Abstract
In vitro fermentations were carried out by using a model of the human colon to simulate microbial activities of lower gut bacteria. Bacterial populations (and their metabolic products) were evaluated under the effects of various fermentable substrates. Carbohydrates tested were polydextrose, lactitol, and fructo-oligosaccharide (FOS). Bacterial groups of interest were evaluated by fluorescence in situ hybridization as well as by species-specific PCR to determine bifidobacterial species and percent-G+C profiling of the bacterial communities present. Short-chain fatty acids (SCFA) produced during the fermentations were also evaluated. Polydextrose had a stimulatory effect upon colonic bifidobacteria at concentrations of 1 and 2% (using a single and pooled human fecal inoculum, respectively). The bifidogenic effect was sustained throughout all three vessels of the in vitro system (P = 0.01 seen in vessel 3), as corroborated by the bacterial community profile revealed by %G+C analysis. This substrate supported a wide variety of bifidobacteria and was the only substrate where Bifidobacterium infantis was detected. The fermentation of lactitol had a deleterious effect on both bifidobacterial and bacteroides populations (P = 0.01) and decreased total cell numbers. SCFA production was stimulated, however, particularly butyrate (beneficial for host colonocytes). FOS also had a stimulatory effect upon bifidobacterial and lactobacilli populations that used a single inoculum (P = 0.01 for all vessels) as well as a bifidogenic effect in vessels 2 and 3 (P = 0.01) when a pooled inoculum was used. A decrease in bifidobacteria throughout the model was reflected in the percent-G+C profiles.
The contribution of the human colonic microbiota towards improved host health and disease is presently a topical area of research. The beneficial effects of probiotic lactic acid bacteria (such as bifidobacteria and lactobacilli) have been documented and have a long history of use in humans (8, 9, 28). Indeed, many clinical trails have been carried out (in both human and animal models) where direct health benefits from probiotic intake has been investigated (16, 23, 33). More recently, interest has turned to the potentially advantageous effects of dietary substrates that can be fermented by populations of indigenous probiotic gut bacteria. These so-called prebiotics selectively stimulate beneficial flora components while suppressing, or having no stimulatory effect upon, less desirable bacteria, such as proteolytic bacteroides and clostridia and gram-negative pathogens such as Escherichia coli, shigellas, and salmonellas (11). Through not relying on the addition of exogenous bacteria, there are no survivability or safety issues. A number of prebiotic substrates already have scientific credibility and widespread acceptance, such as inulin, oligofructose (both fructo-oligosaccharides [FOS]), galacto-oligosaccharides, and lactulose (29, 30, 35). These substrates have been tested in various human trials for their prebiotic effects.
Here we investigate the potential prebiotic effect of polydextrose and lactitol monohydrate, commonly used components in the food industry. In one study, polydextrose has been shown to express prebiotic effects; it decreased fecal pH, increased the residual concentration of short chain fatty acids (SCFA), and increased numbers of bifidobacteria in feces (19). In the present study we further evaluated the prebiotic potential of polydextrose and lactitol (selective fermentation by gut bacteria) by using an in vitro model of the human colon (25). Colonic bacteria supported by these substrates were evaluated by molecular methodologies (fluorescence in situ hybridization [FISH], percent-G+C analysis, and PCR), and the products of their metabolism were determined.
Previous studies have shown that the nature of the glycosidic bonds in polydextrose renders it only partially fermented by intestinal microorganisms and resistant to enzymatic attack (1, 6). It is a water-soluble, randomly bonded, condensation polymer of glucose containing small amounts of sorbitol and citric acid (24). As with accepted prebiotics, polydextrose is not sweet and can be used as a low-calorie bulking agent in a wide range of foods, such as baked goods, confectionery, dairy products, and functional beverages. Similar to polydextrose, lactitol monohydrate (a disaccharide alcohol) is also not absorbed in the human small intestine and thus arrives at the colon as a potential substrate for microbial fermentation (14, 20, 21). The intake of large amounts of nondigestible substrates can have adverse consequences for the host. For example, diarrhea can result from the excessive consumption of such substrates, particularly seen with the intake of high levels of sugar alcohols. However, it must be borne in mind that these effects are dose dependent, and the premise of prebiotic therapy is that quantities are administered so that selectivity and gastrointestinal function are not compromised. Lactitol monohydrate (β-galactosidosorbitol) is highly soluble in water, with a mildly sweet taste (32).
MATERIALS AND METHODS
All chemicals were supplied by Sigma (Poole, Dorset, United Kingdom) unless otherwise stated.
Continuous culture fermentations.
The continuous culture fermentations were carried out with an anaerobic continuous culture system that had been previously validated against colonic contents of sudden death victims (25). The system comprised three glass vessels aligned in series. The first vessel in the system had an operating volume of 280 ml with growth medium being introduced into it. The second vessel had an operating volume of 300 ml and was sequentially fed from the overflow of the first vessel. The third vessel had a volume of 300 ml and was fed sequentially from the overflow of the second vessel. Culture fluid from the final vessel vented into a waste receptacle. Each vessel was continuously stirred and maintained at 37°C by using a circulating water jacket. The pH of the vessels was maintained at 5.5, 6.2, and 7.0 for vessels 1, 2, and 3, respectively, through the addition of 1 N HCl-NaOH as appropriate. The entire system (medium reservoir included) was operated under anaerobic conditions from the continuous bubbling of sterile oxygen-free nitrogen through the liquid (approximately 15 ml/min) into all the vessels. Vessel 1 simulated microbial conditions found in the proximal colon, vessel 2 modelled the transverse colon, and vessel 3 mimicked the distal region of the colon.
Each vessel was half filled with anaerobic fermentation medium (from the medium reservoir) containing (in grams per liter) starch, 5; peptone water (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom), 5; tryptone (Oxoid), 5; yeast extract (Oxoid), 4.5; NaCl (BDH Laboratory Supplies, Poole, Dorset, United Kingdom), 4.5; KCl (BDH), 4.5; pectin, 2; casein, 3; xylan, 2; Larch arabinogalactan, 2; NaHCO3, 1.5; MgSO4, 1.25; guar gum, 1; inulin (Novartis, Neuenegg, Switzerland), 1; cysteine-HCl, 0.8; KH2PO4 (BDH), 0.5; K2HPO4 (BDH), 0.5; bile salts no. 3 (Oxoid), 0.4; CaCl2 (BDH), 0.15; hemin, 0.05; vitamin K, 0.01; and FeSO4 (BDH), 0.005. Tween 80 (BDH) was used at 1 ml/liter. A 4-ml/liter concentration of a 0.025% (wt/vol) solution of resazurin was added to the growth medium to act as an indicator of anaerobicity. The pH of the culture medium was adjusted to 7, after which the medium was autoclaved at 121°C for 15 min and allowed to cool under a nitrogen atmosphere.
The remaining volume of each vessel was made up with a freshly prepared fecal slurry from a healthy human volunteer. The 10% (wt/vol) fresh fecal slurry was prepared with prewarmed phosphate-buffered saline (PBS) (Oxoid) at pH 7.3. Six fermentations were carried out: polydextrose (Litesse Ultra, a high grade form of polydextrose), lactitol monohydrate, lactitol monohydrate:polydextrose (50:50), FOS, polydextrose (using a pooled inoculum), and oligofructose (using a pooled inoculum). Polydextrose and lactitol monohydrate were supplied by Danisco Sweeteners, Redhill, England, and oligofructose (Raftilose P95) was supplied by Orafti, Tienen, Belgium. For the first four fermentations, the same healthy human donated a sample for inoculation into the system. For the last two fermentations, samples from four healthy human donors were pooled and inoculated into both systems, which were then run concomitantly. Following inoculation, the system was left overnight as a batch culture to enable microbial populations to adjust to their new environment. After this stabilization period, the medium flow through the system was switched on at a rate that resulted in a system retention time of approximately 48 h.
The fermentation continued until steady state was reached, after which time the test substrate was added into the medium. All substrates were added at a concentration of 1% (wt/vol) (except in the case of the second fermentation of polydextrose, where 2% [wt/vol] substrate concentration was used). At this stage, samples of culture fluid were removed from each vessel for subsequent bacterial and volatile fatty acid analysis. The fermentation was continued until a further steady state was reached, and again samples of culture fluid were taken from each vessel for subsequent analyses. Steady state was achieved after seven complete turnovers of the system.
Bacterial enumeration via FISH.
The culture-independent procedure, FISH, was used for enumeration of the bacterial populations. Populations enumerated were bacteroides, bifidobacteria, clostridia (Clostridium histolyticum and Clostridium perfringens), and lactobacilli and enterococci. Samples of culture fluid were diluted 1:3 (vol/vol) with 4% (wt/vol) paraformaldehyde at 4°C and were fixed overnight at 4°C. Following pelleting by centrifugation (10,000 × g for 5 min), cells were washed in PBS, resuspended in an ethanol-PBS mixture, and stored at −20°C. Sixteen microliters of this cell suspension was then added to 200 μl of prewarmed (50°C) hybridization buffer (40 mM Tris-HCl, 1.8 M NaCl [pH 7.2]) and 64 μl of filtered high-performance liquid chromatography-grade water. Ninety microliters of the cell mixture was added to 10 μl of the appropriate probe. For Bifidobacterium Bif164, sequence CATCCGGCATTACCACCC (5′ to 3′) was used with a hybridization temperature of 50°C (22). For Bacteroides Bac303, sequence CCAATGTGGGGGACCTT was used with a hybridization temperature of 45°C (26). For Clostridium histolyticum Chis150, sequence TTATGCGGTATTAATCT(C/T)CCTTT was used with a hybridization temperature of 50°C (7). For Lactobacillus/Enterococcus Lab158, sequence GGTATTAGCA(T/C)CTGTTTCCA was used with a hybridization temperature of 45°C (15). The probes were labeled with a fluorescent dye, Cy3 (MWG Biotech, Edersberg, Germany). Cells were incubated with the probes overnight at their appropriate hybridization temperatures. The cells were washed at their hybridization temperature in approximately 5 ml of 1× hybridization buffer for 30 min (20 mM Tris-HCl, 0.9 M NaCl [pH 7.2]). Twenty microliters of 4′,6-diamidino-2-phenylindole (DAPI) was added to this buffer, which stained all DNA to enable a total cell count to be made. Cells were vacuum filtered onto a 0.2-μm-pore-size Isopore membrane filter (Millipore Corporation, Watford, United Kingdom) after washing. The filter was mounted onto a microscope slide, a drop of Slowfade was added (Molecular Probes, Leiden, The Netherlands), and mixture was examined with a Nikon (Kingston upon Thames, Surrey, United Kingdom) Eclipse E400 fluorescent microscope. A 355-nm excitation filter was used to enumerate DAPI-stained cells, and a 550-nm filter was used to enumerate the hybridized cells. Cells from 15 random fields of view were counted, and the average count was used for analysis.
SCFA.
Samples of culture fluid were analyzed for SCFA by gas chromatography, using an internal standard method described previously (18).
Percent-G+C profiling.
At the end point of simulations, bacteria were recovered from 100 ml of the culture medium from each vessel by centrifugation. The resultant pellets were resuspended in 3.1 ml of 50 mM Tris (pH 8) buffer with 50 mM EDTA. Bacterial suspensions were subject to freeze-thaw cycles and were lysed according to a previously described protocol (3), which combines physical (bead beating), chemical (sodium dodecyl sulfate), and enzymatic (lysozyme and proteinase K) steps. This protocol has been shown to lyse more than 99% of the bacteria in chicken cecal samples (3). The recovered total bacterial DNA was purified for percent-G+C profiling as described by Apajalahti et al. (2). The DNA isolated was fractionated according to its G+C percentage by bisbenzimidazole-CsCl equilibrium density gradient centrifugation (3). This approach fractionates the DNA of the component populations of the bacterial community based upon their characteristic percent-G+C content through differential density, which is imposed by the AT-dependent DNA-binding dye bisbenzimidazole (17). Determination of the percent-G+C content represented by each gradient fraction was accomplished by regression analysis (r2 > 0.99) of data obtained from gradients containing standard DNA samples of known percent-G+C content (C. perfringens, E. coli, and Micrococcus lysodeikticus).
Species-specific PCR for bifidobacteria.
DNA recovered from vessel 1 of the different simulations was subjected to a PCR analysis targeted to reveal the presence of various species of Bifidobacterium. The protocol followed that described by Matsuki et al. (27).
RESULTS
Bacterial enumeration via FISH.
Table 1 shows bacterial populations as enumerated via FISH. Data were analyzed by using a Student's t test. The first fermentation showed that the addition of polydextrose resulted in a significant increase in bifidobacteria in all vessels (P = 0.01). Bacteroides levels remained approximately constant, with a decrease in vessel 1 (P = 0.01) and nonsignificant changes seen in vessels 2 and 3. Lactobacilli remained below the detection limit of the FISH technique throughout the entire fermentation. Clostridia were only detected in vessel 1 at the first steady state and were completely undetectable beyond this point.
TABLE 1.
Bacterial populations enumerated via FISH for the fermentation of the test substratesa
| Substrate | Steady state no. | Sample vessel no. | Total Bacteria | Bifidobacteria | Bacteroides | Lactobacilli and enterococci | Clostridia (C. histolyticum and C. perfringens) |
|---|---|---|---|---|---|---|---|
| Polydextrose† | 1 | 1 | 9.15 | 6.91 | 8.92 | <106 | 6.48 |
| 2 | 9.20 | <106 | 8.53 | <106 | <106 | ||
| 3 | 9.17 | <106 | 7.70 | <106 | <106 | ||
| 2 | 1 | 9.35 | 7.99* | 8.53* | <106 | <106 | |
| 2 | 9.17 | 8.04* | 8.53 | <106 | <106 | ||
| 3 | 9.39 | 7.88* | 8.12 | <106 | <106 | ||
| Lactitol monohydrate† | 1 | 1 | 9.25 | 7.80 | 7.66 | <106 | <106 |
| 2 | 9.22 | 7.93 | 7.54 | <106 | <106 | ||
| 3 | 9.24 | 8.44 | 8.04 | <106 | <106 | ||
| 2 | 1 | 8.90 | 6.62* | 6.49* | <106 | 6.51 | |
| 2 | 9.15 | <106* | 7.27* | <106 | <106 | ||
| 3 | 9.15 | <106* | 6.83* | <106 | <106 | ||
| Lactitol monohydrate: | 1 | 1 | 9.20 | 8.17 | 8.42 | 7.00 | <106 |
| polydextrose† | 2 | 9.31 | 8.31 | 8.15 | <106 | 6.84 | |
| 3 | 9.16 | 8.35 | 7.22 | <106 | 6.67 | ||
| 2 | 1 | 9.11 | <106 | 8.36 | <106 | <106 | |
| 2 | 9.22 | 7.07* | 7.81* | 6.73 | 6.38* | ||
| 3 | 9.27 | 6.57* | 6.62* | <106 | <106 | ||
| Oligofructose† | 1 | 1 | 9.18 | <106 | 7.04 | <106 | <106 |
| 2 | 9.24 | <106 | 7.29 | <106 | <106 | ||
| 3 | 9.21 | <106 | 6.72 | <106 | <106 | ||
| 2 | 1 | 9.46 | 8.36* | 7.49 | 7.86* | <106 | |
| 2 | 9.13 | 8.31* | 7.55* | 7.79* | 7.67* | ||
| 3 | 8.98 | 8.11* | 6.87* | 7.39* | 7.80* | ||
| Polydextrose (parallel)‡ | 1 | 1 | 9.40 | 7.84 | 8.13 | 6.62 | <106 |
| 2 | 9.10 | 7.57 | 7.99 | <106 | <106 | ||
| 3 | 9.33 | 6.72 | 8.02 | <106 | <106 | ||
| 2 | 1 | 9.25 | 9.04* | 8.13 | 6.71 | <106 | |
| 2 | 9.30 | 7.64 | 8.35* | 6.50 | <106 | ||
| 3 | 9.23 | 7.94* | 7.65* | <106 | <106 | ||
| FOS (parallel)‡ | 1 | 1 | 9.13 | 8.70* | 6.79 | 6.71 | <106 |
| 2 | 9.28 | 8.56 | 8.13 | 6.46 | <106 | ||
| 3 | 9.28 | 8.71 | 7.98 | <106 | <106 | ||
| 2 | 1 | 9.31 | 8.91 | 6.71 | 6.56 | 6.44 | |
| 2 | 9.27 | 8.68 | 8.66* | <106 | <106 | ||
| 3 | 9.19 | 8.81 | 8.24* | <106 | <106 |
In vitro fermentations were carried out in a three-stage continuous culture system which mimicked conditions in the human colon. Fermentations were first carried out with the test substrate omitted from the medium and were continued until steady state was reached. At this point bacterial populations and their metabolic products were characterized. After this time the test substrate was added and the fermentation continued until a new steady state was reached, following which the same analysis was carried out. All counts refer to log cells/milliliter of culture fluid. A value of <106 is below the detection limit of the FISH technique. A dagger (†) refers to fermentations inoculated with one sample from the same healthy human donor. A double dagger (‡) refers to fermentations inoculated with a pooled sample from four healthy human donors and carried out in parallel. An asterisk indicates statistically significant changes in the bacterial populations enumerated.
For the lactitol monohydrate fermentations, bifidobacterial and bacteroides levels decreased following addition of the test substrate (P = 0.01 for all vessels). Lactobacilli remained below the detection limit of the FISH technique. Clostridia were not detected in any vessel at the first steady state but did appear in vessel 1 following the addition of lactitol monohydrate.
By adding a 50:50 mix of lactitol monohydrate and polydextrose, an intermediary effect was produced. Whereas polydextrose increased bifidobacterial levels and lactitol monohydrate decreased them dramatically, the addition of a 50:50 mixture of the two caused a decrease in this population to a lesser extent than was observed with lactitol monohydrate alone (P = 0.01 for all vessels). Equally, while bacteroides numbers remained constant following the addition of polydextrose and decreased following the addition of lactitol monohydrate, mixing of the two substrates meant that this population was decreased to a lesser extent in all vessels (P = 0.01 for vessels 2 and 3 and a nonsignificant decrease in vessel 1). Lactobacilli were detected at both steady states, although substrate addition can only affect the second steady state. Equally, clostridia were more numerically dominant in this fermentation, with their numbers decreasing upon addition of the substrate mixture (P = 0.01 for vessel 2).
The addition of oligofructose to the fourth fermentation caused a significant increase in bifidobacteria (P = 0.01 for all vessels) to the level where they were the most numerically dominant group in all vessels. Bacteroides also slightly increased upon the addition of FOS (P = 0.05 for vessels 1 and 2 and a nonsignificant increase in vessel 3). Lactobacilli significantly increased by oligofructose (P = 0.01 for all vessels) addition, as were clostridia in the case of vessels 2 and 3 (P = 0.01).
In the first simulation, we found that 55% of glucose in polydextrose was resistant to attack by bacteria, whereas 100% of oligofructose disappeared. To equalize the level of substrate available for bacteria the concentration of polydextrose was doubled in the following fermentation while the oligofructose level was kept the same. The fifth and sixth fermentations were inoculated with a pooled inoculum and were carried out in parallel, using 2% (wt/vol) polydextrose in one model and 1% (wt/vol) oligofructose in the other. Bifidobacterial levels were increased in both fermentations but to a greater extent in the polydextrose model (P = 0.01 for vessels 1 and 3 and a nonsignificant increase in vessel 2 for polydextrose; P = 0.01 for vessel 1 and a nonsignificant increase in vessels 2 and 3 for oligofructose). Bacteroides levels were nonsignificantly decreased in vessel 1 for both models and were increased in vessel 2 (P = 0.01 for both fermentations). For vessel 3, numbers were decreased following the addition of polydextrose (P = 0.01) but increased in this vessel following addition of oligofructose (P = 0.01). Upon the addition of polydextrose, lactobacilli numbers were increased in vessels 1 and 2. Clostridia remained below the detection limit of the FISH technique at both states in the polydextrose fermentation but were detected in vessel 1 after the addition of oligofructose.
Analysis of SCFA.
Addition of polydextrose increased the concentration of SCFA in all three vessels of the simulator (Table 2). Concentration of all the major SCFA was affected in all stages of the simulator, the most pronounced effect being observed in the concentration of acetic acid. The inclusion of lactitol monohydrate also led to an increase in total SCFA concentration in all three vessels. However, the fermentation type was different from that of polydextrose, leading to stimulation of butyric acid (Table 2). Combining polydextrose and lactitol monohydrate appeared to partly neutralize the fermentation, stimulating effects the two carbohydrates had exerted when fed individually. Despite the apparent negative interaction, a combination of the two substrates stimulated fermentation in all stages of the simulation. Oligofructose increased the concentration of total SCFA in a manner similar to that of polydextrose, but the proportion of butyric acid was somewhat higher with oligofructose. When polydextrose and oligofructose were fermented in parallel using the same inoculum, 2% (wt/vol) polydextrose generated a strong stimulation of bacterial fermentation whereas the effect of oligofructose on the residual concentration of SCFA was modest. Profiles of the bacterial metabolites were consistent with previous nonparallel simulations.
TABLE 2.
SCFA detected in fermentation culture fluid via gas chromatographya
| Substrate | Steady state no. | Sample vessel no. | Total SCFA (mM) | Acetic acid (mM) | Propionic acid (mM) | Butyric acid (mM) |
|---|---|---|---|---|---|---|
| Polydextrose† | 1 | 1 | 100 | 29 | 31 | 32 |
| 2 | 135 | 36 | 37 | 50 | ||
| 3 | 149 | 46 | 36 | 52 | ||
| 2 | 1 | 183 | 78 | 51 | 48 | |
| 2 | 224 | 89 | 59 | 64 | ||
| 3 | 245 | 104 | 63 | 57 | ||
| Lactitol monohydrate† | 1 | 1 | 76 | 32 | 16 | 26 |
| 2 | 107 | 35 | 20 | 42 | ||
| 3 | 161 | 50 | 30 | 64 | ||
| 2 | 1 | 190 | 48 | 49 | 89 | |
| 2 | 232 | 57 | 53 | 102 | ||
| 3 | 240 | 63 | 54 | 101 | ||
| Lactitol monohydrate: | 1 | 1 | 133 | 40 | 35 | 47 |
| polydextrose† | 2 | 156 | 43 | 41 | 57 | |
| 3 | 157 | 42 | 40 | 59 | ||
| 2 | 1 | 173 | 82 | 30 | 54 | |
| 2 | 205 | 82 | 41 | 65 | ||
| 3 | 214 | 80 | 42 | 72 | ||
| Oligofructose† | 1 | 1 | 129 | 31 | 38 | 49 |
| 2 | 137 | 34 | 39 | 48 | ||
| 3 | 142 | 37 | 38 | 49 | ||
| 2 | 1 | 174 | 50 | 48 | 69 | |
| 2 | 226 | 79 | 52 | 83 | ||
| 3 | 250 | 93 | 56 | 86 | ||
| Polydextrose (parallel)‡ | 1 | 1 | 79 | 29 | 3 | 39 |
| 2 | 145 | 64 | 12 | 46 | ||
| 3 | 126 | 51 | 7 | 51 | ||
| 2 | 1 | 184 | 69 | 32 | 54 | |
| 2 | 259 | 126 | 40 | 55 | ||
| 3 | 269 | 131 | 43 | 57 | ||
| FOS (parallel)‡ | 1 | 1 | 108 | 43 | 11 | 38 |
| 2 | 151 | 62 | 14 | 45 | ||
| 3 | 165 | 68 | 15 | 48 | ||
| 2 | 1 | 126 | 45 | 9 | 60 | |
| 2 | 189 | 77 | 20 | 67 | ||
| 3 | 196 | 82 | 20 | 66 |
Determination of SCFA in the culture fluid of each fermentation vessel was determined at both the first steady state (before addition of the test substrate) and at the second steady state (after the test substrate had been added). A dagger (†) refers to fermentations inoculated with one sample from the same healthy human donor. A double dagger (‡) refers to fermentations inoculated with a pooled sample from four healthy human donors and carried out in parallel.
Percent-G+C profiling of bacterial communities.
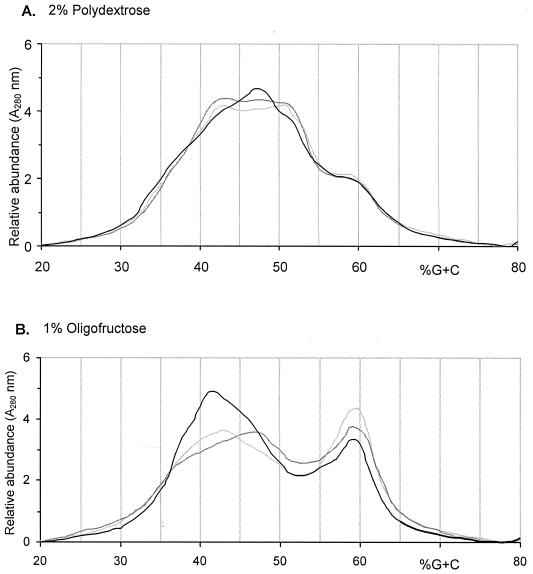
To form a picture of the total bacterial community enriched by the test substrates, percent-G+C profiling of the community DNA was carried out for the steady-state 2 samples of the parallel fermentations. In the simulation with 2% (wt/vol) polydextrose, the bacterial community profiles in vessels 1 and 2 were almost identical (Fig. 1A). In vessel 3 the shoulders apparent at 42 and 52% G+C in profiles from vessels 1 and 3 disappeared and were replaced by a new peak at 47% G+C, indicating an increase in the relative abundance of bacteria with this indicated G+C content in their genomic DNA. The bacterial community after the simulation with oligofructose contained a high abundance of bacteria, with G+C percentages of 58 to 61. This is consistent with the presence of bifidobacteria (34). When moving from vessel 1 to vessel 3 the relative abundance of this apparent bifidobacterial peak decreased (Fig. 1B). Simultaneously, bacteria with G+C percentages of 37 to 47 became more abundant. This emerging peak is consistent with the enrichment of Bacteroides species (31).
FIG. 1.
Percent-G+C profiling of bacterial communities for fermentation of polydextrose (A) and oligofructose (B). Bacterial cells in the fermentation culture fluid were lysed and the DNA was isolated, purified, and then fractionated according to its G+C percentage. The light gray line refers to vessel 1 of steady state 2, the dark gray line refers to vessel 2, and the black line refers to vessel 3.
Major species of bifidobacteria.
The bacteriological analysis described above indicates the presence of bacteria with percent G+C consistent with that of the bifidobacteria (percent-G+C profiling) or the presence of total bifidobacteria (FISH analysis). To determine the major species of bifidobacteria present we used PCR with species-specific primers (27). Table 3 indicates the species of bifidobacteria which were detected in vessel 1 of each fermentation after adaptation to an indicated substrate. Bifidobacterium adolescentis, B. bifidum, B. longum, and B. dentium were detected in all fermentations carried out, and B. catenulatum was detected in all fermentations except that of the combination of polydextrose and lactitol monohydrate. B. infantis was found only in the fermentation with 2% (wt/vol) polydextrose and B. breve in the fermentation with the combination of polydextrose and lactitol (Table 3).
TABLE 3.
Bifidobacterial species detected via species-specific PCRa
| Substrate | B. adolescentis | B. angulatum | B. bifidum | B. breve | B. catenulatum | B. longum | B. infantis | B. dentium |
|---|---|---|---|---|---|---|---|---|
| 1% (wt/vol) Polydextrose | + | + | + | + | + | |||
| 1% (wt/vol) Lactitol | + | + | + | + | + | |||
| 0.5% (wt/vol) Polydextrose + 0.5% lactitol | + | + | + | + | + | |||
| 1% (wt/vol) Oligofructose | + | + | + | + | + | + | ||
| 2% (wt/vol) Polydextrose (p) | + | + | + | + | + | + | + | |
| 1% (wt/vol) Oligofructose (p) | + | + | + | + | + |
DNA recovered from the first vessel of the in vitro fermentation systems was subject to PCR analysis by using species-specific primers for bifidobacteria. (p), parallel fermentations. B. gallicum was not detected with any of the substrates used.
DISCUSSION
A bifidogenic response was observed following the addition of polydextrose, at both 1% (wt/vol) and 2% (wt/vol) concentrations, using single and pooled inocula, respectively. The increase in bifidobacterial levels was statistically significantly in all three vessels of the 1% (wt/vol) fermentation using a single inoculum. In the case of pooled inocula, the increase was statistically significant in two of the three vessels (although an increase was seen in the other vessel, but this was insignificant). Indeed, a more pronounced effect upon bifidobacterial populations was observed in the fermentation using a single inocula, although overall bifidobacterial levels reached their highest in the parallel fermentation than in any other fermentation using any substrate. Bifidobacterial levels showed an increase between vessels 1 and 2 (for the first fermentation) and vessels 2 and 3 (for the second fermentation), demonstrating that polydextrose had a more sustained prebiotic effect throughout the model. Fermentations with polydextrose seemed to maintain a diverse assortment of different bifidobacteria. Indeed, seven different species of bifidobacteria were found, one of them being B. infantis, which was detected only when the medium was supplemented with polydextrose. B. infantis is considered particularly beneficial due to its enhanced ability to inhibit gastrointestinal pathogens through direct antimicrobial action (13). This also indicates that the use of polydextrose together with B. infantis as a probiotic would be a useful synbiotic product (11). Effects on bacteroides seemed approximately equivalent in both fermentations, with significant decreases and increases as well as nonsignificant changes. Generally, bacteroides populations were neither significantly increased nor decreased. The structure of the total bacterial community was studied by percent-G+C profiling of the total chromosomal DNA. After the 2-week enrichment with polydextrose, the peak at about 60% G+C most likely representing bifidobacteria had the same relative abundance throughout the simulator. Clear shifts were observed at G+C percentages of 42 to 52 when moving from vessel 2 to 3.
Lactitol monohydrate appeared to have deleterious effects on both bifidobacterial and bacteroides levels as well as causing an overall decrease in total cell numbers. This may be because under anaerobic conditions sugar alcohols can be inhibitory to fermentative organisms, because they are too reduced to be metabolized in the normal manner. Nevertheless, lactitol monohydrate seemed to stimulate bacterial metabolism when measured as production of SCFA. Indeed, it is possible that lactitol is routed to futile metabolic cycles providing low yield of ATP but high substrate turnover. By mixing lactitol monohydrate and polydextrose in 50:50 combination, bifidobacterial populations responded in a manner approximately the average of the two substrates alone.
In the two oligofructose fermentations, bifidobacteria increased, which has been observed in many other studies, both in vitro and in vivo (5, 12). The bacteriological changes detected by FISH analysis were consistent with the percent-G+C profiling analysis. Both FISH analysis and the percent-G+C analysis of the parallel fermentation with oligofructose indicated that vessel 1 contained the highest level of bifidobacteria. The same two independent analytical techniques also were consistent in detecting increase in bacteroides in the distal vessel when the simulator was fed with oligofructose.
In terms of lactobacilli-enterococci and clostridia (C. histolyticum and C. perfringens) levels, the sizes of most populations were so low as to be below the detection limit of the FISH technique.
With regard to the stimulation of bifidobacteria and other bacteria, lactitol monohydrate (and the mixture containing it) was, undoubtedly, the least promising substrate. However, lactitol strongly shifted bacterial fermentation towards butyric acid, which has been reported to be beneficial for intestinal health (4, 10, 36). If the bacteria stimulated by lactitol do not possess unwanted properties, it could have potential as a novel nonbifidogenic prebiotic. Conversely, oligofructose and polydextrose both proved to have prebiotic potential in all four fermentations carried out with them as the substrate. Significant increases in bifidobacteria were observed in all four fermentations. Although no stimulatory effect upon lactobacilli was observed with polydextrose, it did demonstrate a significantly suppressing effect upon Bacteroides levels in one vessel of all fermentations.
In conclusion, this study confirmed several of the findings previously determined in a human clinical study (19).
Acknowledgments
This work was supported by Danisco Sweeteners, Redhill, England.
We thank Harri Mäkivuokko, Markku Saarinen, Laura Särkilahti, and Jaana Oksanen for skillful analytical work.
REFERENCES
- 1.Achour, L., B. Flourié, F. Briet, P. Pellier, P. Marteau, and J.-C. Rambaud. 1994. Gastrointestinal effects and energy value of polydextrose in healthy nonobese men. Am. J. Clin. Nutr. 59:1362-1368. [DOI] [PubMed] [Google Scholar]
- 2.Apajalahti, J. H. A., H. Kettunen, A. Kettunen, W. E. Holben, P. H. Nurminen, N. Rautonen, and M. Mutanen. 2002. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl. Environ. Microbiol. 68:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apajalahti, J. H., L. K. Sarkilahti, B. R. Maki, J. P. Heikkinen, P. H. Nurminen, and W. E. Holben. 1998. Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl. Environ. Microbiol. 64:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archer, S. Y., S. Meng, J. Wu, J. Johnson, R. Tang, and R. Hodin. 1998. Butyrate inhibits colon carcinoma cell growth through two distinct pathways. Surgery 124:248-253. [PubMed] [Google Scholar]
- 5.Bouhnik, Y., K. Vahedi, L. Achour, A. Attar, J. Salfati, P. Pochart, P. Marteau, B. Flourié, F. Bornet, and J.-C. Rambaud. 1999. Short-chain fructo-oligosaccharide administration dose-dependently increases fecal bifidobacteria in healthy humans. J. Nutr. 129:113-116. [DOI] [PubMed] [Google Scholar]
- 6.Figdor, S. K., and H. H. Rennhard. 1981. Caloric utilization and disposition of [14C]polydextrose in the rat. J. Agric. Food Chem. 29:1181-1189. [DOI] [PubMed] [Google Scholar]
- 7.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 9.Fuller, R. 1999. Probiotics, p. 89-99. In G. R. Gibson and M. B Roberfroid (ed.), Colonic microbiota, nutrition and health. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 10.German, J. B. 1999. Butyric acid: a role in cancer prevention. BNF Nutr. Bull. 24:203-209. [Google Scholar]
- 11.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, G. R., and X. Wang. 1994a. Bifidogenic properties of different types of fructo-oligosaccharides. Food Microbiol. 11:491-498. [Google Scholar]
- 13.Gibson, G. R., and X. Wang. 1994. Regulatory effects of bifidobacteria on the growth of other colonic bacteria. J. Appl. Bacteriol. 77:412-420. [DOI] [PubMed] [Google Scholar]
- 14.Grimble, G. K., D. H. Patil, and D. B. A. Silk. 1988. Assimilation of lactitol, an ′unabsorbed' disaccharide in the normal human colon. Gut 29:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmsen, H. J. M., P. Elfferich, F. Schut, and G. W. Welling. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb. Ecol. Health Dis. 11:3-12. [Google Scholar]
- 16.Hilton, E., P. Kolakowski, C. Singer, and M. Smith. 1997. Efficacy of Lactobacillus GG as a diarrheal preventive in travelers. J. Travel Med. 4:41-43. [DOI] [PubMed] [Google Scholar]
- 17.Holben, W. E., and D. Harris. 1995. DNA based monitoring of total bacterial community structure in environmental samples. Mol. Ecol. 4:627-631. [DOI] [PubMed] [Google Scholar]
- 18.Holben, W. E., P. Williams, M. Saarinen, L. K. Särkilahti, and J. H. A. Apajalahti. 2002. Phylogenetic analysis of intestinal microflora indicates a novel mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 44:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Jie, Z., L. Bang-yao, X. Ming-jie, L. Hai-wei, Z. Zu-kang, W. Ting-song, and S. A. S. Craig. 2000. Studies on the effects of polydextrose intake on physiologic functions in Chinese people. Am. J. Clin. Nutr. 72:1503-1509. [DOI] [PubMed] [Google Scholar]
- 20.Kontula, P., M.-L. Suihko, A. Von Wright, and T. Mattila-Sandholm. 1998. The effect of lactose derivatives on intestinal lactic acid bacteria. J. Dairy Sci. 82:249-256. [DOI] [PubMed] [Google Scholar]
- 21.Koutsou, G. A., D. M. Storey, A. Lee, A. Zumbe, B. Flourié, Y. leBot, and P. Olivier. 1996. Dose-related gastrointestinal response to the ingestion of either isomalt, lactitol or maltitol in milk chocolate. Eur. J. Clin. Nutr. 50:17-21. [PubMed] [Google Scholar]
- 22.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescent in situ hybridisation of Bifidobacterium with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Link-Amster, H., F. Rochat, K. Y. Saudan, O. Mignot, and J. M. Aeschlimann. 1994. Modulation of a specific humoral immune response and changes in intestinal flora mediated through fermented milk intake. FEMS Immunol. Med. Microbiol. 10:55-63. [DOI] [PubMed] [Google Scholar]
- 24.Livesey, G., I. T. Johnson, J. M. Gee, T. Smith, W. E. Lee, K. A. Hillan, J. Meyer, and S. C. Turner. 1993. ‘Determination’ of sugar alcohol and polydextrose absorption in humans by the breath hydrogen (H2) technique: the stoichiometry of hydrogen production and the interaction between carbohydrates assessed in vivo and in vitro. Eur. J. Clin. Nutr. 47:419-430. [PubMed] [Google Scholar]
- 25.Macfarlane, G. T., S. Macfarlane, and G. R. Gibson. 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 35:180-187. [DOI] [PubMed] [Google Scholar]
- 26.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 27.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattila-Sandholm, T., J. Mättö, and M. Saarela. 1999. Lactic acid bacteria with health claims—interactions and interference with gastrointestinal flora. Int. Dairy J. 9:25-35. [Google Scholar]
- 29.Menne, E., N. Guggenbuhl, and M. Roberfroid. 2000. Fn-type chicory inulin hydrolysate has a prebiotic effect in humans. J. Nutr. 130:1197-1199. [DOI] [PubMed] [Google Scholar]
- 30.Rao, V. A. 2001. The prebiotic properties of oligofructose at low intake levels. Nutr. Res. 6:843-848. [Google Scholar]
- 31.Reddy, C. A., and M. P. Bryant. 1977. Deoxyribonucleic acid base composition of certain species of the genus Bacteroides. Can. J. Microbiol. 23:1252-1256. [DOI] [PubMed] [Google Scholar]
- 32.Riggio, O., M. Varriale, G. P. Testore, R. Di Rosa, E. Di Rosa, M. Merli, A. Romiti, C. Candiani, and L. Capocaccia. 1990. Effect of lactitol and lactulose administration on the fecal flora in cirrhotic patients. J. Clin. Gastroenterol. 12:433-436. [DOI] [PubMed] [Google Scholar]
- 33.Schiffrin, E. J., F. Rochat, H. Link-Amster, J. M. Aeschlimann, and A. Donnet-Hughes. 1995. Immunomodulation of human blood cells following the ingestion of lactic acid bacteria. J. Dairy Sci. 78:491-497. [DOI] [PubMed] [Google Scholar]
- 34.Sgorbati, B., B. Biavati, and D. Palenzona. 1995. The genus Bifidobacterium, p. 279-306. In B. J. B. Wood and W. H. Holzapfel (ed.), The genera of lactic acid bacteria. Chapman & Hall, Glasgow, United Kingdom.
- 35.Tuohy, K. M., C. J. Ziemer, A. Klinder, Y. Knöbel, B. L. Pool-Zobel, and G. R. Gibson. 2002. A human volunteer study to determine the prebiotic effects of lactulose powder on human colonic microbiota. Microb. Ecol. Health Dis. 14:165-173. [Google Scholar]
- 36.Wang, J., and E. A. Friedman. 1998. Short-chain fatty acids induce cell cycle inhibitors in colonocytes. Gastroenterology 114:940-946. [DOI] [PubMed] [Google Scholar]