplast |
| TaClpD2 | RFL_Contig3864 | AK333318 | 946 | 102.67 | 8.74 | Chloroplast |
aa, amino acids; pI, isoelectric point; Mw, Molecular weight. The subcellular localization of Clps were predicted through WoLF PSORT tool and TargetP 1.1 server.
Table 2.
Functional domains identified in wheat ClpB, ClpC, and ClpD proteins.
| Gene | Signal peptide length | Clp_N (PF02861) | ATPase domain | Coiled coil domain (M domain) | ClpB_D2 small domain | Uvr domain |
|---|---|---|---|---|---|---|
| TaClpB1 | 71 | 97–149, 174–225 | 278–442, 681–856 | 518–613 | 855–949 | |
| TaClpB2 | – | 17–69, 98–148 | 202–347, 600–759 | 407–503 | 770–860 | |
| TaClpB3 | – | 17–69, 98–148 | 201–346, 599–742 | 406–502 | 768–859 | |
| TaClpB4 | – | 17–66, 104–154 | 209–354, 609–752 | 421–511 | 780–871 | |
| TaClpB5 | 40 | 52–104, 129–180 | 233–378, 636–778 | 446–568 | 811–900 | |
| TaClpC1 | 70 | 104–156, 179–231 | 290–430, 633–814 | 510–535 | 813–904 | 507–542 |
| TaClpC2 | 51 | 119–171, 194–246 | 307–447, 651–832 | 530–550 | 831–931 | 524–559 |
| TaClpD1 | 43 | 81–127, 157–208 | 292–440, 640–820 | 829–919 | ||
| TaClpD2 | 89 | 94–144, 172–224 | 305–454, 659–806 | 839–929 |
SMART tool was used to predict and locate the position of conserved domains and COILS Server was used to predict Coiled coil domain (M domain).
Figure 1.
Diagrammatic illustration of conserved domains position present in wheat TaClp genes. The conserved domains were predicted using SMART tool.
Phylogenetic relationship of class I clpatpases of family
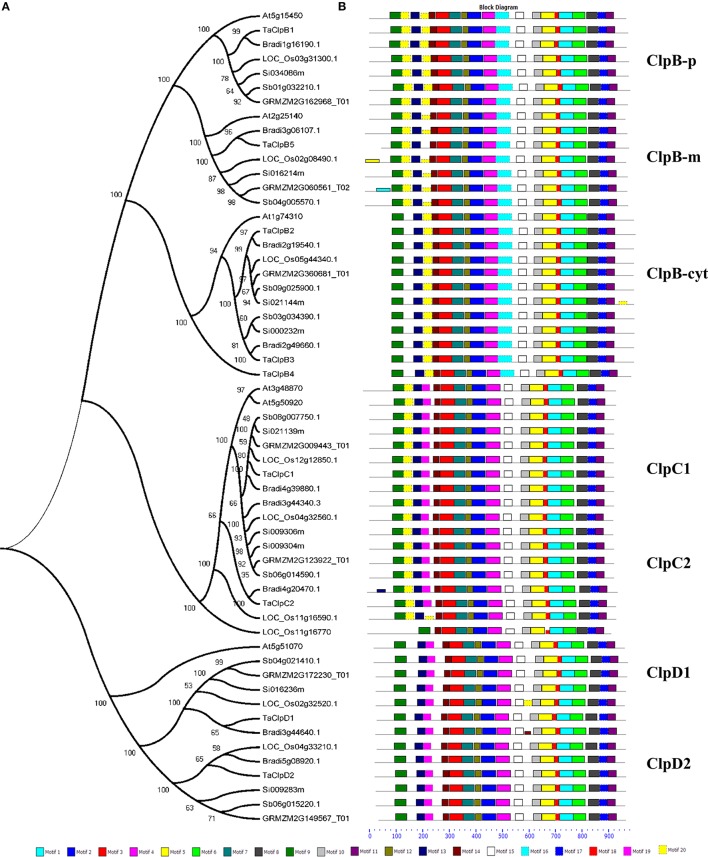
Blastp search was performed using rice Class I ClpATPases proteins against publicly available grass genomes (maize, sorghum, brachypodium, and setaria) in Phytozome database and the respective grass Clp protein homologs were downloaded. Our analysis led to the identification of seven Clp proteins from maize, eight Clp proteins from sorghum and nine Clp proteins each from brachypodium and setaria (Supplementary Table S1). Maize encode for 3, 2, and 2 ClpB, ClpC, and ClpD genes, respectively, sorghum encode for 4, 2, and 2 ClpB, ClpC, and ClpD genes, respectively, while brachypodium and setaria encode 4, 3, and 2 ClpB, ClpC, and ClpD genes, respectively (Supplementary Table S1). The multiple sequence alignment of wheat Clp proteins with their respective grass Clp protein homologs revealed that the proteins TaClpB1, TaClpB2, TaClpB3, and TaClpC1 were highly conserved across the grass genomes (Supplementary Figures 1A–C). We also studied the phylogenetic relationship of wheat class I ClpATPases proteins with their respective grass Clp protein homologs and Arabidopsis homologs using MEGA 5 software (Tamura et al., 2011). The analysis showed that Clp proteins from grass genomes and Arabidopsis genome falls into three groups designated as ClpB, ClpC, and ClpD (Figure 2), in accordance with rice and Arabidopsis Clp proteins classification (Singh et al., 2010). Within each subclass, Clp proteins from grass genome clustered together suggesting that they were highly conserved across grass genomes (Figure 2).
Figure 2.
Phylogenetic relationship and conserved motif compositions of wheat Clp proteins and their respective grass homologs. (A) Multiple sequence alignment of 57 full-length grass Clp proteins was done using MUSCLE and phylogenetic tree was constructed using MEGA5. The number at each node represents the bootstrap value from 2000 replicate. Protein IDs—At, Arabidopsis thaliana; LOC_Os, Oryza sativa; Sb, Sorghum bicolor; GRMZM, Zea mays; Bradi, Brachypodium distachyon; Si, Setaria italica. (B) Schematic representation of the conserved motifs in the Clp proteins as revealed by MEME analysis. Each motif is represented by a box numbered at the bottom.
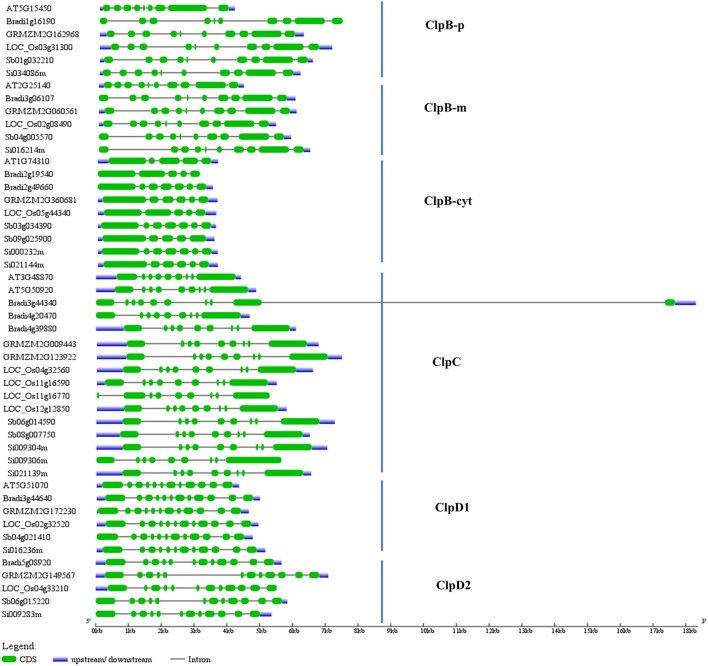
We also studied the exon/intron organization of Arabidopsis and grass Clp genes. For these we downloaded the grass Clp genomic and CDS sequences from Phytozome database and these sequences were compared in gene structure display server program to infer the exon/intron organization. Interestingly ClpB-m, ClpC1, ClpD1, and ClpD2 family genes of Arabidopsis and grass genomes displayed similar exon-intron structures (Figure 3). ClpB-m family genes contain 10 exons, and grass ClpD1 and ClpD2 family genes contain 12 exons each (Figure 3). Arabidopsis ClpB-p has 9 exons, whereas grass ClpB-p family genes contain 10 exons (Figure 3).
Figure 3.
Exon/intron organization of grass Clp genes. The blue lines represent 5′-UTR or 3′-UTR, green boxes represent exons and black lines indicate introns. The Clp genomic sequences and CDS sequences were downloaded from Phytozome and these sequences were compared in gene structure display server program to infer the exon/intron organization of grass Clp genes. Gene IDs—At, Arabidopsis thaliana; LOC_Os, Oryza sativa; Sb, Sorghum bicolor; GRMZM, Zea mays; Bradi, Brachypodium distachyon; Si, Setaria italica.
Expression analysis of class I Clps
Digital expression analysis
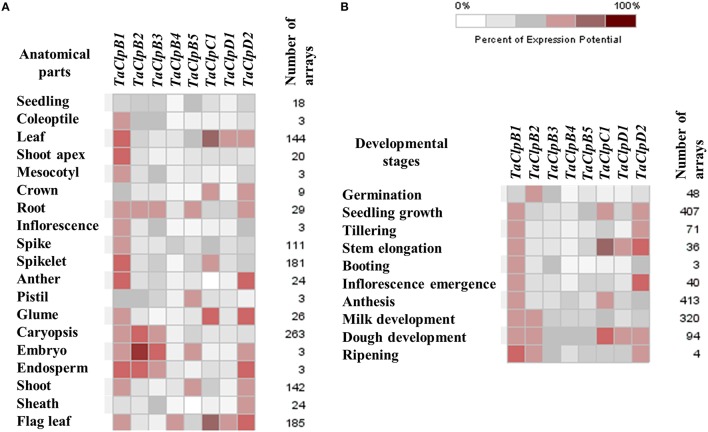
UniGene ID information of wheat Clp genes was retrieved from NCBI database (Table 3). These UniGene IDs were used to retrieve the respective Clp gene EST sequences along with their respective cDNA library information from NCBI UniGene database. Based on this, expression of these genes were assigned into five tissues namely leaf, root, stem, inflorescence, and developing seeds (Table 3). We also studied the spatio-temporal expression pattern, abiotic, and biotic stress inducible expression pattern of identified wheat Clp genes (except for TaClpC2, for which the probe was not available in affymetrix wheat genome array) using wheat genome array expression datasets through Genevestigator v3 database (Hruz et al., 2008). We found that all the nine wheat Clp genes were supported by expression data (Table 3, Figures 4A,B). All the TaClp genes except TaClpB4 expressed in most of the anatomical tissues and in all 10 developmental stages (Figures 4A,B). However, there were significant differences in the level of expression of these genes. The expression of TaClpB1 and TaClpD2 was relatively higher and ubiquitously detected in all the tissues and developmental stages as compared to expression level of TaClpB2, TaClpB3, TaClpB5, TaClpC1, and TaClpD1 (Figures 4A,B). TaClpB4 showed highest expression in flag leaf while TaClpD1 was highly expressed in leaf and flag leaf (Figure 4A).
Table 3.
Digital expression analysis.
| Genes | NCBI UniGene Id | EST and cDNA source from various wheat tissues | No of ESTs available in NCBI | No of mRNA seq available in NCBI | ||||
|---|---|---|---|---|---|---|---|---|
| Leaf | Root | Stem | Inflorescence | Developing seed | ||||
| TaClpB1 | Ta.54573 | ✓ | ✓ | ✓ | ✓ | 400 | 41 | |
| TaClpB2 | Ta.256 | ✓ | ✓ | ✓ | ✓ | 24 | 8 | |
| TaClpB3 | Ta.261 | ✓ | ✓ | ✓ | ✓ | 15 | 14 | |
| TaClpB4 | Ta.51477 | ✓ | 4 | 3 | ||||
| TaClpB5 | Ta.48427 | ✓ | ✓ | ✓ | ✓ | 41 | 19 | |
| TaClpC1 | Ta.49781 | ✓ | ✓ | ✓ | ✓ | ✓ | 293 | 3 |
| TaClpC2 | Ta.68721 | ✓ | 9 | |||||
| TaClpD1 | Ta.54390 | ✓ | ✓ | ✓ | ✓ | ✓ | 220 | 28 |
| TaClpD2 | Ta.48556 | ✓ | ✓ | ✓ | ✓ | ✓ | 23 | 27 |
NCBIUniGene ID was used to retrieve the respective Clp gene ESTs sequence information along with their respective cDNA library information from NCBI UniGene database (✓, Expressed; blank, not expressed).
Figure 4.
Array based expression profiles of TaClp genes in various tissues and developmental stages of wheat. (A) Anatomical tissues and (B) Developmental stages. The expression levels of TaClp genes are presented as heat maps generated using meta-analysis tool at Genevestigator (V3) http://www.genevestigator.ethz.ch. These expression analyses were performed using normalized wheat genome array expression datasets.
The array based expression analysis also revealed that ClpB-cyt genes (TaClpB2 and TaClpB3) and ClpB-m (TaClpB5) transcripts were highly upregulated in leaf tissues under high temperature stress conditions in both heat-tolerant and heat-sensitive wheat genotypes (Figure 5A). TaClpD1 expression was increased during drought stress in flag leaf tissues (Figure 5A). Two genes, TaClpB4 and TaClpD1 were found to be most responsive to biotic stresses analyzed in this study. More than two fold increase in expression of TaClpB4 was observed in response to fungal pathogen Blumeria graminis and Puccinia triticina treatments (Figure 5B) while TaClpD1 was highly upregulated by Tilletia caries and Mayetiola destructor treatments (Figure 5B). In contrast, TaClpB2 and TaClpB3 expression levels were decreased by more than 2.5 fold in response to fungal pathogen B. graminis treatment (Figure 5B).
Figure 5.
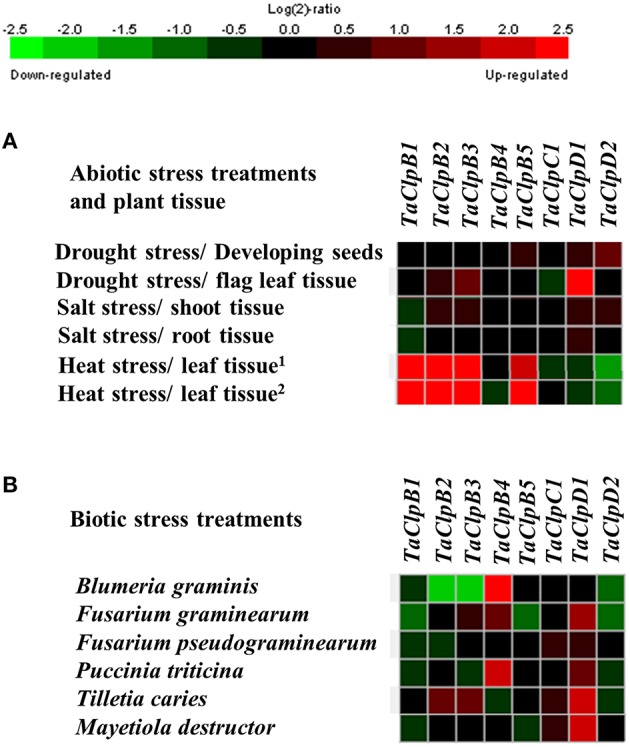
Array based expression of TaClp genes under stress conditions. (A) Expression of Clp genes under abiotic stress conditions. 1Expression profile of TaClp genes in heat sensitive genotype Chinese spring. 2Expression profile of TaClp genes in heat tolerant genotype TAM. (B) Expression of Clp genes under biotic stress conditions. The expression levels of TaClp genes are presented as heat maps generated using meta-analysis tool at Genevestigator (V3) http://www.genevestigator.ethz.ch. Details of wheat abiotic and biotic array experiments used in this study are given in Supplementary Table S2.
qRT-PCR expression analysis of wheat class I Clps
In crop plants, class I ClpATPases are expressed in most of the tissues, at different developmental stages and get induced under various abiotic stress conditions (Keeler et al., 2000; Singh and Grover, 2010; Singh et al., 2010). To investigate the response of TaClp genes to different abiotic stresses, a comprehensive analysis of their expression was performed in 15 days old seedlings of seven wheat genotypes (T. aestivium cvs DBW16, NIAW34, WR544, C306, HD2687, PBW343, and HW2019) differing in thermotolerance. These genotypes were subjected to various abiotic stress conditions such as, high temperature, cold, oxidative, and salt stress (Figures 6–9).
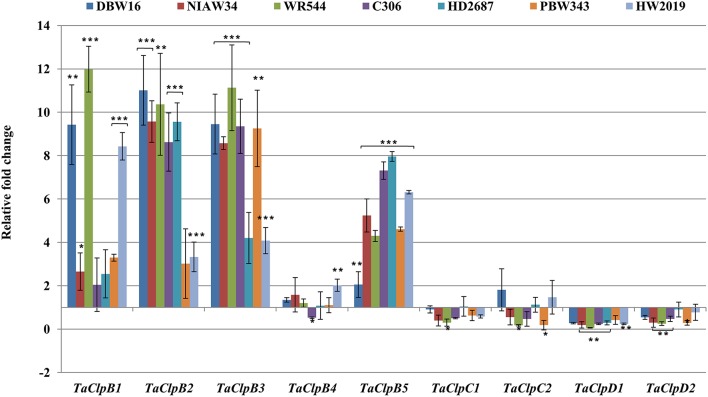
Figure 6.
Expression levels of TaClpgenes under high temperature stress. High temperature stress was imposed to 15 days old seedlings at 42 ± 2°C for 3 h. Error bar represents standard deviation of three biological replicates. p value is calculated using two tailed unpaired t-test, *p < 0.05, **p < 0.01, ***p < 0.001.
The expression levels of TaClpB1, TaClpB2, TaClpB3, and TaClpB5 were either unchanged or up regulated under high temperature stress in all the wheat genotypes (Figure 6). However, the level of expression varied among the genotypes. There was 8–12 fold increase in expression of TaClpB1 in wheat genotypes, DBW16, WR544, and HW2019 whereas the genotypes NIAW34, C306, HD2687 showed two fold or lesser increase in expression of TaClpB1 (Figure 6). Similarly 8–11 fold increased in expression of TaClpB2 and TaClpB3 genes was recorded in five out of seven genotypes under high temperature stress condition (Figure 6). TaClpB5 expression was enhanced by 4–8 folds in all the genotypes except DBW16 (Figure 6). High temperature stress did not change the expression pattern of TaClpB4, TaClpC1, TaClpC2, TaClpD1, and TaClpD2 genes among wheat genotypes (Figure 6).
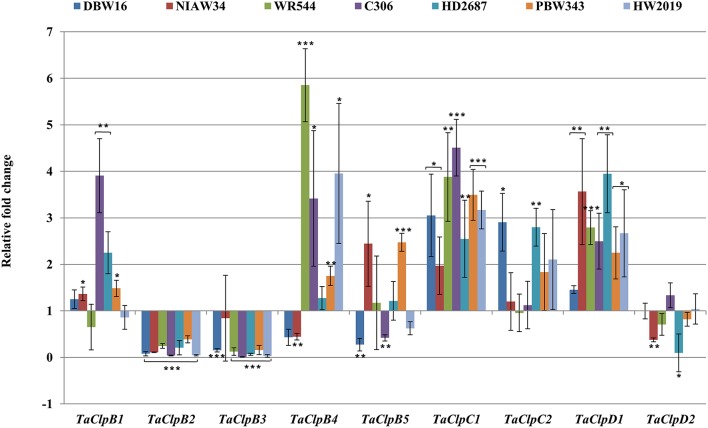
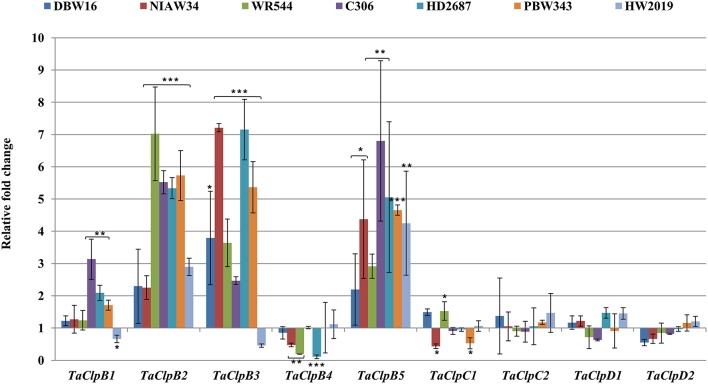
TaClpC1 and TaClpD1 were upregulated while TaClpB2 and TaClpB3 were significantly downregulated in all the genotypes under cold stress conditions (Figure 7). Expression of TaClpB1, TaClpB4, and TaClpB5 were upregulated in some genotypes and downregulated in other genotypes (Figure 7). The expression profile of TaClpB2, TaClpB3, and TaClpB5 under oxidative stress was similar to that recorded under high temperature stress. Expression of these three genes was significantly enhanced in all wheat genotypes except for down regulation of TaClpB3 in HW2019 (Figure 8). TaClpB1 expression was either unchanged as in DBW16, NIAW34, and WR544 or showed induction of 1.7–3 fold in C306, HD2687, and PBW343 (Figure 8). There was no significant difference in the expression of TaClpC1, TaClpC2, TaClpD1, and TaClpD2 under oxidative stress in all the genotypes while TaClpB4 was significantly down regulated in NIAW34, WR544, and HD2687 (Figure 8).
Figure 7.
Expression levels of TaClp genes under cold stress. Cold stress was imposed to 15 days old seedlings at 2 ± 2°C for 3 h. Error bar represents standard deviation of three biological replicates. p value is calculated using two tailed unpaired t-test, *p < 0.05, **p < 0.01, ***p < 0.001.
Figure 8.
Expression levels of TaClp genes under oxidative stress. For oxidative stress, 15 days old seedlings were treated with 10 mM H2O2 and kept in dark conditions for 6 h. Error bar represents standard deviation of three biological replicates. p value is calculated using two tailed unpaired t-test, *p < 0.05, **p < 0.01, ***p < 0.001.
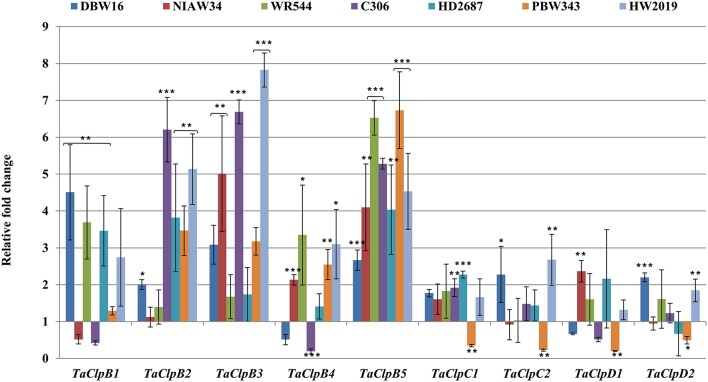
All class I TaClp genes showed upregulation at least in some of the genotypes under salt stress. The level of induction by salt stress was higher (up to eight fold) for the TaClpB genes as compared to that of TaClpC and TaClpD genes (up to 2.7 fold) (Figure 9). Interestingly TaClpC and TaClpD genes were consistently downregulated in stress susceptible wheat genotype PBW343.
Figure 9.
Expression levels of TaClp genes under salt stress. Salt stress was imposed to 15 days old seedlingsby saturating the cotton bed with 150 MmNaCl for 6 h. Error bar represents standard deviation of three biological replicates. p value is calculated using two tailed unpaired t-test, *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Class I ClpATPases (ClpB/HSP100, ClpC, and ClpD) function as molecular chaperones and play vital role in thermotolerance of plants. Arabidopsis and rice genomes encode 6 and 9 Class I Clps, respectively (Zheng et al., 2002; Singh et al., 2010). As in case of rice, we identified 9 Class I ClpATPases genes in wheat which include five ClpB genes, two ClpC genes, and two ClpD genes (Table 1). Multiple sequence alignment and phylogenetic analyses of wheat, Arabidopsis, rice, maize, sorghum, brachypodium, and setaria Clp sequences revealed that the Class I ATPases are highly conserved across the grass genome (Figure 2). The phylogenetic tree branched into three major clades corresponding to ClpB, ClpC, and ClpD protein family, respectively (Figure 2). The ClpB proteins were categorized into cytoplasmic, mitochondrial and chloroplastic isoforms based on their sub-cellular localization predicted by TargetP and WoLF PSORT tool (Supplementary Table S1). The Class I ATPase protein family is highly conserved molecular chaperones in most organisms from bacteria to plants (Trösch et al., 2015). It was evident from the similar exon-intron structures of TaClpB5, ClpC1, ClpD1, and ClpD2 family genes of Arabidopsis and grass genomes (Figure 3). TaClpB5 family genes contain 10 exons, and ClpD1 and ClpD2 family genes contain 12 exons each (Figure 3).
The array based expression analysis revealed significant spatio-temporal transcriptional regulation of TaClps by developmental and stress cues (Figures 4A,B; 5A,B). In addition to chaperone activity, ClpATPase proteins are also involved in various cellular process related to plant growth and development (Trösch et al., 2015). The ubiquitous presence of TaClpB1 and TaClpD2 and their abundance in most of the tissues and developmental stages indicate the significance of these genes in growth and development of wheat. The higher expression of TaClpB2 and TaClpB3 in embryo, endosperm, caryopsis and root reflect their specific role during seed development and in roots (Figure 4A). These observations are further corroborated in array analysis which exhibited higher expression levels of TaClpB2 during milk, dough, and ripening stages of wheat grain development (Figure 4B). In Arabidopsis, loss of function of AtClpC1 resulted in reduced plant growth, lowered protein translocation and chloroplast development (Constan et al., 2004; Olinares et al., 2011). In this study, the expression of TaClpC1 was not only found in green tissues but also in non-green tissues such as root and dough stage of grain development (Figures 4A,B) suggesting its potential role beyond chloroplast development and function in wheat.
Cytoplasm localized ClpBs (ClpB-cyt) act as molecular chaperones, and mediate disaggregation of denatured proteins during high temperature stress (Singh and Grover, 2010). In Arabidopsis (Lee et al., 2007), rice (Singh et al., 2010), maize (Nieto-Sotelo et al., 2002), and Lima bean (Keeler et al., 2000), ClpB-cyt are involved in both basal and acquired thermotolerance. Arabidopsis ClpB-cyt, ClpB-p, and ClpB-m transcripts accumulate dramatically at high temperature stress (Lee et al., 2007). Transgenic tomato lines with antisense suppression of chloroplast localized ClpB-P (LeHSP100/ClpB) failed to recover from heat shock treatment (Yang et al., 2006). Overexpression of Arabidopsis ClpB-cyt (AtHSP101) in an indica rice variety Pusa basmati 1 resulted in increased basal thermotolerance in transgenic rice (Katiyar-Agarwal et al., 2003). Wheat ClpB-cyt genes, TaHSP101B and TaHSP101C, were found to be induced by high temperature stress, dehydration and ABA, but not affected by chilling or wounding (Campbell et al., 2001). Expression of TdHSP101B and TdHSP101C were increased under high temperature stress in Triticum durum (Gullì et al., 2007; Rampino et al., 2009, 2012). In maize, ClpB-cyt null mutants were defective in both induced and basal thermotolerance (Nieto-Sotelo et al., 2002). Our array based analysis also showed that wheat ClpB-p (TaClpB1), and ClpB-cyt (TaClpB2 and TaClpB3), and ClpB-m (TaClpB5) genes were upregulated under high temperature stress conditions in both heat-tolerant and heat-sensitive genotypes (Figure 5A). We further confirmed this by real-time RT-PCR expression analysis wherein high temperature stress was found to upregulate the expression of ClpB-p (TaClpB1), ClpB-cyt (TaClpB2 and TaClpB3), and ClpB-m (TaClpB5) in all wheat genotypes (Figure 6). This suggests their potential role in imparting basal thermotolerance to wheat genotypes. In contrast, TaClpD2 showed downregulation in both array based analysis and qRT-PCR analysis (Figures 5A, 6). Sumesh et al. (2008) observed higher amount of TaClpB/HSP100 content at elevated temperature in a relatively tolerant wheat variety. In our study, high temperature stress enhanced the expression of TaClpB2 and TaClpB3 genes in all the genotypes. However, TaClpB2 and TaClpB3 expression levels were higher in the all four thermotolerant genotypes (DBW16, NIAW34, WR544, and C306) and lower in at least two out of three sensitive genotypes (Figure 6). Among thermotolerant genotypes, DBW16 and WR544 displayed higher expression of ClpB-p (TaClpB1), and ClpB-cyt (TaClpB2 and TaClpB3) genes than C306 and NIAW34 (Figure 6). All the genotypes showed 2–8 fold increase in ClpB-m (TaClpB5) expression under heat stress (Figure 6). In Arabidopsis, all the ClpB transcripts were upregulated by heat while ClpC and ClpD were not regulated by heat. Moreover, the T-DNA mutants of ClpB-p and ClpB-m were not defective in acquired thermotolerance as compared to heat sensitive null mutant of cytosolic/nuclear AtHsp101 (ClpB-cyt) (Lee et al., 2007). In our study also transcript levels of ClpB-cyt (TaClpB2 and TaClpB3) were up regulated in four of the thermotolerant genotypes which emphasizes the critical role of ClpB-cyt in thermotolerance (Figure 6). The relatively heat sensitive genotype HD2687 and PBW343 demonstrated higher TaClpB2 and TaClpB3 expression levels, respectively (Figure 6). This suggests that simultaneous expression of both the ClpB-cyt genes (TaClpB2 and TaClpB3) might be required for better thermotolerance. Thus, TaClpB/HSP100 might be playing important role than ClpB-p (TaClpB1) and ClpB-m (TaClpB5) genes in imparting thermotolerance in wheat.
Cold stress is known to upregulate rice ClpC transcripts (Singh et al., 2010) and protein (Cui et al., 2005). ClpC1 is associated with the chloroplast protein translocation machinery and might play important role in importing stress responsive protein under stress conditions (Adam et al., 2006; Kovacheva et al., 2007). AtClpD expression was increased during cold stress (Zheng et al., 2002). ZmHsp101 (ortholog of TaClpB2 and TaClpB3) expression was increased only during high temperature stress not under cold, osmotic and ABA induced stress conditions (Nieto-Sotelo et al., 1999). In our study also cold stress induced the expression of TaClpC1 and TaClpD1 (Figure 7), while the expression of TaClpB2 and TaClpB3 was enhanced by heat stress but downregulated by cold stress in most of the wheat genotypes (Figure 7). However, we also observed the induced expression of TaClpB1 in C306 and HD2687, TaClpB4 expression in WR544, HW2019 and C306, TaClpB5 expression in NIAW34 and PBW343, and TaClpC2 expression in DBW16, HD2687, PBW343, and HW2019 under cold stress (Figure 7). These results suggest that TaClpC1 and TaClpD1 may play specific protective role under low temperature stress, while TaClpB2 and TaClpB3 may be important specifically for high temperature stress response of wheat. This also highlights the highly conserved role of these proteins across plant species. OsClpB-cyt and OsClpB-m expression was increased under oxidative stress treatment (Singh et al., 2010). In our study also oxidative stress upregulated the expression of Clp-cyt (TaClpB2 and TaClpB3) and Clp-m (TaClpB5) in all the genotypes (except for TaClpB3 in HW2019) (Figure 8). TaClpB1 expression was induced in C306 and HD2687 under oxidative stress (Figure 8). In rice, oxidative stress induced the expression of OsClpC2, OsClpD1, and OsClpD2 (Singh et al., 2010). Arabidopsis ClpD also showed upregulation under short-term severe oxidative stress (Zheng et al., 2002). However, we did not find significant difference in the expression levels of TaClpC1, TaClpC2, TaClpD1, and TaClpD2 under oxidative stress in the wheat genotypes (Figure 8).
Drought and salt stress inducible AtClpD was shown to impart drought tolerance to Arabidopsis (Nakashima et al., 1997; Tran et al., 2004). We also found that drought stress upregulates the expression of TaClpD1 (Figure 5A). In rice, ClpB proteins were enhanced during salt stress (Pareek et al., 1995). AtClpC expression was increased under short-term severe salt stress (Zheng et al., 2002). In this study, TaClpB genes were upregulated in most of the wheat genotypes under salt stress (Figure 9). However, there was no distinct pattern of expression difference between thermotolerant and susceptible genotypes. All the genotypes showed induction of TaClpB5 expression in the range of ~2.75–6.75 fold under salt stress (Figure 9). TaClpB1 expression was induced in DBW16, WR544, HD2687, and HW2019 whereas, TaClpB2 expression was induced in all the genotypes (except in NIAW34 andWR544). TaClpB4 expression was induced in NIAW34, WR544, PBW343, and HW2019 (Figure 9). TaClpC1 was induced in all the genotypes (except in PBW343), whereas TaClpC1 and TaClpD2 expression was induced in DBW 16 and HW2019 (Figure 9). TaClpD1 was induced in NIAW34 and HD2687 (Figure 9). One of the gene encoding plastid Clp, TaClpD2 was upregulated only under salt stress (Figure 9). In our study, the TaClps were differentially expressed among the genotypes under abiotic stress conditions (Figures 6–9). Further studies on mining the regulatory sequences of TaClps in contrasting genotypes and their regulatory mechanisms will enhance the understanding of TaClps in wheat.
In tomato, prolonged infection of tomato yellow leaf curl virus resulted in decline in the amounts of HSPs, which was more pronounced in susceptible than in resistant plants (Gorovits and Czosnek, 2007). In our study, Clps were found to be regulated by various biotic stresses (Figure 5B). TaClpD1 was upregulated by both fungal pathogens and insect pests, while TaClpB4 was highly upregulated under bacterial and fungal pathogen treatments (Figure 5B). These results suggest potential biotic-stress specific roles of Clp genes in wheat.
Conclusion
This study elucidated the genomic organization of TaClps, evolutionary relationship with the grass orthologous and developmental and stress regulated expression of TaClp gene family in wheat. We identified nine class I Clp proteins in wheat and showed high conservation of ClpB/Hsp100, ClpC and ClpD proteins across grass genomes. We noted that several TaClps genes are expressed constitutively in vegetative tissues and various developmental stages. Through wheat array transcript profiling and qRT-PCR analysis, we have shown that the class I Clp genes are differentially regulated by abiotic stresses. High temperature and oxidative stresses upregulated TaClpB1 (ClpB-p), TaClpB2 and TaClpB3 (ClpB-cyt), and TaClpB5 (ClpB-m) expression in most of the genotypes except that of TaClpB3 in HW2019 under oxidative stress. Cold stress specifically upregulated TaClpC and TaClpD1 genes. The TaClpB5 gene for mitochondrial Clp showed upregulation under heat, salt and oxidative stresses. The differential expression regulation suggests a distinct role of TaClpB2, TaClpB3, and TaClpB5 in high temperature stress response, and TaClpC1, TaClpC2, and TaClpD1 in low temperature stress response of wheat. Some of the TaClps showed considerable upregulation under fungal pathogen treatment and insect pest attack. Our results showed wheat ClpB/Hsp100, ClpC, and ClpD genes play distinct roles in plant growth, development and response to abiotic and biotic stress conditions.
Author contributions
SM, VC, and KB conceived the idea and designed the experiments. SM performed all the experiments, analyse the data, and drafted the manuscript. MD contributed to data analysis. MD, VC, and KB edited the manuscript. All authors have read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SM gratefully acknowledges Department of Science and Technology (DST) for DST-INSPIRE Fellowship and Indian Agricultural Research Institute (IARI) for Senior Research fellowship (IARI-SRF). The project work was supported by grants from ICAR-NRCPB, ICAR-IARI and ICAR-NBPGR, New Delhi.
Glossary
Abbreviations
- Clp
Caseinolytic protease
- HSP
Heat shock proteins
- ClpB-cyt
Cytoplasmic ClpB
- ClpB-p
Chloroplastic ClpB
- ClpB-m
Mitochondrial ClpB.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00929
References
- Adam Z., Rudella A., van Wijk K. J. (2006). Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr. Opin. Plant Biol. 9, 234–240. 10.1016/j.pbi.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Aggarwal P. K., Bandyopadhyay S. K., Pathak H., Kalra N., Chander S., Kumar S. (2000). Analysis of yield trends of the rice-wheat system in north-western India. Outlook Agric. 29, 259–268. 10.5367/000000000101293329 [DOI] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton M. D., Kolmer J. A., Xu W. W., Garvin D. F. (2008). Lr34-mediated leaf rust resistance in wheat: transcript profiling reveals a high energetic demand supported by transient recruitment of multiple metabolic pathways. Mol. Plant Microbe Interact. 21, 1515–1527. 10.1094/MPMI-21-12-1515 [DOI] [PubMed] [Google Scholar]
- Brisson N., Gate P., Gouache D., Charmet G., Oury F. X., Huard F. (2010). Why are wheat yields stagnating in Europe? A comprehensive data analysis for France. Field Crop. Res. 119, 201–212. 10.1016/j.fcr.2010.07.012 [DOI] [Google Scholar]
- Campbell J. L., Klueva N. Y., Zheng H., Nieto-Sotelo J., Ho T.-H., Nguyen H. T. (2001). Cloning of new members of heat shock protein HSP101 gene family in wheat (Triticum aestivum (L.) Moench) inducible by heat, dehydration, and ABA. Biochim. Biophys. Acta 1517, 270–277. 10.1016/S0167-4781(00)00292-X [DOI] [PubMed] [Google Scholar]
- Carroni M., Kummer E., Oguchi Y., Wendler P., Clare D. K., Sinning I., et al. (2014). Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife 3:e02481. 10.7554/eLife.02481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain F., Côté-Beaulieu C., Belzile F., Menzies J. G., Bélanger R. R. (2009). A comprehensive transcriptomic analysis of the effect of silicon on wheat plants under control and pathogen stress conditions. Mol. Plant Microbe Interact. 22, 1323–1330. 10.1094/MPMI-22-11-1323 [DOI] [PubMed] [Google Scholar]
- Constan D., Froehlich J. E., Rangarajan S., Keegstra K. (2004). A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol. 136, 3605–3615. 10.1104/pp.104.052928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S., Huang F., Wang J., Ma X., Cheng Y., Liu J. (2005). A proteomic analysis of cold stress responses in rice seedlings. Proteomics 5, 3162–3172. 10.1002/pmic.200401148 [DOI] [PubMed] [Google Scholar]
- DeSantis M. E., Shorter J. (2012). The elusive middle domain of Hsp104 and ClpB: location and function. Biochim. Biophys. Acta 1823, 29–39. 10.1016/j.bbamcr.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmond O. J., Manners J. M., Schenk P. M., Maclean D. J., Kazan K. (2008). Gene expression analysis of the wheat response to infection by Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 73, 40–47. 10.1016/j.pmpp.2008.12.001 [DOI] [Google Scholar]
- Doyle S. M., Wickner S. (2009). Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 34, 40–48. 10.1016/j.tibs.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. 10.1038/nprot.2007.131 [DOI] [PubMed] [Google Scholar]
- Ergen N. Z., Thimmapuram J., Bohnert H. J., Budak H. (2009). Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct. Integr. Genomics 9, 377–396. 10.1007/s10142-009-0123-1 [DOI] [PubMed] [Google Scholar]
- Frickey T., Lupas A. N. (2004). Phylogenetic analysis of AAA proteins. J. Struct. Biol. 146, 2–10. 10.1016/j.jsb.2003.11.020 [DOI] [PubMed] [Google Scholar]
- Godfray H. C. J., Beddington J. R., Crute I. R., Haddad L., Lawrence D., Muir J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327, 812–818. 10.1126/science.1185383 [DOI] [PubMed] [Google Scholar]
- Gorovits R., Czosnek H. (2007). Biotic and abiotic stress responses in tomato breeding lines resistant and susceptible to tomato yellow leaf curl virus, in Tomato Yellow Leaf Curl Virus Disease SE - 13, ed Czosnek H. (Dordrecht: Springer; ), 223–237. [Google Scholar]
- Gullì M., Corradi M., Rampino P., Marmiroli N., Perrotta C. (2007). Four members of the HSP101 gene family are differently regulated in Triticum durum Desf. FEBS Lett. 581, 4841–4849. 10.1016/j.febslet.2007.09.010 [DOI] [PubMed] [Google Scholar]
- Horton P., Park K.-J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. 10.1093/nar/gkm259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008:420747. 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A.-Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B. J., Park W. J., Chung C. H., Goldberg A. L. (1987). Escherichia coli contains a soluble ATP-dependent protease (Ti) distinct from protease La. Proc. Natl. Acad. Sci. U.S.A. 84, 5550–5554. 10.1073/pnas.84.16.5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Agarwal M., Grover A. (2003). Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol. Biol. 51, 677–686. 10.1023/A:1022561926676 [DOI] [PubMed] [Google Scholar]
- Keeler S. J., Boettger C. M., Haynes J. G., Kuches K. A., Johnson M. M., Thureen D. L., et al. (2000). Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiol. 123, 1121–1132. 10.1104/pp.123.3.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J., Schlothauer T., Dougan D. A., Lilie H., Tischendorf G., Mogk A., et al. (2006). Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 25, 1481–1491. 10.1038/sj.emboj.7601042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T., Yamaguchishinozaki K., Shinozaki K. (1993). Characterization of cDNA for a dehydration-inducible gene that encodes a CLP A, B-like protein in Arabidopsis thaliana L. Biochem. Biophys. Res. Commun. 196, 1214–1220. 10.1006/bbrc.1993.2381 [DOI] [PubMed] [Google Scholar]
- Kovacheva S., Bédard J., Wardle A., Patel R., Jarvis P. (2007). Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50, 364–379. 10.1111/j.1365-313X.2007.03060.x [DOI] [PubMed] [Google Scholar]
- Krugman T., Chagué V., Peleg Z., Balzergue S., Just J., Korol A. B., et al. (2010). Multilevel regulation and signalling processes associated with adaptation to terminal drought in wild emmer wheat. Funct. Integr. Genomics 10, 167–186. 10.1007/s10142-010-0166-3 [DOI] [PubMed] [Google Scholar]
- Krzewska J., Konopa G., Liberek K. (2001). Importance of two ATP-binding sites for oligomerization, ATPase activity and chaperone function of mitochondrial Hsp78 protein. J. Mol. Biol. 314, 901–910. 10.1006/jmbi.2001.5190 [DOI] [PubMed] [Google Scholar]
- Lee S., Sowa M. E., Choi J.-M., Tsai F. T. F. (2004). The ClpB/Hsp104 molecular chaperone-a protein disaggregating machine. J. Struct. Biol. 146, 99–105. 10.1016/j.jsb.2003.11.016 [DOI] [PubMed] [Google Scholar]
- Lee U., Rioflorido I., Hong S.-W., Larkindale J., Waters E. R., Vierling E. (2007). The Arabidopsis ClpB/Hsp100 family of proteins: chaperones for stress and chloroplast development. Plant J. 49, 115–127. 10.1111/j.1365-313X.2006.02940.x [DOI] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lobell D. B., Sibley A., Ivan Ortiz-Monasterio J. (2012). Extreme heat effects on wheat senescence in India. Nat. Clim. Change 2, 186–189. 10.1038/nclimate1356 [DOI] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Mishra R. C., Grover A. (2015). ClpB/Hsp100 proteins and heat stress tolerance in plants. Crit. Rev. Biotechnol. [Epub ahead of print]. 10.3109/07388551.2015.1051942. [DOI] [PubMed] [Google Scholar]
- Mochida K., Yoshida T., Sakurai T., Ogihara Y., Shinozaki K. (2009). TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol. 150, 1135–1146. 10.1104/pp.109.138214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott I. W., Wang R. R.-C. (2007). Comparative transcriptome analysis of salt-tolerant wheat germplasm lines using wheat genome arrays. Plant Sci. 173, 327–339. 10.1016/j.plantsci.2007.06.005 [DOI] [Google Scholar]
- Myouga F., Motohashi R., Kuromori T., Nagata N., Shinozaki K. (2006). An Arabidopsis chloroplast-targeted Hsp101 homologue, APG6, has an essential role in chloroplast development as well as heat-stress response. Plant J. 48, 249–260. 10.1111/j.1365-313X.2006.02873.x [DOI] [PubMed] [Google Scholar]
- Nakashima K., Kiyosue T., Yamaguchi-Shinozaki K., Shinozaki K. (1997). A nuclear gene, erd1, encoding a chloroplast-targeted Clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. Plant J. 12, 851–861. 10.1046/j.1365-313X.1997.12040851.x [DOI] [PubMed] [Google Scholar]
- Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. (1999). AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43. 10.1101/gr.9.1.27 [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo J., Kannan K. B., Martínez L. M., Segal C. (1999). Characterization of a maize heat-shock protein 101 gene, HSP101, encoding a ClpB/Hsp100 protein homologue. Gene 230, 187–195. 10.1016/S0378-1119(99)00060-8 [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo J., Martínez L. M., Ponce G., Cassab G. I., Alagón A., Meeley R. B., et al. (2002). Maize HSP101 plays important roles in both induced and basal thermotolerance and primary root growth. Plant Cell 14, 1621–1633. 10.1105/tpc.010487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., van Wijk K. J. (2015). Organization, function and substrates of the essential Clp protease system in plastids. Biochim. Biophys. Acta 1847, 915–930. 10.1016/j.bbabio.2014.11.012 [DOI] [PubMed] [Google Scholar]
- Olinares P. D. B., Kim J., van Wijk K. J. (2011). The Clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta 1807, 999–1011. 10.1016/j.bbabio.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Ortiz R., Sayre K. D., Govaerts B., Gupta R., Subbarao G. V., Ban T., et al. (2008). Climate change: can wheat beat the heat? Agric. Ecosyst. Environ. 126, 46–58. 10.1016/j.agee.2008.01.019 [DOI] [Google Scholar]
- Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M., Childs K., et al. (2007). The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 35, D883–D887. 10.1093/nar/gkl976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek A., Singla S. L., Grover A. (1995). Immunological evidence for accumulation of two high-molecular-weight (104 and 90 kDa) HSPs in response to different stresses in rice and in response to high temperature stress in diverse plant genera. Plant Mol. Biol. 29, 293–301. 10.1007/BF00043653 [DOI] [PubMed] [Google Scholar]
- Park S., Rodermel S. R. (2004). Mutations in ClpC2/Hsp100 suppress the requirement for FtsH in thylakoid membrane biogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 12765–12770. 10.1073/pnas.0402764101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porankiewicz J., Wang J., Clarke A. K. (1999). New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32, 449–458. 10.1046/j.1365-2958.1999.01357.x [DOI] [PubMed] [Google Scholar]
- Qin D., Wu H., Peng H., Yao Y., Ni Z., Li Z., et al. (2008). Heat stress-responsive transcriptome analysis in heat susceptible and tolerant wheat (Triticum aestivum L.) by using Wheat Genome Array. BMC Genomics 9:432. 10.1186/1471-2164-9-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampino P., Mita G., Fasano P., Borrelli G. M., Aprile A., Dalessandro G., et al. (2012). Novel durum wheat genes up-regulated in response to a combination of heat and drought stress. Plant Physiol. Biochem. 56, 72–78. 10.1016/j.plaphy.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Rampino P., Mita G., Pataleo S., De Pascali M., Di Fonzo N., Perrotta C. (2009). Acquisition of thermotolerance and HSP gene expression in durum wheat (Triticum durum Desf.) cultivars. Environ. Exp. Bot. 66, 257–264. 10.1016/j.envexpbot.2009.04.001 [DOI] [Google Scholar]
- Schlothauer T., Mogk A., Dougan D. A., Bukau B., Turgay K. (2003). MecA, an adaptor protein necessary for ClpC chaperone activity. Proc. Natl. Acad. Sci. U.S.A. 100, 2306–2311. 10.1073/pnas.0535717100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42, 264–273. 10.1093/pcp/pce031 [DOI] [PubMed] [Google Scholar]
- Singh A., Grover A. (2010). Plant Hsp100/ClpB-like proteins: poorly-analyzed cousins of yeast ClpB machine. Plant Mol. Biol. 74, 395–404. 10.1007/s11103-010-9682-8 [DOI] [PubMed] [Google Scholar]
- Singh A., Singh U., Mittal D., Grover A. (2010). Genome-wide analysis of rice ClpB/HSP100, ClpC and ClpD genes. BMC Genomics 11:95. 10.1186/1471-2164-11-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L. L. E., MacDonald T. M., Sutinen S., Clarke A. K. (2004). Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol. 136, 4114–4126. 10.1104/pp.104.053835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren L. L. E., Tanabe N., Lymperopoulos P., Khan N. Z., Rodermel S. R., Aronsson H., et al. (2014). Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J. Biol. Chem. 289, 11318–11330. 10.1074/jbc.M113.534552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider J., Thibault G., Houry W. A. (2008). The AAA+ superfamily of functionally diverse proteins. Genome Biol. 9:216. 10.1186/gb-2008-9-4-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumesh K. V., Sharma-Natu P., Ghildiyal M. C. (2008). Starch synthase activity and heat shock protein in relation to thermal tolerance of developing wheat grains. Biol. Plant. 52, 749–753. 10.1007/s10535-008-0145-x [DOI] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.-S. P., Nakashima K., Sakuma Y., Simpson S. D., Fujita Y., Maruyama K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. 10.1105/tpc.104.022699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trösch R., Mühlhaus T., Schroda M., Willmund F. (2015). ATP-dependent molecular chaperones in plastids - More complex than expected. Biochim. Biophys. Acta 1847, 872–888. 10.1016/j.bbabio.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Viswanathan C., Khanna-Chopra R. (2001). Effect of heat stress on grain growth, starch synthesis and protein synthesis in grains of wheat (Triticum aestivum L.) varieties differing in grain weight stability. J. Agron. Crop Sci. 186, 1–7. 10.1046/j.1439-037x.2001.00432.x [DOI] [Google Scholar]
- Wan Y., Poole R. L., Huttly A. K., Toscano-Underwood C., Feeney K., Welham S., et al. (2008). Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9:121. 10.1186/1471-2164-9-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Mei Z., Qi Y., Yan C., Hu Q., Wang J., et al. (2011). Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature 471, 331–335. 10.1038/nature09780 [DOI] [PubMed] [Google Scholar]
- Wang J.-R., Wang L., Gulden S., Rocheleau H., Balcerzak M., Hattori J., et al. (2010). RNA profiling of fusarium head blight-resistant wheat addition lines containing the Thinopyrum elongatum chromosome 7E. Can. J. Plant Pathol. 32, 188–214. 10.1080/07060661003740512 [DOI] [Google Scholar]
- Weaver L. M. (1999). Chloroplast-targeted ERD1 protein declines but its mRNA increases during senescence in Arabidopsis. Plant Physiol. 119, 1209–1216. 10.1104/pp.119.4.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., et al. (2004). Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665. 10.1016/j.cell.2004.11.027 [DOI] [PubMed] [Google Scholar]
- Wu H., Ji Y., Du J., Kong D., Liang H., Ling H.-Q. (2010). ClpC1, an ATP-dependent Clp protease in plastids, is involved in iron homeostasis in Arabidopsis leaves. Ann. Bot. 105, 823–833. 10.1093/aob/mcq051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Sun Y., Sun A., Yi S., Qin J., Li M., et al. (2006). The involvement of chloroplast HSP100/ClpB in the acquired thermotolerance in tomato. Plant Mol. Biol. 62, 385–395. 10.1007/s11103-006-9027-9 [DOI] [PubMed] [Google Scholar]
- Zheng B., Halperin T., Hruskova-Heidingsfeldova O., Adam Z., Clarke A. K. (2002). Characterization of chloroplast Clp proteins in Arabidopsis: localization, tissue specificity and stress responses. Physiol. Plant. 114, 92–101. 10.1034/j.1399-3054.2002.1140113.x [DOI] [PubMed] [Google Scholar]
- Zhu L., Liu X., Wang H., Khajuria C., Reese J. C., Whitworth R. J., et al. (2012). Rapid mobilization of membrane lipids in wheat leaf sheaths during incompatible interactions with hessian fly. Mol. Plant Microbe Interact. 25, 920–930. 10.1094/MPMI-01-12-0022-R [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.