Abstract
Vertical distributions of dominant bacterial populations in saline meromictic Lake Kaiike were investigated throughout the water column and sediment by quantitative oligonucleotide probe membrane hybridization. Three oligonucleotide probes specific for the small-subunit (SSU) rRNA of three groups of Chlorobiaceae were newly designed. In addition, three general domain (Bacteria, Archaea, and Eukarya)-specific probes, two δ-Proteobacteria-specific probes, a Chlorobiaceae-specific probe, and a Chloroflexi-specific probe were used after optimization of their washing conditions. The abundance of the sum of SSU rRNAs hybridizing with probes specific for three groups of Chlorobiaceae relative to total SSU rRNA peaked in the chemocline, accounting for up to 68%. The abundance of the δ-proteobacterial SSU rRNA relative to total SSU rRNA rapidly increased just below the chemocline up to 29% in anoxic water and peaked at the 2- to 3-cm sediment depth at ca. 34%. The abundance of SSU rRNAs hybridizing with the probe specific for the phylum Chloroflexi relative to total SSU rRNA was highest (31 to 54%) in the top of the sediment but then steeply declined with depth and became stable at 11 to 19%, indicating the robust coexistence of sulfate-reducing bacteria and Chloroflexi in the top of the sediment. Any SSU rRNA of Chloroflexi in the water column was under the detection limit. The summation of the signals of group-specific probes used in this study accounted for up to 89% of total SSU rRNA, suggesting that the DGGE-oligonucleotide probe hybridization approach, in contrast to conventional culture-dependent approaches, was very effective in covering dominant populations.
Lake Kaiike is a small saline meromictic lake on the seaward side of Kamikoshiki Island, in southwest Japan. A dense population of phototrophic purple sulfur bacteria has been observed in the chemocline of this lake (19, 25, 30) as well as in other meromictic lakes (32, 33, 51). A previous denaturing gradient gel electrophoresis (DGGE) analysis of PCR-amplified small-subunit (SSU)-rRNA genes in the water column and sediment demonstrated the occurrence of drastic depth-related changes in the microbial community at the chemocline and the sediment-water interface (19). Some conspicuous DGGE bands whose sequences were related to Chlorobiaceae (also known as green sulfur bacteria) and sulfate-reducing bacteria (SRB) affiliated with the δ-Proteobacteria were observed in anoxic water. An analysis of chloropigments also confirmed the predominance of members of the Chlorobiaceae in anoxic water and classified them as the brown variety of Chlorobiaceae, which were considered better adapted to lower light intensity than the green variety (54), owing to their possession of bacteriochlorophyll e (30). On the basis of phylogenetic analysis, Chlorobiaceae observed in Lake Kaiike could be divided into three groups, of which two were distinct from any isolated strains found to date. A comparison of DGGE banding patterns of the water column and sediment revealed that Chlorobiaceae accumulated on the bottom from the overlying anoxic water, and that indigenous SRB and Chloroflexi (formerly known as green nonsulfur bacteria) were present in the sediment.
For over a decade, quantification of microbial communities by molecular biological methods such as fluorescence in situ hybridization and oligonucleotide probe membrane hybridization has been widely conducted in natural environments (4, 5, 21, 28, 38, 39, 48, 53). Although fluorescence in situ hybridization has proved to be an effective technique for directly identifying microorganisms, highly concentrated autofluorescent bacteria such as cyanobacteria together with an unknown high background made it difficult to apply this technique to the sediment of Lake Kaiike. On the other hand, membrane hybridization of total SSU rRNA extracted from environmental samples provides a reliable estimate of the relative abundance of active populations with good sensitivity regardless of the sample type, i.e., water or sediment. Community structure analyses by these methods are more quantitative than those by PCR-based approaches, since biases introduced during DNA amplification can be avoided (40, 49). Nevertheless, PCR-DGGE analysis prior to membrane hybridization is effective in determining which probe set should be used, enabling the design of probes specific for interesting populations. In this study, we designed three novel oligonucleotide probes based on the DGGE fragment sequences of three distinct groups within Chlorobiaceae.
The aim of the present study was to evaluate dominant populations determined by a previous PCR-DGGE analysis and to investigate their relative contributions and vertical distributions throughout the water column and sediment of Lake Kaiike. We discuss the potential ecological importance of those dominant populations with regard to their relative abundance and distribution and the potential occurrence of symbiotic relationships.
MATERIALS AND METHODS
Sampling site and procedures.
Lake Kaiike is located on the seaward side of Kamikoshiki Island, southwest Japan. It has a surface area of 0.15 km2 and a maximum depth of approximately 11 m. Sampling was carried out at a central point (30°51′N, 129°52′E) on 13 March and 3 June 2003. Water samples were obtained with a convertible Niskin sampler at 0.25- to 1.0-m intervals down to 10 m. Large sediment cores (12 cm in diameter and 50 cm in length) were collected with a core sampler (type MT-2; Rigosya, Tokyo, Japan). The multilayered microbial mats and underlying sediments were sectioned at 0.3- to 2.0-cm intervals to an approximately 20-cm depth. Sliced samples were packed into sterile plastic bags (Fisher Scientific, Pittsburgh, Pa.). Packed samples were then put into anaerobic gas pack pouches (Becton Dickinson Co., Md.) and transported to the laboratory in an ice-cooled box. More detailed descriptions of the sampling site, sample characteristics, and physicochemical properties of the water and sediment have been presented in previous reports (19, 30, 31).
RNA sample preparation.
For intact RNA from the microorganisms listed in Table 1, RNA was extracted from cell pellets by the low-pH bead-beating method (27, 48). Reference SSU rRNAs from the major DGGE bands were generated by in vitro transcription as described previously (17).
TABLE 1.
Oligonucleotide probes used in this study
| Probe or microorganisma | Target sequence | % GC | Td (°C) | Tw (°C) | % Washed off at Tw | Reference |
|---|---|---|---|---|---|---|
| S-*-Univ-1390-a-A-18 (Univ1390)b | 5′GACGGGCGGTGTGTACAA3′ | 61.1 | 43 | 57 | ||
| S-D-Bact-0338-a-A-18 (Bact338) | 5′GCTGCCTCCCGTAGGAGT3′ | 66.7 | 55 | 57 | ||
| S-D-Arch-0915-a-A-20 (Arch915) | 5′GTGCTCCCCCGCCAATTCCT3′ | 65.0 | 58 | 57 | ||
| S-D-Euka-0502-a-A-16 (Euka502) | 5′ACCAGACTTGCCCTCC3′ | 62.5 | 52 | 57 | ||
| S-F-GSB-0528-a-A-19 (GSB528)c | 5′TGCCACCCCTGTATCACCG3′ | 63.2 | 67 | 53 | ||
| Pelodictyon luteolum DSM 273 | 3′ACGGUGGGGACAUAGUGGC5′ | 67.5 | 67 | 49.0 | 53 | |
| Chlorobium vibrioforme DSM 262 | 3′-------------------5′ | 66.5 | 67 | 53.0 | 53 | |
| 21ws, 24ws, 25s, 26s | 3′-------------------5′ | NDd | 67 | ND | 53 | |
| S-*-Chlorb I-0440-a-A-20 (Chlorb I 440) | 5′CCGACATGTTCGTCCCTGAC3′ | 60.0 | ||||
| 21ws (in vitro) | 3′GGCUGUACAAGCAGGGACUG5′ | 57.5 | 62 | 86.5 | This study | |
| Chlorobium vibrioforme DSM 262 | 3′-------------C---A--5′ | 55.0 | 62 | 98.5 | This study | |
| Pelodictyon luteolum DSM 273 | 3′----------------GAC-5′ | 53.5 | 62 | 99.0 | This study | |
| S-*-Chlorb II-0842-a-A-20 (Chlorb II 842) | 5′AGCTACGACACTGATCACGA3′ | 50.0 | ||||
| 24ws (in vitro) | 3′UCGAUGCUGUGACUAGUGCU5′ | 57.0 | 57 | 50.0 | This study | |
| Chlorobium phaeovibrioides DSM 1678 | 3′--------------------5′ | ND | 57 | ND | This study | |
| Chlorobium vibrioforme DSM 262 | 3′----A---------------5′ | 44.5 | 57 | 100.0 | This study | |
| Pelodictyon luteolum DSM 273 | 3′----C---------------5′ | 44.5 | 57 | 100.0 | This study | |
| S-*-Chlorb III-0441-a-A-20 (Chlorb III 441) | 5′ATCAGCATGTTCGTCCCTGA3′ | 50.0 | ||||
| 25s, 26s (in vitro) | 3′UAGUCGUACAAGCAGGGACU5′ | 58.5 | 59 | 60.0 | This study | |
| 21ws | 3′-G-CU---------------5′ | ND | 59 | ND | This study | |
| S-C-dProt-0495-a-A-18 (DELTA495a) | 5′AGTTAGCCGGTGCTTCCT3′ | 55.6 | ||||
| Desulfovibrio desulfuricans DSM 642 | 3′UCAAUCGGCCACGAAGGA5′ | 57.5 | 58e | 56.0 | 22 | |
| 10w, 11s, 12ws, 13s, 14s | 3′------------------5′ | ND | 58e | ND | 22 | |
| Desulfobacter latus DSM 3381 | 3′-----------G------5′ | 54.0 | 58e | 83.0 | 22 | |
| S-C-dProt-0495-b-A-18 (DELTA495b) | 5′AGTTAGCCGGCGCTTCCT3′ | 61.1 | ||||
| Desulfobacter latus DSM 3381 | 3′UCAAUCGGCCGCGAAGGA5′ | 59.5 | 59.5 | 50.0 | 22 | |
| 15s | 3′------------------5′ | ND | 59.5 | ND | 22 | |
| Desulfovibrio desulfuricans DSM 642 | 3′----------A-------5′ | 48.0 | 59.5 | 98.0 | 22 | |
| S-*-GNSB-0941-a-A-17 (GNSB941) | 5′AAACCACACGCTCCGCT3′ | 58.8 | ||||
| Chloroflexus aurantiacus DSM 635 | 3′UUUGGUGUGCGAGGCGA5′ | 56.0 | 56 | 50.0 | 13 | |
| Clostridium aceticum DSM 1496 | 3′-----------------5′ | 47.0 | 56 | 100.0 | 13 |
Probes are named according to the work of Alm et al. (2). Names in parentheses are the short names used in the text. Designations with “w,” “ws,” and “s” are names of DGGE bands defined in a previous study (19).
This probe was used only for normalization of reference SSU rRNA.
This probe was modified from the S-F-GSB-0532-a-A-15 (53).
ND, not determined.
This probe needed a correction because of a lack of clear discrimination between target and nontarget organisms with one mismatch.
The water samples were filtered with Sterivex filter cartridges (0.22-μm pore size; Millipore, Billerica, Mass.) and stored frozen until nucleic acid extraction. Filters were removed from broken cartridges and then transferred into 2-ml screw-cap tubes together with 0.5 g of baked glass beads. Nucleic acid extraction from cells on the filters was carried out as described previously (19). The sediment samples were washed three times with 120 mM sodium phosphate buffer (pH 8.0) to remove any extracellular nucleic acids (52) and then stored at −80°C until nucleic acid extraction. Extraction and purification of nucleic acids from sediment samples were performed by the hydroxyapatite spin-column method (35) with some modification (17). After total nucleic acid extraction from both water and sediment samples, DNA was degraded by DNase I as previously described (29).
Oligonucleotide probe design and determination of Tw.
Three oligonucleotide probes were newly designed using the Probe_Design tool of the ARB software package (http://www.arb-home.de/) in order to quantify three distinct Chlorobium-like groups (Chlorb I, including DGGE bands 21ws, 22w, and 23ws; Chlorb II, including band 24ws, and Chlorb III, including bands 25s and 26s), which were determined by PCR-DGGE analysis (Fig. 1 and 2) (19). The Probe_Match program, provided by the Ribosomal Database Project II (23), and a BLAST search (3) were both used to evaluate probe specificity. The dissociation temperature (Td), the washing temperature (Tw), and the specificities of the oligonucleotide probes were assessed by the general method described by Zheng et al. (57) (Fig. 3; Table 1).
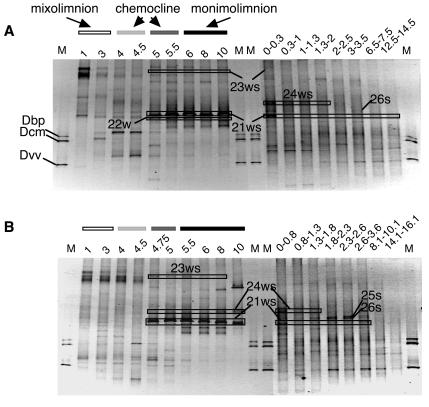
FIG. 1.
DGGE analyses of PCR-amplified 16S rRNA gene fragments of bacterial communities in the water columns and sediments of Lake Kaiike (modified from reference 19 with permission of the publisher). Sequences of the labeled bands were affiliated with the family Chlorobiaceae. The bands were designated as described in a previous report (19). Numbers above each lane indicate depths (meters in the water columns [left]; centimeters in the sediment [right]) where the samples were obtained. Amplified 16S rRNA gene fragments derived from Desulfobulbus propionicus (Dbp), Desulfococcus multivorans (Dcm), and Desulfovibrio vulgaris (Dvv) were used as markers. (A) March 2003; (B) June 2003. M, molecular size markers.
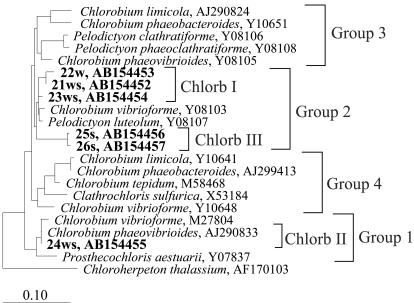
FIG. 2.
Phylogenetic affiliation of Chlorb I, II, and III groups within the family Chlorobiaceae. Environmental clones indicated by a number plus “w,” “ws,” and “s” were the sequenced DGGE fragments from Lake Kaiike. Identification of DGGE bands is defined in Fig. 1. Bar, estimated 10% divergence. Chlorobiaceae are divided into four groups according to Alexander et al. (1).
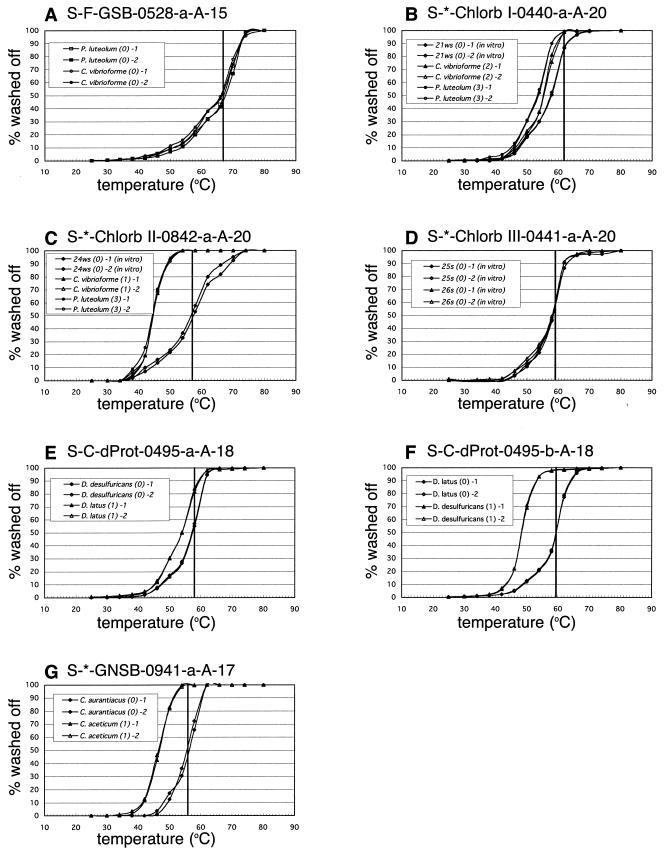
FIG. 3.
Normalized Td curves determined by using intact RNA or transcribed RNA (positions 338 to 907, E. coli numbering) of target and nontarget microorganisms. (A) GSB528; (B) Chlorb I 440; (C) Chlorb II 842; (D) Chlorb III 441; (E) DELTA495a; (F) DELTA495b; (G) GNSB941. Vertical lines indicate the final Tws of individual probes.
Quantitative membrane hybridization and data analysis.
Quantitative membrane hybridization was performed as described previously (18, 48). The concentrations of all samples and reference RNA were normalized by using the probes Univ1390 and Bact338 for intact rRNA and in vitro-transcribed RNA, respectively. A known concentration of intact Escherichia coli SSU rRNA determined spectrophotometrically and correlated by the signal intensity ratio of 23S and 16S rRNA on an ethidium bromide-stained agarose gel was used for normalization. RNA extracted from water and sediment (1.0 ± 0.04 g [wet weight]) samples and a dilution series of reference RNAs were immobilized in duplicate on a nylon membrane (Hybond-XL; Amersham Biosciences UK Ltd., Little Chalfont, Buckinghamshire, United Kingdom) using a slot blotter (Minifold II; Schleicher & Schuell GmbH, Postfach, Germany) after denaturation with 2% (vol/vol) glutaraldehyde. Blotted RNAs on the membrane were air dried, baked at 80°C for 10 min, and fixed by UV exposure (50 mJ) (GS Gene Linker; Bio-Rad Laboratories). All oligonucleotide probes (Table 1) were synthesized commercially (Invitrogen Co., Carlsbad, Calif.), end labeled with 32P ([γ-32P]ATP; ICN Biomedicals, Costa Mesa, Calif.), and purified by using columns (Nuc-Trap probe purification columns; Stratagene, La Jolla, Calif.) according to the manufacturer's protocol. The membranes were prehybridized at 40°C for 2 h, hybridized for over 9 h after addition of a 32P-labeled probe, and washed at the Tw given for each probe (Table 1). The hybridization signals were measured with a phosphorimager (Bio-Imaging analyzer, BAS2000; FUJIX, Tokyo, Japan). Measured signal intensities were converted to an amount of rRNA for each sample by comparison with a pure-culture control standard curve (r2 > 0.997). The standardized results were expressed as a percentage of the combined signals of general bacterial, archaeal, and eukaryal probes. Maximum and minimum ratios of specific probes to total SSU rRNA were calculated by all combinations as follows: aA−1, aB−1, bA−1, and bB−1, where the lowercase letters represent the signal intensities of each specific probe in duplicate and the capital letters represent the combined signal intensities of Bact338, Arch915, and Euka502 in duplicate.
RESULTS
Probe design and evaluation of probe specificity.
In order to determine levels of the dominant populations in Lake Kaiike, which were investigated by DGGE of PCR-amplified SSU-ribosomal-DNA (rDNA) fragments from the water column and sediment, we used seven probes together with three general domain-specific probes (Table 1). A previous PCR-DGGE analysis recognized six phylotypes affiliated with the family Chlorobiaceae in Lake Kaiike (19), consisting of three distinct groups, designated Chlorb I (21ws, 22w, and 23ws), II (24ws), and III (25s and 26s) (Fig. 1 and 2). Three (Chlorb I 440, Chlorb II 842, and Chlorb III 441) of seven probes were newly designed to be specific for these groups.
Since four (GSB528, DELTA495a and -b, and GNSB941) of the seven probes were previously designed for fluorescent in situ or DNA microarray hybridization (13, 22, 53), we optimized the final Tws of these probes as well as those of three novel probes for membrane hybridization by a Td study (Fig. 3). Normalized Td curves of those probes were obtained with high reproducibility. Clear discrimination between target and nontarget organisms was possible with probes Chlorb II 842, DELTA495b, and GNSB941 by washing at their Td (Fig. 3C, F, and G). For probes Chlorb I 440 and DELTA495a, however, the Tds for nontarget organisms containing one to three mismatches were only 2.5 to 4.0°C lower than that for the target organisms (Table 1; Fig. 3B and E). Nevertheless, two- or three-mismatch discrimination of probe Chlorb I 440 could be achieved by washing at 62°C, although its detection limit declined and a slight residual probe-nontarget rRNA duplex still remained (Table 1; Fig. 3B). One-mismatch discrimination of probe DELTA495a was impossible to achieve by washing at any temperature. Thus, the data obtained with probe DELTA495a by washing at 58°C were corrected by deducting the residual signals derived from one-mismatch organisms. The residual signals from one mismatch organism were calculated by multiplying the signal with probe DELTA495b by 0.172, which was the ratio of the remaining duplex of probe and target containing one mismatch by washing at 58°C (Table 1).
Vertical profiles of domain compositions.
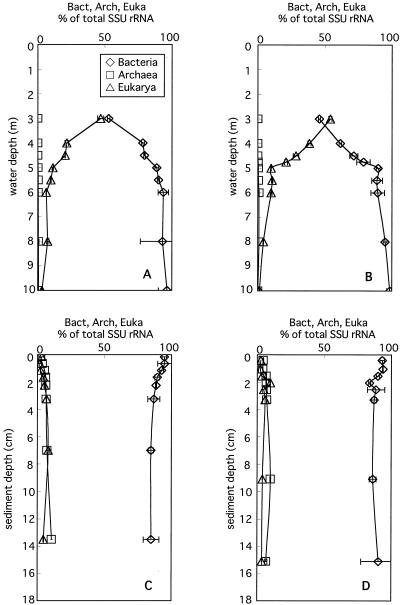
The vertical profiles of the relative abundance of Bacteria, Archaea, and Eukarya determined with three general domain-specific probes are shown in Fig. 4. The vertical profiles obtained on the two different sampling dates showed similar tendencies. The abundances of bacterial and eukaryal SSU rRNA relative to total SSU rRNA (combined signals of Bact338, Arch915, and Euka502) were almost equal at ca. 50% in the 3-m layer of the water column but subsequently showed opposite gradients along with depth. In anoxic layers, the relative contributions of Bacteria gradually increased from 89 to 90% to 96 to 98% (those contributions are expressed by the ratio of specific SSU rRNAs to total SSU rRNA unless otherwise indicated), and that of Eukarya decreased from 9.8 to 11% to 1.4 to 3.1%. Although archaeal rRNA was almost negligible, ranging from 0 to only 0.09% throughout the water column, it was detected in the sediment and gradually increased from 2 to 4% to 10 to 11% with depth. Bacterial rRNA decreased from 93 to 95% to 85 to 90% with depth in the sediment. Eukaryal rRNA peaked at 6.7 to 9.6% in the 2- to 3-cm layer of the sediment.
FIG. 4.
Vertical profiles of relative abundance of Bacteria, Archaea, and Eukarya to total SSU rRNA in the water column (A and B) and sediment (C and D) in March 2003 (A and C) and June 2003 (B and D). The total SSU-rRNA amount was calculated by totaling the signals of three domain-specific probes. Horizontal bars indicate maximum and minimum ratios of specific probes to total SSU rRNA.
Vertical distributions of specific bacterial groups.
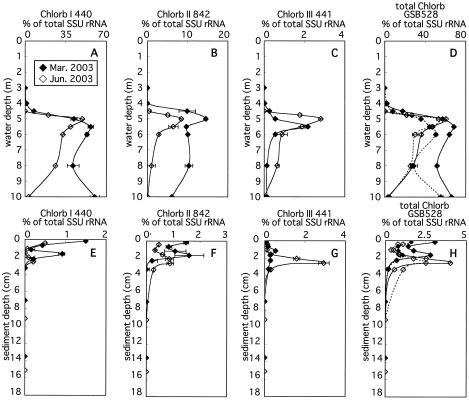
(i) Chlorobiaceae. SSU rRNAs from organisms affiliated with Chlorb I, II, and III groups were negligible in oxic water (3 m) but then noticeably increased with depth and peaked at the chemocline (5- to 5.5-m water depth) in anoxic water (Fig. 5). SSU rRNA of Chlorb I was predominant in the chemocline, accounting for 49 to 57% (55 to 63% of bacterial SSU rRNA) (Fig. 5A). The vertical profiles of the fraction of Chlorb I differed below the chemocline depending on the sampling date. In March 2003, this fraction slightly decreased from 57% at 5.5 m to 41% at 8 m but increased again to 60% at 10 m. In June 2003, this fraction steadily decreased from 49% at 5 m to 3.2% at 10 m. In the sediment, this fraction was highest in the uppermost layers, accounting for 0.5 to 1.5%, but decreased down to 1 to 1.5 cm in depth (Fig. 5B). Secondary peaks of this fraction were observed at 1.5 to 2.5 cm, accounting for 0.3 to 0.9%. Below these secondary peaks, the fraction of Chlorb I decreased to below the detection limit.
FIG. 5.
Vertical profiles of abundance of SSU rRNA hybridizing with Chlorb I (A and E), II (B and F), and III (C and G) group-specific probes, with the cumulative signals obtained with three specific probes (D and H, solid line) and SSU rRNA of Chlorobiaceae hybridizing with general probe GSB528 (D and H; broken line), relative to total SSU rRNA in the water column (A to D) and sediment (E to H). Horizontal bars indicate maximum and minimum ratios of specific probes to total SSU rRNA.
The fraction of Chlorb II was smaller than that of Chlorb I in the water column, accounting for 8.4 to 14% at the chemocline (Fig. 5B), but was comparable to it, accounting for 0.5 to 1.5% in the uppermost sediment layers, and actually exceeded that of Chlorb I, accounting for 0.9 to 1.6% at the secondary peaks in the sediment (Fig. 5C). The fraction of Chlorb III was smaller than that of each of the other two groups in the water column, accounting for 2.2 to 2.8% at the chemocline (Fig. 5C). This fraction peaked at 1.5 to 2.5 cm in the sediment. In June 2003, the fraction of this peak was significantly larger than those of Chlorb I and II, accounting for 2.9% (Fig. 5G). Any SSU rRNA derived from Chlorobiaceae was below the detection limit at sediment depths greater than 6 cm (Fig. 5H).
GSB528 was used as a nested probe (37) to enhance the reliability of the detection of target organisms within the family Chlorobiaceae by newly designed probes. The vertical profiles of the relative signal intensity of probe GSB528 were highly comparable with that of the sum of signals from probes specific for Chlorb I, II, and III (Fig. 5D and H). However, the sum of SSU rRNAs hybridized with individual probes tended to be higher than SSU rRNAs hybridized with probe GSB528 specific for the family Chlorobiaceae in March 2003 (Fig. 5D and H). This was probably due to the use of in vitro-transcribed RNAs as references when probes specific for Chlorb I, II, and III were hybridized (26).
(ii) δ-Proteobacteria.
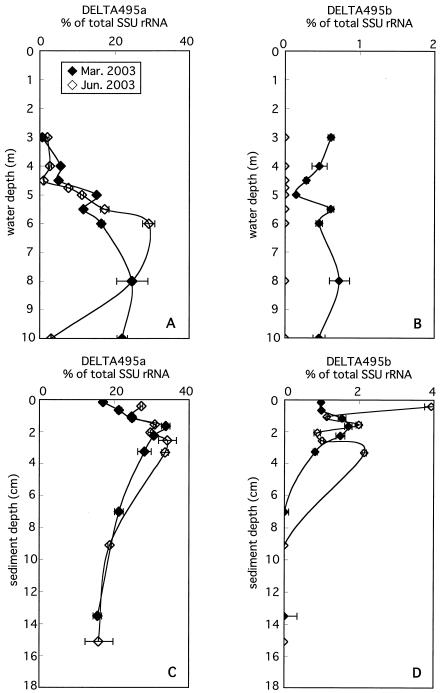
Probes DELTA495a and -b were used to detect almost all δ-Proteobacteria, which included dominant SRB in the anoxic settings of Lake Kaiike. SSU rRNAs hybridized with probe DELTA495a accounted for 0.6 to 7.6% in the oxic and microaerophilic layers of the water column and rapidly increased with depth just below the chemocline (Fig. 6A). The vertical profiles of the relative contribution of SSU rRNAs hybridized with probe DELTA495a were different in anoxic water depending on the sampling date. In March 2003, the fraction of DELTA495a gradually increased from 5.0% at 4.5 m to 25% at 8 m and slightly declined to 22% at 10 m. In June 2003, this fraction markedly increased from 7.6% at 4.75 m to 29% at 6 m but then decreased to 3.0% at 10 m. In the sediment, this fraction peaked at 2 to 3 cm, accounting for 34% irrespective of the sampling date. Below this peak, the fraction gradually decreased to 16% at 14 to 15 cm.
FIG. 6.
Vertical profiles of the abundance of SSU rRNA hybridizing with probes DELTA495a (A and C) and DELTA495b (B and D) relative to total SSU rRNA in the water column (A and B) and sediment (C and D). Horizontal bars indicate maximum and minimum ratios of specific probes to total SSU rRNA.
SSU rRNAs hybridized with probe DELTA495b were detected at a level of less than 1% throughout the water column (3 to 10 m) only in March 2003 (Fig. 6B). In the sediment, the fraction of DELTA495b peaked at 2 to 3 cm, accounting for 1.7 to 2.1%, and then declined to near or below the detection limit. In June 2003, it reached its maximum of 3.9% in the uppermost layer (Fig. 6D).
(iii) The phylum Chloroflexi.
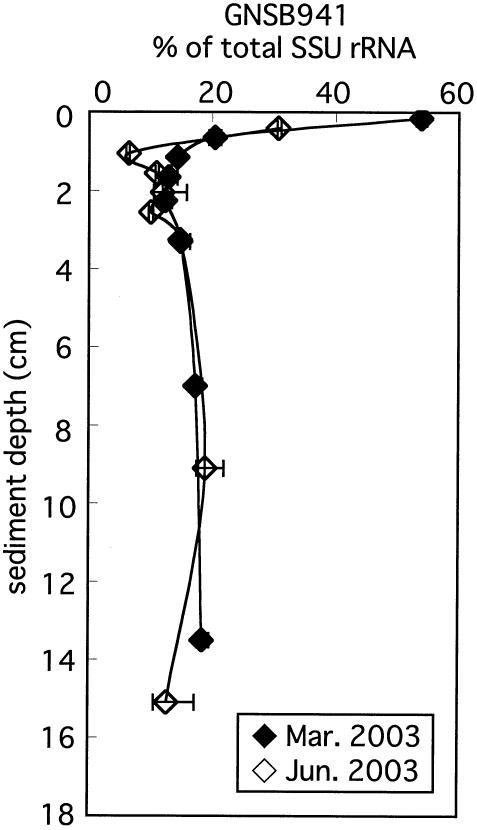
Since DGGE fragments (338 to 907 of the E. coli numbers) do not include the target region of probe GNSB941, we could not verify whether this probe perfectly matched with the environmental clone sequences. Nevertheless, we used this probe to detect organisms affiliated with the phylum Chloroflexi in Lake Kaiike, assuming that their sequences perfectly matched probe GNSB941 on the basis of comparative SSU-rRNA sequence analysis. Any SSU rRNA that hybridized with probe GNSB941 was under the detection limit in the water column, whereas the SSU rRNAs were predominant in the top of the sediment, accounting for 31 to 54% (Fig. 7). Their fraction notably declined to 6 to 14% within 1 cm but then gradually increased up to 19% with depth.
FIG. 7.
Vertical profiles of abundance of SSU rRNA hybridized with probe GNSB941 relative to total SSU rRNA in the sediment. Horizontal bars indicate maximum and minimum ratios of specific probes to total SSU rRNA.
DISCUSSION
Microbial community structure revealed by quantitative membrane hybridization.
The sum of the SSU rRNAs hybridized with the probes used in this study were 1.2 to 7.2% of total SSU rRNA in oxic and microaerophilic waters (3 to 4 m), 53 to 89% in anoxic water, except at 10 m in June 2003 (3.2%), and 30 to 75% in the sediments. The detection rates were higher in the anoxic than in the oxic settings, since all specific probes used in this study targeted strictly anaerobic bacteria. SSU rRNAs that hybridized with probe DELTA495a were, however, detected in oxic water. This result was supported by the fact that members of the genera Desulfovibrio and Desulfobulbus are capable of oxygen respiration (11, 12, 24) and that SRB are present in the anaerobic microniches within cell and sediment aggregates (15, 36, 45, 46).
We conclude that the rates of detection in the anoxic water column were up to 89% of total SSU rRNA (91% of total prokaryotic SSU rRNA, which was the combined signals of Bact338 and Arch915) in this study, indicating that microbial communities in the anoxic water were very simple. They consisted merely of some Chlorobiaceae and the δ-proteobacterial SRB. Although species heterogeneity within a probe target group might further complicate the picture (38), the simplicity of the microbial communities was corroborated by the simple DGGE banding patterns in anoxic water (Fig. 1) (19). Thus, sulfur and carbon cycles in the anoxic water of Lake Kaiike seem to be carried out exclusively by these two functional groups. As for the other microbial populations in anoxic water, members of the genus Arcobacter, which is affiliated with the ɛ-Proteobacteria, and the class Spirochaetes have been recognized by SSU-rDNA-based DGGE analysis (19). We, however, did not quantify the fraction of their SSU rRNA by membrane hybridization, since probes encompassing each group could not be designed.
The sum of SSU rRNAs hybridizing with the probe specific for Chlorb I, II, and III was abundant throughout anoxic water (5 to 10 m), accounting for up to 68%, whereas in the top of the sediment these groups accounted for only up to 4.0%. However, the absolute amount of this SSU rRNA in the top of the sediment might correspond to that in anoxic water, since the numbers of DAPI-stained cells in the surface sediment were about 2 orders of magnitude higher than those in anoxic water (data not shown). Furthermore, the total amounts of extracted SSU rRNA were very high, at 32,000 to 35,000 ng g of sediment−1 in the uppermost layers of the sediment, and markedly declined to 2,600 to 3,200 ng g of sediment−1 in about the 2-cm layer (data not shown). One of the possible explanations for the presence of Chlorobiaceae in the sediment surface is that they survived by using an alternative metabolism. Sirevåg and Ormerod (47) demonstrated that “Chlorobium thiosulfatophilum” could survive in the dark by obtaining energy from the degradation of intracellular polyglucose as a storage material. This kind of investigation, however, has not been performed for other strains of Chlorobiaceae. Thus, we do not know whether fermentation of intracellular polyglucose in the dark is a common metabolism among members of the Chlorobiaceae. Another possible explanation is that these Chlorobiaceae are similar to those observed by Overmann et al. in the Black Sea (33), which were adapted to a very low light intensity at a depth of 68 m (0.003 μmol m−2 s−1). In Lake Kaiike, light intensities in the bottom water were negligible or very low, at a level of 0.01 μmol m−2 s−1, in November 2002. However, light may reach the bottom of Lake Kaiike during other periods of the year. Thus, we do not rule out the possibility that they may survive by photosynthesizing at the sediment surface. Nakajima et al. (30) speculated that Chlorobiaceae might survive at the sediment surface of Lake Kaiike by adapting to low light, which became evident from chloropigment analyses. Bacteriochlorophyll e, which is a specific chloropigment of Chlorobiaceae, was hardly degraded even at the sediment surface. Furthermore, enrichment in highly alkylated bacteriochlorophyll e homologues at the sediment surface indicated a physiological response of Chlorobiaceae to growth at extremely low-light intensities. The fractions of SSU rRNA hybridized with probes Chlorb II 842 and Chlorb III 441 were larger than that of SSU rRNA in the sediment hybridized with probe Chlorb I 440 (Fig. 5E to G). Furthermore, the secondary peaks of the fraction of those SSU rRNAs were recognized at 1.5 to 2.5 cm. In particular, it is unlikely that organisms affiliated with Chlorb III survive photosynthetically at a depth of approximately 2 cm in the sediment, although its clone sequences are obviously clustered into Chlorobiaceae. We could not determine whether this abundant SSU rRNA of Chlorb III was caused by SSU rRNA remaining after the blooming and sedimentation of Chlorb III in the past or by intrinsically active growth. Nevertheless, the existence of Chlorb III at a 2-cm depth was cross-checked by both DNA- and RNA-based approaches (19). The isolation of the organisms corresponding to these clones and their physiological characterization are needed to determine which environmental factors affect the Chlorobiaceae community structure and to check the dark metabolisms of the members of Chlorb III.
SSU rRNA hybridized with probe GNSB941 predominated in the surface sediment. On the basis of a PCR-DGGE analysis of SSU rDNA, all clone sequences retrieved from Lake Kaiike were affiliated with subphylum I within the phylum Chloroflexi defined by Hugenholtz et al. (14). Subphylum I contains diverse environmental clones obtained mainly from anaerobic environments such as hot springs (7, 14), aerobic and anaerobic wastewater treatment sludges (6, 16, 43), and sediments (20, 56). Although they have been found to be widespread in various environments at relatively high levels, their roles in such environments remain ambiguous because of a failure to isolate these organisms and carry out their physiological characterizations. Recently, two strains (UNI-1T and STL-6-O1T) within subphylum I were isolated, respectively, from a thermophilic upflow anaerobic sludge blanket reactor and a hot spring in Japan (44). Neither of them was photosynthetic, but they were able to grow under anaerobic and fermentative conditions. In particular, strain UNI-1T was a strictly anaerobic organism utilizing only some sugars and no organic acids or H2/CO2. In addition, it did not utilize nitrate, sulfate, sulfite, or thiosulfate as electron acceptors. If these physiological characteristics are applicable to the clones retrieved from the top of the sediment of Lake Kaiike, they are unlikely to contribute to sulfur metabolism but instead probably grow heterotrophically by degrading macromolecules such as sugars and starch originating from organic debris. This speculation is supported by a previous report that no clone sequences affiliated with subphylum I could be obtained from sediment slurries enriched with various organic acids or H2/CO2 (Koizumi et al., submitted for publication).
Vertical distributions of the δ-Proteobacteria and sulfur metabolisms in Lake Kaiike.
Since SRB phylotypes detected in Lake Kaiike were sporadic in a complete oxidizing SRB cluster (19), general probes DELTA495a and -b were used to encompass most SRB within the δ-Proteobacteria cluster. Probe DELTA495a could not detect all δ-Proteobacteria based on comparative SSU rRNA sequence analysis. Thus, probe DELTA495b, which hybridizes to SSU rRNA of the genera Desulfobacter and Desulfotignum, was used together with probe DELTA495a to enhance the coverage. In Lake Kaiike, only the sequence 15s is clustered in the genus Desulfotignum (19).
In the top of the sediment, the robust coexistence of SRB and Chloroflexi was observed. However, the SSU-rRNA peaks of SRB and Chloroflexi appeared to be vertically separated. Substrate utilization seems to be different between SRB and microbes belonging to subphylum I of Chloroflexi. In general, SRB utilize fatty acids as electron donors and sulfate as an electron acceptor (55), whereas the recently isolated strain UNI-1T belonging to subphylum I of Chloroflexi grows by fermentation of sugars such as sucrose, yielding acetate and hydrogen as the main end products (44). Thus, we suppose a novel syntrophic relationship between SRB and Chloroflexi, i.e., the members clustered into subphylum I of Chloroflexi might be initial degraders of macromolecules, providing fatty acids and hydrogen as electron donors for SRB. Acetate, formate, and lactate were detected at a level of ca. 10 μmol liter−1 as major fatty acids in the interstitial water of the uppermost sediment layers and had become seriously depleted within ca. 5 cm (19). Furthermore, the addition of those fatty acids accelerated sulfide production in the sediment slurries, indicating that substrate concentrations, which might be determined by the activities of fermentative organisms such as Chloroflexi, limit sulfate reduction. Thus, we expect to find a syntrophic relationship between SRB and Chloroflexi in organic-rich, permanently anoxic environments such as meromictic lake sediments. If that is actually the case, a relatively ancient syntrophic relationship between them may have been discovered in Lake Kaiike. To validate this hypothesis, a study using enrichment by various sugars followed by a determination of microbial species components and tracer experiments using radioisotope-labeled sugars would be needed to investigate their syntrophic relationship.
The population within the ɛ-Proteobacteria was also considered an important player in the sulfur cycle, since all ɛ-Proteobacteria isolated have been considered to be involved in the sulfur cycle by either reducing elemental sulfur to sulfide, oxidizing sulfide to sulfur, or both, depending on environmental conditions (8). In Lake Kaiike, one ɛ-proteobacterial clone was consistently detected in the anoxic water by PCR-DGGE analysis, and the signal intensity of its DGGE band was especially prominent at 10 m in June 2003, where the cumulative detection rate was very low, at 6.3% of total SSU rRNA. Thus, SSU rRNA derived from this organism might compensate for the gap between total SSU rRNA and summed SSU rRNA hybridizing with specific probes. Although it was difficult to detect the ɛ-Proteobacteria strain in the sediment, it was frequently obtained from enrichment cultures. Thus, members of the ɛ-Proteobacteria might be important in the sulfur cycle in Lake Kaiike and deserve further attention.
Comparison of DGGE and oligonucleotide probe hybridization analysis—methodological considerations.
The cumulative detection ratios were lower in the sediment than in the water column, suggesting the existence of a higher diversity among the microbial community in the sediment. On the basis of previous SSU-rDNA-based PCR-DGGE analysis, the probe set used in this study should have covered almost all populations in the sediment. This result would indicate that PCR-DGGE analysis tends to overlook minor populations in highly diverse environments (18). Furthermore, the decrease in detection rates with depth contradicted the simplicity of DGGE banding patterns. One reason for this observation was the increase in archaeal and eukaryal SSU rRNA with depth in the sediment (Fig. 4C and D). DGGE analysis of archaeon-specific PCR-amplified SSU rDNA revealed that the archaeal community in the sediment did not change with increasing depth and that one conspicuous band whose sequence was most similar (95%) to environmental clone J4.75-12 obtained from Solar Lake (10) was observed throughout the sediment (data not shown). This sequence was affiliated with an unidentified euryarchaeal cluster unrelated to methanogens. The increase in archaeal SSU rRNA, however, does not compensate for the gap between the sum of SSU rRNAs hybridized with the probe set used in this study and bacterial SSU rRNA. We compared the DNA-based DGGE profiles with the RNA-based DGGE profiles of bacterial communities in the sediment of Lake Kaiike in a previous study (19). Some bands were observed to be more prominent on the RNA-based DGGE gel than on the DNA-based DGGE gel. One of their sequences was most similar to that of a clone affiliated with the Holophaga group that was obtained from a hydrothermal vent. In addition, there were bands that were observed only on the RNA-based DGGE gel, though their sequences were not determined. Microbial community analysis using a clone library might also be effective in revealing the existence of a minor population diversity.
Nevertheless, PCR-DGGE analysis prior to oligonucleotide probe hybridization was effective in determining which probe set should be used and in designing probes specific for remarkable populations. Using this strategy, up to 89% (92%) and 75% (79%) of total SSU rRNA (bacterial SSU rRNA) in the anoxic water and sediment, respectively, could be detected. These detection rates are much higher than the 0.001 to 15% detection rates yielded by culture-dependent methods (5), though the results of membrane hybridization could not be translated into cell numbers. One advantage of combined DGGE-membrane hybridization is that species heterogeneity within a probe target can be recognized, to some extent, by DGGE banding patterns. For example, GSB528, DELTA495, and GNSB941 are probes specific for a relatively wide range of phylogenetic groups. As mentioned above, SSU rRNAs hybridized with probe GSB528 were assigned to three groups in Lake Kaiike. One of the DGGE bands corresponding to the population detected with Chlorb I 440 was most conspicuous, and its signal intensity did not drastically change throughout the anoxic water on either sampling date (19). However, the fraction of SSU rRNAs hybridized with probe Chlorb I 440 gradually decreased from 49% at 5 m to 3.2% at 10 m in June 2003. This discrepancy between PCR-DGGE and membrane hybridization results may be mainly attributable to the difference between DNA- and RNA-based microbial community analysis described elsewhere (18, 19). Thus, we conclude that the organisms in Chlorb I were distributed evenly throughout the anoxic water but that their activities decreased with depth concomitant with the decrease in intercellular SSU rRNA in June 2003.
The diversity of SRB populations was higher in sediment than in anoxic water. SSU-rDNA-based PCR-DGGE analysis found two phylotypes of SRB in anoxic water that were distributed separately, whereas five phylotypes of SRB were found in the sediment, and their vertical distributions varied (19). Changes in the signal intensities of the DGGE bands whose sequences were affiliated with the SRB cluster were relatively comparable to the vertical profile of the fraction of SSU rRNA that hybridized with probe DELTA495a, in contrast to the case with probe Chlorb I 440. In order to investigate more complicated SRB community structures, both PCR-DGGE analysis using an SRB-specific primer set and membrane hybridization using more specific probes are effective in focusing on the diversity and community structure of SRB (38, 41, 42).
One drawback to the DGGE-membrane hybridization approach is that some organisms which exist only in small populations but possess important ecological functions might be overlooked. This drawback is due to the limited capability of detecting minor microbial fractions and the low coverage of microbial diversity by PCR-DGGE analysis when it is applied to complex samples (19). For example, it was difficult to detect purple sulfur bacteria, Chromatium sp., within the γ-Proteobacteria by PCR-DGGE even though they seemed to be dominant in the chemocline due to their large cell volume. However, their cell numbers were at most 1% of total cell counts (19). Nevertheless, they play an important role in the sulfur cycle of the chemocline, since they possess capabilities for both anaerobic photosynthetic sulfide oxidation and lithotrophic sulfide oxidation using oxygen (34). In addition, they can move to a zone in the chemocline that is more favorable for the acquisition of light or oxygen (50). In order to overcome this limitation, PCR-DGGE analysis using a primer set specific for narrower groups (e.g., the Chromatiaceae) might be effective in investigating minor but ecologically important populations (9).
Acknowledgments
We are indebted to all of the members of IFREE 4 for their sampling assistance during research. We are grateful to Y. Nakajima, N. Ohkouchi, K. Oguri, and H. Kitazato for intensive discussion. We thank S. Takii for his critical reading of the manuscript and helpful comments. Chloroflexus aurantiacus strain J10 (DSM 635) was kindly provided by Cell Energetics Laboratory, Tokyo Metropolitan University. We also thank S. Higashi, Planning Division of Kamikoshiki Village Office, for arrangement of a boat and a space for experiments during research.
This work was supported by a grant from the Ministry of Education, Culture, Sport, Science, and Technology, Japan, to M.F. (no. 16370014).
REFERENCES
- 1.Alexander, B., J. H. Andersen, R. P. Cox, and J. F. Imhoff. 2002. Phylogeny of green sulfur bacteria on the basis of gene sequences of 16S rRNA and of the Fenna-Matthews-Olson protein. Arch. Microbiol. 178:131-140. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R. I., J. Stromley, R. Devereux, R. Key, and D. A. Stahl. 1992. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl. Environ. Microbiol. 58:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Björnsson, L., P. Hugenholtz, G. W. Tyson, and L. L. Blackall. 2002. Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology 148:2309-2318. [DOI] [PubMed] [Google Scholar]
- 7.Boomer, S. M., D. P. Lodge, B. E. Dutton, and B. Pierson. 2002. Molecular characterization of novel red green nonsulfur bacteria from five distinct hot spring communities in Yellowstone National Park. Appl. Environ. Microbiol. 68:346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, B. J., C. Jeanthon, J. E. Kostka, G. W. Luther III, and S. C. Cary. 2001. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coolen, M. J., and J. Overmann. 1998. Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment. Appl. Environ. Microbiol. 64:4513-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cytryn, E., D. Minz, R. S. Oremland, and Y. Cohen. 2000. Distribution and diversity of archaea corresponding to the limnological cycle of a hypersaline stratified lake (Solar Lake, Sinai, Egypt). Appl. Environ. Microbiol. 66:3269-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dannenberg, S., M. Kroder, W. Dilling, and H. Cypionka. 1992. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Arch. Microbiol. 158:93-99. [Google Scholar]
- 12.Dilling, W., and H. Cypionka. 1990. Aerobic respiration in sulfate-reducing bacteria. FEMS Microbiol. Lett. 71:123-128. [Google Scholar]
- 13.Gich, F., J. Garcia-Gil, and J. Overmann. 2001. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 177:1-10. [DOI] [PubMed] [Google Scholar]
- 14.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jørgensen, B. B. 1977. Bacterial sulfate reduction within reduced microniches of oxidized marine sediment. Mar. Biol. 41:7-17. [Google Scholar]
- 16.Juretschko, J., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 17.Koizumi, Y., J. J. Kelly, T. Nakagawa, H. Urakawa, S. El-Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa, and D. A. Stahl. 2002. Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl. Environ. Microbiol. 68:3215-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koizumi, Y., S. Takii, M. Nishino, and T. Nakajima. 2003. Vertical distributions of sulfate-reducing bacteria and methane-producing archaea quantified by oligonucleotide probe hybridization in the profundal sediment of a mesotrophic lake. FEMS Microbiol. Ecol. 44:101-108. [DOI] [PubMed] [Google Scholar]
- 19.Koizumi, Y., H. Kojima, K. Oguri, H. Kitazato, and M. Fukui. 2004. Vertical and temporal shifts of microbial communities in the water column and sediment of saline meromictic Lake Kaiike (Japan), as determinated by a 16S rDNA-based analysis, and related to physicochemical gradients. Environ. Microbiol. 6:622-637. [DOI] [PubMed] [Google Scholar]
- 20.Li, L., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 21.Llobet-Brossa, E., R. Rossello-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maidak, B. L., N. Larsen, M. J. McCaughey, R. Overbeek, G. J. Olsen, K. Fogel, J. Blandy, and C. R. Woese. 1994. The ribosomal database project. Nucleic Acids Res. 22:3485-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marschall, C., P. Frenzel, and H. Cypionka. 1993. Influence of oxygen on sulfate reduction and growth of sulfate-reducing bacteria. Arch. Microbiol. 159:168-173. [Google Scholar]
- 25.Matsuyama, M., and E. Shirouzu. 1978. Importance of photosynthetic sulfur bacteria, Chromatium sp. as an organic matter producer in Lake Kaiike. Jpn. J. Limnol. 39:103-111. [Google Scholar]
- 26.McMahon, K. D., D. A. Stahl, and L. Raskin. 1998. A comparison of the use of in vitro-transcribed and native rRNA for the quantification of microorganisms in the environment. Microb. Ecol. 36:362-371. [DOI] [PubMed] [Google Scholar]
- 27.Miller, D. N., J. E. Bryant, E. L. Madsen, and W. C. Ghiorse. 1999. Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl. Environ. Microbiol. 65:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minz, D., S. Fishbain, S. J. Green, G. Muyzer, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eucaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran, M. A., V. L. Torsvik, T. Torsvik, and R. E. Hodson. 1993. Direct extraction and purification of rRNA for ecological studies. Appl. Environ. Microbiol. 59:915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima, Y., H. Okada, K. Oguri, S. Suga, H. Kitazato, Y. Koizumi, M. Fukui, and N. Ohkouchi. 2003. Distribution of chloropigments in suspended particulate matter and benthic microbial mat of a meromictic lake, Lake Kaiike, Japan. Environ. Microbiol. 5:1103-1110. [DOI] [PubMed] [Google Scholar]
- 31.Oguri, K., M. Itou, S. Hirano, T. Hisamitsu, S. Sakai, M. Murayama, H. Kitazato, Y. Koizumi, M. Fukui, and A. Taira. 2002. Environmental characteristics of water and sediments, and microbial activities at Lake Kaiike, Kamikoshiki Island, Kagoshima Prefecture. J. Geol. Soc. Jpn. 108:12. (In Japanese). [Google Scholar]
- 32.Overmann, J., J. T. Beatty, K. J. Hall, N. Pfennig, and T. G. Northcote. 1991. Characterization of a dense, purple sulfur bacterial layer in a meromictic salt lake. Limnol. Oceanogr. 36:846-859. [Google Scholar]
- 33.Overmann, J., H. Cypionka, and N. Pfennig. 1992. An extremely low-light adapted phototrophic sulfur bacterium from the Black Sea. Limnol. Oceanogr. 37:150-155. [Google Scholar]
- 34.Pfennig, N., and H. G. Trüper. 1992. The family Chromatiaceae, p. 3200-3221. In A. Balows, M. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., Springer-Verlag, New York, N.Y.
- 35.Purdy, K. J., T. M. Embley, S. Takii, and D. B. Nedwell. 1996. Rapid extraction of DNA and rRNA from sediments by a novel hydroxyapatite spin-column method. Appl. Environ. Microbiol. 62:3905-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsing, N. B., M. Kühl, and B. B. Jørgensen. 1993. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl. Environ. Microbiol. 59:3840-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravenschlag, K., K. Sahm, C. Knoblauch, B. B. Jørgensen, and R. Amann. 2000. Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine arctic sediments. Appl. Environ. Microbiol. 66:3592-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine Arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahm, K., B. J. MacGregor, B. B. Jørgensen, and D. A. Stahl. 1999. Sulphate reduction and vertical distribution of sulphate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ. Microbiol. 1:65-74. [DOI] [PubMed] [Google Scholar]
- 42.Sass, H., E. Wieringa, H. Cypionka, H. D. Babenzien, and J. Overmann. 1998. High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch. Microbiol. 170:243-251. [DOI] [PubMed] [Google Scholar]
- 43.Sekiguchi, Y., Y. Kamagata, K. Syutsubo, A. Ohashi, H. Harada, and K. Nakamura. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655-2665. [DOI] [PubMed] [Google Scholar]
- 44.Sekiguchi, Y., T. Yamada, S. Hanada, A. Ohashi, H. Harada, and Y. Kamagata. 2003. Anaerolinea thermophila gen. nov., sp. nov., and Caldilinea aerophila gen. nov., sp. nov., two novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol.. 53:1843-1851. [DOI] [PubMed] [Google Scholar]
- 45.Sigalevich, P., E. Meshorer, Y. Helman, and Y. Cohen. 2000. Transition from anaerobic to aerobic growth conditions for the sulfate-reducing bacterium Desulfovibrio oxyclinae results in flocculation. Appl. Environ. Microbiol. 66:5005-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigalevich, P., M. V. Baev, A. Teske, and Y. Cohen. 2000. Sulfate reduction and possible aerobic metabolism of the sulfate-reducing bacterium Desulfovibrio oxyclinae in a chemostat coculture with Marinobacter sp. strain MB under exposure to increasing oxygen concentrations. Appl. Environ. Microbiol. 66:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirevåg, R., and J. G. Ormerod. 1977. Synthesis, storage and degradation of polysulfide in Chlorobium thiosulfatophilum. Arch. Microbiol. 111:239-244. [DOI] [PubMed] [Google Scholar]
- 48.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thar, R., and M. Kühl. 2001. Motility of Marichromatium gracile in response to light, oxygen, and sulfide. Appl. Environ. Microbiol. 67:5410-5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trüper, H., G., and S. Genovese. 1968. Characterization of photosynthetic sulfur bacteria causing red water in Lake Faro (Messina, Sicily). Limnol. Oceanogr. 13:225-232. [Google Scholar]
- 52.Tsai, Y., and B. H. Olson. 1991. Rapid method for direct extraction of DNA from soil and sediments. Appl. Environ. Microbiol. 57:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuschak, C., J. Glaeser, and J. Overmann. 1999. Specific detection of green sulfur bacteria by in situ hybridization with a fluorescently labeled oligonucleotide probe. Arch. Microbiol. 171:265-272. [DOI] [PubMed] [Google Scholar]
- 54.Veldhuis, M. J. W., and H. van Germerden. 1986. Competition between purple and brown phototrophic bacteria in stratified lakes; sulfide, acetate, and light as limiting factors. FEMS Microbiol. Ecol. 38:31-38. [Google Scholar]
- 55.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, M. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., Springer-Verlag, New York, N.Y.
- 56.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 1997. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl. Environ. Microbiol. 63:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng, D., E. W. Alm, D. A. Stahl, and L. Raskin. 1996. Characterization of universal small-subunit rRNA hybridization probe for quantitative molecular microbial ecology studies. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]