Abstract
The α-proteobacteria phylogenetically related to the Roseobacter clade are predominantly responsible for the degradation of organosulfur compounds, including the algal osmolyte dimethylsulfoniopropionate (DMSP). Silicibacter sp. strain TM1040, isolated from a DMSP-producing Pfiesteria piscicida dinoflagellate culture, degrades DMSP, producing 3-methylmercaptopropionate. TM1040 possesses three lophotrichous flagella and is highly motile, leading to a hypothesis that TM1040 interacts with P. piscicida through a chemotactic response to compounds produced by its dinoflagellate host. A combination of a rapid chemotaxis screening assay and a quantitative capillary assay were used to measure chemotaxis of TM1040. These bacteria are highly attracted to dinoflagellate homogenates; however, the response decreases when homogenates are preheated to 80°C. To help identify the essential attractant molecules within the homogenates, a series of pure compounds were tested for their ability to serve as attractants. The results show that TM1040 is strongly attracted to amino acids and DMSP metabolites, while being only mildly responsive to sugars and the tricarboxylic acid cycle intermediates. Adding pure DMSP, methionine, or valine to the chemotaxis buffer resulted in a decreased response to the homogenates, indicating that exogenous addition of these chemicals blocks chemotaxis and suggesting that DMSP and amino acids are essential attractant molecules in the dinoflagellate homogenates. The implication of Silicibacter sp. strain TM1040 chemotaxis in establishing and maintaining its interaction with P. piscicida is discussed.
In the marine environment, unicellular algae and other eukaryotic microorganisms, such as dinoflagellates, coexist with a diverse community of heterotrophic bacteria. Much of the carbon and nitrogen required for the growth of these bacteria is supplied by the unicellular bloom-forming eukaryotes (10). During feeding, stress, and lysis, nutrients are released from these microbes and made available to the bacteria. The nutrient plumes emitted occur on a microscale and are quickly dispersed and diluted by diffusion and turbulence (3). Over space and time not all species of bacteria are equally likely to obtain these nutrients (8). While some bacterial species rely upon high-affinity uptake mechanisms, the ability to sense and move towards increasing chemical gradients, known as chemotaxis, provides motile bacteria with a distinct advantage over their nonmotile counterparts (8). Knowledge of how marine bacteria sense and respond to algal cells is therefore important to our understanding of bacterial physiology and the interactions between these prokaryotes and their eukaryotic hosts and ultimately impacts our greater understanding of nutrient cycling in marine ecosystems.
Algae-derived compounds that are likely to serve as chemoattractants for marine heterotrophic bacteria include sugars, amino acids, nucleosides, and other small diffusible organic compounds. Many of these molecules are common to all living cells; however, some are algae specific. For example, most marine unicellular algae, especially dinoflagellates and prymnesiophytes, produce large quantities of the organosulfur compound dimethylsulfoniopropionate (DMSP) (21). This compound is degraded by both algae and bacteria, resulting in the production of dimethylsulfide and acrylate, or demethylated by bacteria only, producing demethylation products including 3-methylmercaptopropionate (MMPA) and 3-mercaptopropionate (MPA) (30). All of these compounds can be utilized as a source of bacterial carbon and sulfur and are candidates for algae-specific chemoattractants (15, 22, 29, 33).
The α-, β-, and γ-proteobacteria, as well as the Cytophaga-Flexibacter-Bacteroides spp., are important members of the marine picoplankton and are often found within algal communities (13). Of these groups, the α-Proteobacteria genera phylogenetically related to the Roseobacter clade are of particular interest. These bacteria are abundant in algal cultures as well as within blooms of algal species (2, 17, 35). They are also predominantly responsible for the degradation of DMSP (15), and their activity has been correlated with blooms of high-DMSP-producing phytoplankton, including dinoflagellates and prymnesiophytes (35). Interestingly, Roseobacter clade species are involved in both physical (attached or intracellular) and physiological (toxin production and growth-enhancing) interactions with dinoflagellates (2, 7, 12). Mechanisms for these interactions may rely upon bacterial motility and chemotaxis behavior that allow these bacteria to sense and move towards their eukaryotic host, where they derive beneficial nutrients, once in close physical proximity to the dinoflagellate. While much is known about the cooccurrence of motile Roseobacter species and bloom-forming dinoflagellates, little or no data describing chemotactic behavior in any of the Roseobacter clade bacteria have been reported.
We have used monocultures of the DMSP-producing dinoflagellates Pfiesteria piscicida, Pfiesteria shumwayae, and Pfiesteria-like (Cryptoperidiniopsis) dinoflagellates as our model system for studying Roseobacter-dinoflagellate interactions (2). Within these cultures, bacteria native to the algal niche assimilate dinoflagellate-derived nutrients and are intrinsically propagated with the dinoflagellates in continuous subcultures. The bacterial community inhabiting Pfiesteria cultures is diverse, containing at least 19 different species depending upon the strain examined (2). Some of these bacteria can be found physically attached to and aiding in the growth of the dinoflagellates, and a major component of this bacterial community consists of bacteria related to Roseobacter species (2).
In a previous report (23), we isolated and identified four DMSP-degrading Roseobacter species from a P. piscicida culture. All of the Roseobacter species isolated from these dinoflagellates degrade DMSP (23), and one, strain Silicibacter sp. strain TM1040, is highly motile in semisolid agar and in liquid media (15). In this report, we tested the hypothesis that strain TM1040 is chemotactic towards specific molecules produced by the dinoflagellate.
MATERIALS AND METHODS
Bacteria and media.
Silicibacter sp. strain TM1040 was isolated from a culture of the dinoflagellate P. piscicida CCMP1830 (Provasoli-Guillard National Center for Culture of Marine Phytoplankton) and was maintained on either HIASW agar (25 g of heart infusion broth [Difco], 15 g of artificial seawater [ASW; Instant Ocean], 16 g of Bacto Agar per liter) or half-strength 2216 marine agar (2). Liquid broth cultures were made with half-strength 2216 marine broth. Marine motility agar was prepared by supplementing half-strength 2216 marine broth with 3.0 g of Bacto Agar per liter. For the chemotaxis plate assay, a basal minimal (BM) medium (12.1 g of Tris HCl, 1.0 g of NH4Cl, 0.0075 g of K2HPO4, 15 g of ASW, 3.0 g of Bacto Agar per liter; pH 7.6) was used. After autoclaving, the BM medium was cooled to 50°C and supplemented with 0.2 g of FeSO3 and 1 ml of Balch's vitamins (5), and a single carbon source (glycerol, glucose, succinate, or alanine) was added to a final concentration of 10 mM.
Dinoflagellates and cultivation.
P. piscicida CCMP1830 was grown as previously described (2). Dinoflagellates were fed a diet of the cryptomonad prey alga Rhodomonas sp. CCMP768, supplied as described by Alavi et al. (2).
Chemotaxis plate screening assay.
A qualitative chemotaxis screening method, described by DeLoney-Marino et al. (11), was used with only slight modification. An isolated colony of TM1040 was inoculated in the center of a BM-glycerol motility agar plate. After 3 days, a point where the bacteria had swam outwards 5 cm from the site of inoculation, an inoculum taken from the outer ring of the motile colony was used to inoculate the center of a second fresh BM-glycerol motility agar plate. Following 36 h of incubation, a putative attractant was placed approximately 5 cm from the site of inoculation (or 2.5 cm in front of the periphery of motile cells). Possible chemoattractant compounds were administered as either a sterile solid or concentrated stock solutions. The cells were further incubated for 16 h to allow for additional outward movement, at which time measurement of diameter, shape, and chemotaxis rings internal to the motile colony were made. The resulting data were scored on a plus-minus scale, where minus indicates no change compared to distilled water or no attractant controls and either one or two pluses indicates moderate to strong alteration in motile colony phenotype (respectively). All chemotaxis assays were performed in a 30°C walk-in incubator at 65% relative humidity.
Quantitative capillary chemotaxis assay.
The capillary method of Adler (1), as modified by Palleroni (26), was used to quantitatively measure the chemotactic response of TM1040 toward a subset of compounds screened by the plate method. A broth culture of TM1040 was grown overnight in half-strength 2216 marine broth at 30°C to an optical density at 600 nm (OD600) of 0.3 to 0.4, which corresponds to the mid-exponential phase of the Silicibacter sp. strain TM1040 growth cycle. The cells were pelleted by centrifugation at 4,000 × g in a tabletop centrifuge (Centra-CL2; International Equipment Company), the supernatant was discarded, and the pellet was resuspended in chemotaxis buffer (CB) (15 g of ASW, 6 g of Tris-HCl per liter [pH 7.6]) to the same OD. Five hundred microliters of washed cells was placed in one of the wells of the Palleroni chamber, followed by a 1-μl capillary (Microcaps; Drummond) filled with a putative attractant diluted in CB. This process was repeated for each of the four wells in the chamber. A capillary filled with CB only was included in each experiment as a negative control. Depending on the experiment, capillaries remained in the chambers for 0.5 to 2 h, after which time they were removed, and the contents of each capillary were serially diluted in CB. Final dilutions were then spread on HIASW agar plates and incubated for 16 h at 30°C. The concentration of TM1040 in each capillary was derived by counting the CFU on each plate and multiplying by the appropriate dilution factor. To normalize all data, a “response factor” as described by Wei and Bauer (31) was calculated by dividing the mean number of bacteria in the attractant-filled capillaries by the mean number of bacteria in control capillaries.
The capillary assay was also used to measure the reduction in chemotaxis through competition when a known attractant was supplied exogenously (i.e., outside the capillary). In this variation, the quantitative capillary method was used, but a known attractant was included in the CB used to suspend the washed cells. A capillary containing a mixture of potential chemoattractant molecules was then placed in the chamber and incubated as described earlier. At the completion of the assay, the percent reduction in chemotaxis was calculated using the following formula: % reduction = [1 − (CFU ml−1exogenous/CFU ml−1control)] × (100).
Preparation of cell homogenates.
Dinoflagellates and Rhodomonas sp. were grown to late exponential phase (ca. 105 to 106 cells per ml), and the cell density was measured using a Coulter Counter (model M/SZRII; Beckman Coulter). Dinoflagellates were harvested after starvation of the culture to remove Rhodomonas prey algae, which was confirmed by microscopy. The cell densities in P. piscicida and Rhodomonas cultures were normalized to 105 cells per ml, and each culture was then pelleted by centrifugation at 4,000 × g and 4°C for 10 min. The supernatant was removed, and the pellets were separately resuspended with either 1 ml of ice-cold distilled water or CB. The cells were homogenized by sonication (Sonic Dismembrator; Fisher), and cell disruption was assessed by microscopy. To examine the heat sensitivity of chemotaxis elicitors, an aliquot of each homogenate was heated to 80°C for 15 min prior to the assay. All homogenates were then stored at −20°C. When required, the homogenates were thawed, held on ice, and used in the capillary assay as described.
Bacterial homogenates were prepared in a similar manner. Briefly, an aliquot from several 10-fold dilutions of the P. piscicida culture was inoculated onto half-strength 2216 marine agar and incubated until bacterial colonies were evident. Approximately 1,000 mixed colonies were then resuspended from the agar surface using 10 ml of CB, and the cells were collected by centrifugation at 4,000 × g for 10 min. The bacterial pellet was resuspended to an OD600 of 0.3 in CB. Aliquots of the bacterial suspension were sonicated, and a portion was heated, as described above.
Video microscopy.
To compare the motility of TM1040 with and without an attractant, cells were grown and prepared as described for in the capillary assay. A sample of the washed cells was incubated at 30°C, with and without the addition of 10 mM succinate (in CB, pH 7.0). Cell swimming and behavior over a 5-h period were measured using phase-contrast microscopy (Nikon Optiphot BX60) equipped with a digital video camera (Canon Elura, Lake Success, N.Y.). After examination of the entire video, three separate fields were chosen, and 1-min intervals of each were transferred to a computer for further analysis, using Adobe (San Jose, Calif.) Premier version 6.0 software. Individual 0.33-s frames were exported as a tagged image file format sequence and reassembled with IPLab version 3.55 (Scanalytics, Fairfax, Va.) as a sequence of ordered frames (t-series). The numbers of motile and nonmotile cells per field were determined to determine the average percentage of motile cells per field.
Transmission electron microscopy (TEM).
Silicibacter sp. strain TM1040, taken from the periphery of a motile colony growing in semisolid BM glycerol motility agar, was inoculated in half-strength 2216 marine broth and incubated at 30°C to an OD600 of 0.3. A 400-μm-mesh carbon-coated parlodion copper grid was floated over a 30-μl aliquot of this culture for 1 to 2 min and blotted dry. Bacteria adhering to the grid were stained two times for 30 s with 1% uranyl acetate and 0.04% tylose in distilled water. Negatively stained cells were viewed using a Philips BioTwin CM120 transmission electron microscope at an operating voltage of 20 kV. The resulting images were recorded on film and scanned into a computer, and the brightness and contrast were changed for optimum viewing using Adobe Photoshop 7.
Analytical techniques.
The concentration of DMSP in 1 ml-samples of heated or boiled 200 μM DMSP and in heated and untreated P. piscicida homogenates was measured using gas chromatography with flame ionization detection, as previously described (23).
Chemicals.
DMSP was synthesized from acrylate and dimethylsulfide as previously described (9). MMPA was synthesized by alkaline hydrolysis of its methyl ester, methyl-3-(methylthio)propionate (Aldrich, Milwaukee, Wis.) (19). All other compounds were of at least reagent grade quality and were purchased from Sigma-Aldrich (Milwaukee, Wis.).
RESULTS
TM1040 possesses three lophotrichous flagella.
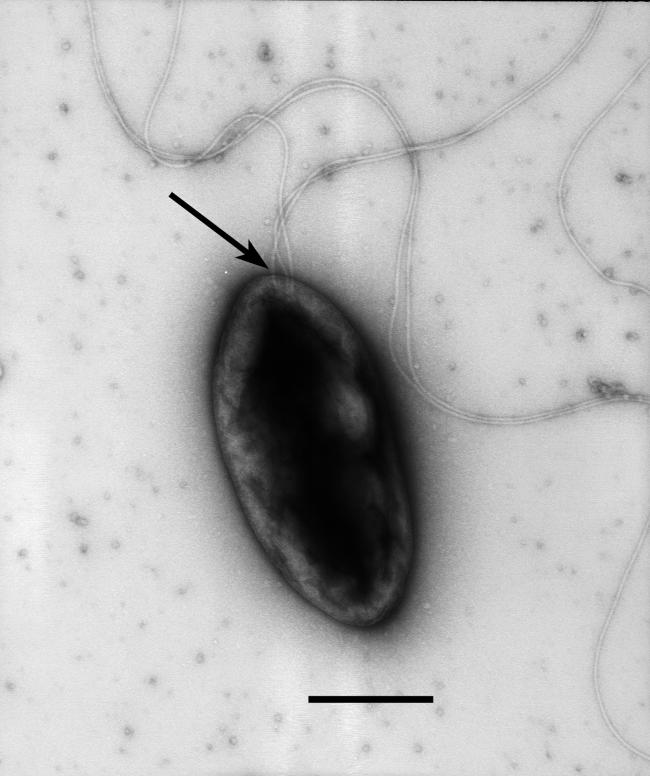
Prior to beginning studies of chemotaxis, motile cells of TM1040 were visualized by TEM to confirm the presence, number, and location of flagella. As shown in Fig. 1, TM1040 is a small (ca. 1.0- to 1.5-μm) rod- or oval-shaped bacterium with at least three lophotrichous flagella. These flagella are located at one end of the cell, slightly off-center from the cell pole. An analysis of the structure of filaments suggests that they are simple filaments, rather than the complex forms found in Silicibacter pomeroyi DSS-3 (14) or other α-proteobacteria (27).
FIG. 1.
TEM of Silicibacter sp. strain TM1040. A culture of TM1040 was prepared in half-strength 2216 marine broth and grown for 20 h at 30°C without agitation. Motile cells were blotted onto copper disks, stained with 1% uranyl acetate, and visualized by TEM. Three simple lophotrichous flagella attached to the pole of the cell are apparent (arrow). Bar = 0.5 μm.
Chemotaxis of TM1040 is enhanced by prior starvation.
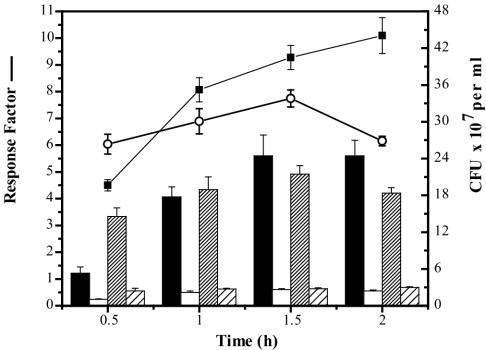
Initial observations suggested that TM1040 cells held for 1 to 2 h in CB (a starvation condition) were more responsive to methionine than cells that had not been starved (data not shown). To examine this more closely, chemotaxis of starved and unstarved TM1040 toward methionine was measured by using a quantitative capillary assay. As shown in Fig. 2, starving the bacteria of nutrients prior to the assay resulted in a greater maximal response to the attractant, which was maintained for at least 2 h. This can be seen by comparing the starved-cell response to methionine to the unstarved response to methionine in Fig. 2. After 0.5 h, the mean concentration of bacteria in methionine-filled capillaries was 5.3 × 107 CFU per ml and increased thereafter, with increasing exposure time rising from 1.8 × 108 CFU per ml at 1 h to a maximum of 2.4 × 108 CFU per ml at 1.5 and 2 h. By contrast, without starvation the maximum mean concentration of bacteria in methionine-filled capillaries reached a peak of 2.1 × 108 CFU per ml at 1.5 h and decreased thereafter. The bacterial concentration in control capillaries for either starved or unstarved cells reached an equilibrium of 2 × 107 CFU per ml after 0.5 h.
FIG. 2.
The effects of starvation on the chemotactic response of Silicibacter sp. strain TM1040. The chemotaxis of unstarved versus starved cells to 200 μM methionine was measured using the capillary assay (Materials and Methods). Chemotaxis of starved cells to either methionine (black bars) or buffer lacking nutrient (white bars) is compared to the response of unstarved cells either to methionine (dense cross-hatched bars) or to the buffer lacking nutrient (sparse cross-hatched bars). The response factor, calculated by dividing the mean number of bacteria in methionine-filled capillaries by the mean number of bacteria in buffer-filled capillaries, is significantly increased in the starved cells (▪) compared to that in the unstarved cells (○). The values shown are the mean and standard deviation (n = 12).
The difference between the chemotactic responses of starved and unstarved cells is also shown in Fig. 2. The response factor is calculated by dividing the concentration of bacteria in attractant-filled capillaries by the concentration of bacteria in control capillaries (31). The response factor of starved cells to methionine is higher than the response of unstarved cells at all times except 0.5 h (where equilibrium has yet to be reached). Significantly, a comparison of the response factors of starved cells and unstarved cell confirms that starvation improves the chemotactic response of TM1040 and maintains it longer.
Motility is affected by starvation.
The motility of the starved TM1040 cells was measured by light microscopy. While starvation enhanced the chemotactic response, it also reduced swimming motility, and prolonged starvation resulted in a majority of the cells becoming nonmotile. After 2 h, the percentage of motile cells under starvation conditions decreased drastically to a mean of 5.93% (n = 69), and by 4 h all of the cells were nonmotile (n = 66). In comparison, the percentage of motile cells under nonstarvation conditions did not change after 5 h and was similar to the percentage of motile cells after 1 h (mean, 26.64%; n = 69). Thus, while starvation stimulates chemotaxis, it also depletes the energy supplies required to rotate the flagella. Based on these results, a starvation period of 1.5 h was considered optimal and was used in all further capillary assays.
TM1040 is attracted to dinoflagellate homogenates.
Does TM1040 sense and respond to dinoflagellates? To determine this, chemotaxis of TM1040 toward cell homogenates of P. piscicida and other constituents of the dinoflagellate culture was measured using the capillary assay. It is important to emphasize that the P. piscicida culture normally contains three different types of organisms: the dinoflagellates, the prey algae (Rhodomonas sp.), and a diverse bacterial population that includes TM1040 (2). Rhodomonas can be virtually eliminated from the cultures through attrition from dinoflagellate feeding, but the bacterial community cannot be removed without adversely affecting the dinoflagellates themselves (2). So, a strategy was developed to measure the chemotaxis of TM1040 towards three cell homogenates: dinoflagellates plus associated bacteria, Rhodomonas, and a mixture of heterotrophic bacteria obtained from the same dinoflagellate culture (see Materials and Methods). In this manner, the relative contribution of each population towards eliciting a chemotactic response from TM1040 could be assessed.
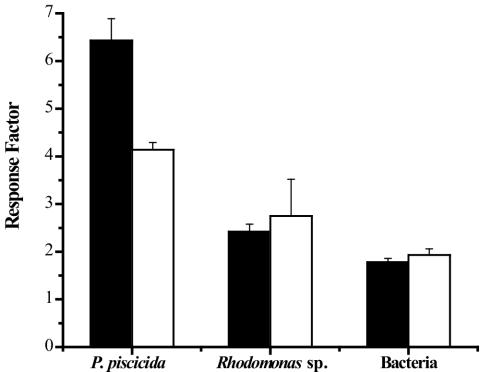
As is shown in Fig. 3, TM1040 responded strongly to P. piscicida homogenates, producing a response factor of 6.4. This response is mediated in part by heat-labile compounds because heating of the same homogenates prior to the assay reduced the response by 36% (response factor = 4.1) (Fig. 3). TM1040 is substantially less chemotactic to Rhodomonas homogenates, and this response did not change upon heating of the algal homogenate, remaining at a mean of ca. 2.5 (Fig. 3). Equally, bacterial homogenates were also poor elicitors of TM1040 chemotaxis, giving a mean response factor of only 1.9 and 1.7, with or without heating, respectively. These results indicate that TM1040 is more strongly attracted to components present in P. piscicida cell homogenates, some of which are heat labile.
FIG. 3.
Chemotaxis of Silicibacter sp. strain TM1040 toward cell homogenates. Chemotaxis of TM1040 toward heated (80°C) (white bars) and untreated (black bars) homogenates was measured using the capillary chemotaxis assay (Materials and Methods). The response factor is calculated by dividing the mean number of bacteria in homogenate-filled capillaries by the mean number of bacteria in buffer-filled capillaries. The chemotactic response of TM1040 to the untreated dinoflagellate homogenate is >2.5 times greater than the response to either algal or bacterial homogenates, and heating the dinoflagellate homogenate reduces the response factor by ca. 40%. The values shown are the mean and standard deviation (n = 8).
DMSP compounds and amino acids are strong chemoattractants of TM1040.
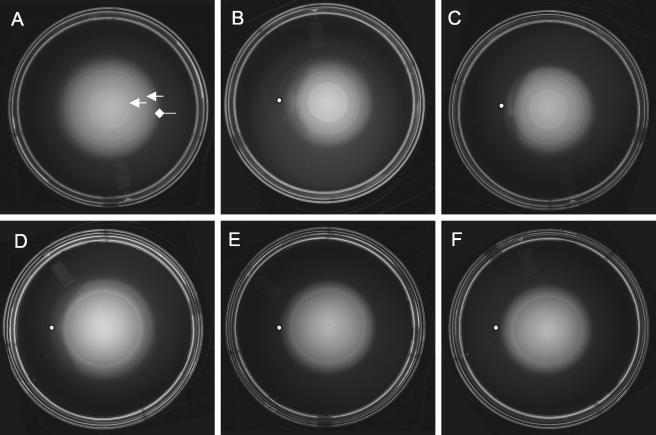
A chemotaxis plate assay (11) was used to screen a large number of pure compounds thought to be in dinoflagellate cell homogenates for their ability to affect the chemotactic behavior of TM1040. This assay utilizes a minimal medium with 0.3% agar (BM glycerol motility agar) that allows the bacteria to swim through the agar matrix. Bacteria inoculated into the center of the agar consume nutrients and create a concentration gradient that increases outward from the point of inoculation. The bacteria sense the increasing gradient and swim outwards, seeking higher concentrations of nutrients, which is manifested as a symmetrical “motile” colony. If an attractant is placed at a short distance in front of the advancing motile colony, it forms a second gradient moving towards the oncoming cells that will affect the symmetry of the colony by reducing the net outward swimming of the bacteria. This is shown in Fig. 4. When inoculated in BM glycerol motility agar (Fig. 4A), TM1040 cells swim outward from the point of inoculation to form a colony that often contains one or two internal bands of cells (Fig. 4A). These bands have been associated with subpopulations of bacteria that are responding to different attractants (32).
FIG. 4.
Screening of putative attractant compounds using the qualitative chemotaxis assay. Chemotaxis behavior of TM1040 was assessed using BM glycerol motility agar on which a compound to be tested is placed (indicated by the dot to the left of the swimming colony). (A) Chemotaxis of Silicibacter sp. strain TM1040 through BM glycerol motility agar results in an ever-increasing colony of motile bacteria that is punctuated by one or more internal bands of bacteria (indicated by the arrows) and the periphery (diamondhead arrow). Shown is the response of TM1040 to DMSP (B), methionine (C), valine (D), arabinose (E), and fructose (F). In the presence of either DMSP, methionine, or valine, the outer ring is disturbed and becomes asymmetrical, indicating a chemotactic response from the cells. In contrast, neither arabinose nor fructose causes a change in the symmetry of the colony, suggesting that TM1040 does not respond to these compounds.
As measured by this method, TM1040 is chemotactically responsive to a number of different chemicals, most notably DMSP and its catabolites, as well as amino acids (Fig. 4). For example, addition of DMSP (Fig. 4B) caused a marked deformation in the periphery of the motile colony, highlighted by flare of bacteria and disruption of the outer ring. Responses to methionine (Fig. 4C) and valine (Fig. 4D) also had significant effects on the periphery of the motile colony. In comparison, there was no detectable change in the appearance of the motile colony when either arabinose (Fig. 4E) or fructose (Fig. 4F) was tested, suggesting that these two sugars do not affect TM1040 chemotaxis.
The response to other chemicals was scored on a plus-minus scale (Table 1). TM1040 is attracted to all amino acids, similar to the response observed with methionine or valine. The bacterium responded strongly to the DMSP metabolites, acrylate and MMPA, while only weakly to MPA. These responses were similar to that of methionine and valine. Of the 12 sugars that were tested, TM1040 responded positively to (in order of response) sucrose, N-acetylglucosamine (NAG), galactose, glucose, and maltose. The tricarboxylic acid cycle (TCA) intermediates, citrate and fumarate, also elicited a strong positive response from TM1040, while only a mild response to succinate and no response to α-ketoglutarate was seen.
TABLE 1.
Change in motile colony morphology of TM1040 in response to attractants, compared to the control
| Attractant(s) | Responsea |
|---|---|
| Amino acids | |
| Alanine, arginine, asparagine, aspartic acid, glutamic acid, methionine, phenylalanine, proline, threonine, valine | ++ |
| Glycine, histidine, cysteine, isoleucine, leucine, lysine, serine, tryptophan, tyrosine | + |
| DMSP metabolite(s) | |
| DMSP, acrylate, MMPA | ++ |
| MPA | + |
| DMSO | − |
| Sugars | |
| Galactose, N-acetylglucosamine, sucrose | ++ |
| Glucose, maltose, glycerol | + |
| Lactose, arabinose, fructose, fucose, mannose, ribose, xylose | − |
| TCA Intermediate(s) | |
| Citrate, fumurate | ++ |
| Succinate | + |
| Alpha-ketoglutarate | − |
The strength of the response indicates the change in motile colony morphology when a chemical is spotted near the colony periphery compared to the control, no spotted chemical.
As described, BM glycerol medium was used because TM1040 can utilize glycerol as a sole carbon source, as has been observed for other roseobacters (28). However, it is known that chemotaxis behavior in other bacterial species can be affected by the availability of background nutrients (11). Indeed, when glucose was used in place of glycerol as the sole background carbon source, TM1040 chemotaxis was severely affected, and many of the chemicals that were attractants using BM glycerol failed to produce an effect in BM glucose (Table 2). The two exceptions were acrylate and succinate, which both produced a moderate response in the glucose background (Table 2). In contrast to glucose, when either succinate or alanine was used as the sole carbon source, TM1040 chemotactic behavior was similar to what had been observed in BM glycerol medium, albeit the response was often less intense. Not surprisingly, since chemotaxis relies on the establishment of chemical gradients, TM1040 did not respond to any chemical when the same chemical was incorporated into the BM motility medium (Table 2).
TABLE 2.
Change in motile colony morphology of TM1040 in response to putative attractants when grown in BM motility agar with different carbon sources
| Chemical | Change in morphology with carbon sourcea
|
|||
|---|---|---|---|---|
| Glycerol | Glucose | Succinate | Alanine | |
| Acrylate | ++ | + | ++ | + |
| Alanine | ++ | − | + | − |
| Alpha-ketoglutarate | − | − | + | − |
| DMSO | − | − | − | − |
| DMSP | ++ | − | + | + |
| Galactose | ++ | − | + | − |
| Glucose | + | − | + | + |
| Glutamic Acid | ++ | − | + | − |
| Lactose | − | − | − | − |
| Lysine | + | − | − | − |
| Maltose | + | − | ++ | ++ |
| N-acetylglucosamine | ++ | − | + | + |
| Succinate | + | + | − | ++ |
| Valine | ++ | − | + | + |
TM1040 was inoculated into the center of a BM motility agar plate containing a single carbon source (glycerol, glucose, succinate or alanine) and allowed to grow and move outwards. Chemicals were then spotted near the periphery of the motile colony, and the change in colony appearance compared to the control (no spotted chemical) was recorded on a plus/minus scale.
Quantitative chemotaxis toward pure compounds.
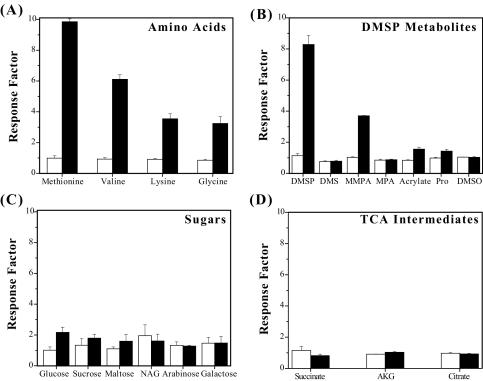
The results of the chemotaxis screening assay were used to select a subset of chemicals for further testing, using a quantitative capillary assay. As the data in Fig. 5 show, the response of TM1040 to a given attractant is concentration dependent, e.g., concentrations below 2 μM failed to elicit a significant response, while a 200 μM concentration of an attractant produce strong chemotaxis (Fig. 5A). Among the amino acids tested, methionine and valine gave the strongest response factors (means of 9.8 and 6.1, respectively), while lysine and glycine produced a similar response (mean, ca. 3.5) from TM1040. TM1040 also showed strong chemotaxis toward DMSP, which produced a response factor of 8.2 (Fig. 5B). The only breakdown product of DMSP that is a significant chemoattractant for TM1040 is MMPA (mean response factor of 3.8). In comparison, all the sugars tested produced only a weak to mild chemotactic response from TM1040 (means of <3 [Fig. 5C]). Of these sugars, TM1040 was most attracted to glucose, producing a mean response factor of 2.1. Figure 5 also shows the response of TM1040 to TCA intermediates. Of the three TCA intermediates that were tested (succinate, α-ketoglutarate, and citrate), none were found to be significant chemoattractants for TM1040, suggesting that these chemicals play a minor role in the overall response of TM1040 toward dinoflagellates. Overall, the results from the capillary assay agree with the data obtained from the qualitative assay using BM motility agar.
FIG. 5.
Quantitative measurement of the chemotactic response of Silicibacter sp. strain TM1040 to pure compounds. Chemotaxis of TM1040 was assessed using the capillary assay with a subset of potential attractants discovered using the qualitative assay. Capillaries were filled with either buffer (as a control) or 2 μM (white bars) or 200 μM (black bars) attractant. The response factor was calculated by dividing the mean number of bacteria in attractant-filled capillaries by the mean number of bacteria in buffer-filled capillaries. DMSP and amino acids produced the strongest chemotactic response from TM1040. NAG, N-acetylglucosamine; Pro, propionate; AKG, α-ketoglutarate. The values shown are the means and standard deviations (n = 12).
Chemotaxis toward P. piscicida homogenates is inhibited by externally supplied attractants.
As was noted in the data presented in Table 2, in the presence of an externally supplied attractant distributed homogeneously throughout the medium, chemotaxis toward a point source of the same attractant is inhibited because the gradient is deflated.
Using this knowledge, experiments were designed to identify the heat-labile attractants found in the dinoflagellate homogenates. As shown in Table 3 and in accord with the data in Fig. 3, the response of TM1040 to untreated dinoflagellate homogenate in the absence of any externally supplied attractant was greater than its response when the homogenate was preheated to 80°C. This relationship was altered, however, when DMSP, methionine, or valine was supplied externally in the buffer (Table 3). In these cases, the mean number of bacteria in the untreated capillaries was 3.33 × 104, 3.03 × 104, or 1.53 × 104 CFU per ml, giving a reduction in chemotaxis of 54, 58, or 78%, respectively (Table 3). An externally added attractant also reduced TM1040 chemotaxis to heated homogenates, with external valine producing the greatest reduction (74%), followed by methionine (48%) and DMSP (11%).
TABLE 3.
Reduction in chemotaxis of TM1040 toward untreated and heated P. piscicida homogenates by external addition of known attractants
| Treatmenta | No. of bacteria per capillary (CFU, 104)b with:
|
||||||
|---|---|---|---|---|---|---|---|
| Controld | Attractant added to external bufferc
|
||||||
| DMSP | % Reductione | Methionine | % Reduction | Valine | % Reduction | ||
| None | 7.24 (± 1.39) | 3.33 (± 0.15) | 54.01 | 3.03 (± 0.32) | 58.15 | 1.53 (± 0.15) | 78.87 |
| Heat | 3.51 (± 0.35) | 3.10 (± 0.16) | 11.68 | 1.80 (± 0.11) | 48.72 | 0.91 (± 0.24) | 74.07 |
Chemotaxis of TM1040 toward heated (80°C for 15 min) or untreated P. piscicida homogenates was measured using the capillary assay (see Materials and Methods).
Standard deviation is given in parentheses.
Prior to the assay, DMSP, methionine, or valine was added to external buffer containing motile cells of TM1040 at a final concentration of 200 μM and mixed to homogeneity.
The control was buffer only.
% Reduction, the percent reduction in CFU per capillary compared to the control.
DMSP in P. piscicida homogenates is destroyed by heating.
These results suggest that while DMSP is an attractant in untreated homogenates, its attractant quality is reduced upon heating, possibly due to degradation of the molecule. Destruction of 200 μM DMSP in CB was assessed after heating the samples to either 80 or 100°C for 15 min. At both temperatures, negligible (<2%) loss of DMSP was observed. These results indicate that pure DMSP is not a heat-labile molecule.
An alternative explanation for why heat reduced the chemoattractant quality of the homogenates is that heat acts indirectly through an intermediate to inactivate DMSP. This hypothesis predicts that the concentration of DMSP decreases when a homogenate is heated. The DMSP concentration in untreated and heated homogenates was measured. In untreated P. piscicida homogenates the mean concentration of DMSP was 8.21 ± 1.2 μM. After the homogenate was heated, the concentration of DMSP fell below the level of detectability (<1 μM). These data indicate that heat-activated components of P. piscicida homogenates enhance the degradation of DMSP and may explain the reduction in chemotaxis of TM1040 toward heated homogenates.
DISCUSSION
The results of the present study show that Silicibacter sp. strain TM1040, originally isolated from P. piscicida dinoflagellate cultures, senses and responds chemotactically to compounds produced by the dinoflagellate cells. Thus, this represents the first report of chemotaxis behavior of a Roseobacter clade bacterial species. Since Roseobacter species are prevalent in marine environments and abundant within blooms of DMSP-producing phytoplankton, chemotaxis to DMSP by these bacteria is likely to be an important mechanism in establishing close interactions with the dinoflagellate at both the physical and physiological levels.
Several conclusions may be drawn from these data. First, Silicibacter sp. strain TM1040 senses and responds chemotactically to dinoflagellate cell homogenates, and the response is reduced when the homogenate undergoes heat treatment prior to testing. Second, DMSP, MMPA, and amino acids elicit a strong, positive chemotactic response from Silicibacter sp. strain TM1040. Third, the concentration of DMSP in dinoflagellate cell homogenates is significantly reduced upon heating. Fourth, addition of DMSP (or amino acids) into the buffer suspending the bacteria inhibits chemotaxis to unheated dinoflagellate cell homogenates but not heat-treated homogenates. Fifth, prior starvation of the bacteria enhances their chemotactic response but also results in an ultimate decrease in swimming motility.
Cell homogenates of P. piscicida produce a strong chemotactic response from Silicibacter sp. strain TM1040 (Fig. 3). This response is specific to the dinoflagellate homogenate, which elicits a response factor that is >2.5 times greater than the response factor from either Rhodomonas algal cell homogenates or homogenates of a mixture of bacteria obtained from the dinoflagellate culture. The simplest interpretation of these results is that the dinoflagellate cell homogenates contain chemoattractant molecules that are specific to the dinoflagellate, such as DMSP. This suggestion is reinforced by the results from the chemotaxis assays, showing a strong response from TM1040 when pure DMSP is used (Fig. 4 and 5). Moreover, when DMSP is homogenously applied in the chemotaxis assays, thus eliminating the concentration gradient of this attractant required for chemotactic sensing, its presence results in a significant reduction in the chemotaxis of TM1040 to a DMSP point source (in the capillary or a spot on BM motility agar). The ability to reduce the response of TM1040 to DMSP by deflating the DMSP gradient is strong evidence for chemotaxis of this roseobacter to the dinoflagellate product.
DMSP is not the only compound that elicits a response from Silicibacter sp. strain TM1040. MMPA and amino acids such as methionine and valine also produce a strong response, which is also reduced by exogenous application of the respective attractant molecule, suggesting that TM1040 is capable of sensing more than just DMSP. This is reasonable, since most motile bacteria can sense and chemotactically respond to multiple elicitors (4) and having this capacity increases the survival of the bacteria by providing the cell with multiple inputs to control chemotaxis towards complex nutrient sources.
The response of TM1040 to heat-treated dinoflagellate cell homogenates was significantly reduced compared to the response to untreated homogenates (Fig. 3 and Table 3). One explanation for the reduced response is that heat treatment causes the loss of an important attractant molecule, such as DMSP. This explanation, however, is complicated by the knowledge that in vitro pure DMSP is not degraded by elevated temperatures (>80°C), yet heating a dinoflagellate cell homogenate reduces the concentration of DMSP in the homogenate to below detectable levels. Thus, some other component in the homogenates must, when heat treated, act to destroy DMSP. Since most enzymes are sensitive to heating, it is not likely that the loss of DMSP at 80°C is due to enzymatic cleavage, but at elevated temperatures DMSP does react with halides to produce volatile methyl halides (18). This process occurs naturally in algal cultures and is significantly enhanced by heating (18). Such a mechanism would have the overall effect of lowering the concentration of DMSP in heated homogenates and could help explain the reduced chemotaxis of TM1040 toward heated homogenates.
The loss of DMSP in the dinoflagellate cell homogenates upon heating also provides clues as to why exogenously added DMSP reduces TM1040 chemotaxis to untreated homogenates but not to heat-treated ones. In considering this, it is important to keep in mind that TM1040 responds to several attractants, not just DMSP, and bacteria respond to a gradient of the respective attractant. Anything that reduces that attractant gradient also reduces chemotaxis. Therefore, the application of exogenous DMSP adversely affects TM1040 chemotaxis to the untreated cell homogenates (Table 3), which contain DMSP (23), because the DMSP gradient is disrupted. In contrast, the heat-treated dinoflagellate cell homogenates lack DMSP, so the exogenous application of DMSP has no effect because the cells are not responding to DMSP attraction. We speculate that the bacteria are instead responding to other attractants that are still present and active in the heat-treated samples. This is borne out by the data in Table 3 showing the effect of methionine and valine addition on chemotaxis of TM1040 to the heat-treated homogenate. In both cases, addition of the exogenous amino acid results in a significant reduction in chemotaxis, suggesting that the bacteria are sensing methionine and/or valine gradients, and not DMSP gradients, when they swim towards heat-treated homogenates.
The chemotactic response of Silicibacter sp. strain TM1040 is increased when the cells are starved prior to measuring chemotaxis (Fig. 2). These results are interesting and potentially significant to the survival of TM1040, since they suggest that chemotactic behavior of this roseobacter in the oligotrophic marine environment may be modulated by the concentration of nutrients surrounding the cells. This is not unusual, and similar results have been reported with other bacteria. As an example, Wei and Bauer (31) observed a sixfold increase in the chemotactic response of Rhizobium meliloti L5-30 to 10 mM glutamine when the cells were starved for 3 h. Similarly, chemotaxis of Pseudomonas aeruginosa to inorganic phosphate is dependent upon starvation of the cells for phosphate (20).
The enhancement of chemotaxis when TM1040 is starved is a two-edged sword, however, because prolonged starvation ultimately results in the loss of swimming motility. While the mechanism responsible for the loss of motility is not known, starvation almost certainly causes the energy reserves in the cell to be depleted, and this is likely to adversely affect the rotation of the flagella on TM1040. This phenomenon, i.e., loss of motility under prolonged starvation, is frequently observed with motile marine bacteria. While most marine bacteria isolates are motile in culture (34), only a small fraction (<10%) of bacteria in natural assemblages are motile at any given time (25). This motile fraction increases to ca. 80% after 15 to 30 h of enrichment with suitable nutrients (24), presumably through prolonging the percentage of time the bacteria are motile (16). This phenomenon may be an adaptation to save energy when marine bacteria experience nutrient-depleted conditions, while also enhancing chemotactic sensitivity to nutrients if and when they become available.
Many physiological characteristics of the Roseobacter clade of bacteria, such as Silicibacter sp. strain TM1040, make them well suited for life in close proximity to dinoflagellates and algal cells. Since the area immediately surrounding a dinoflagellate is hypothesized to be a habitable niche for some marine bacteria (6), it is not surprising to find that a dinoflagellate-associated bacterium like TM1040 has mechanisms to exploit this niche. At present, these mechanisms include swimming motility and the chemotactic response of the cells to DMSP and other dinoflagellate molecules, as well as the enzymatic mechanisms to degrade and utilize DMSP (23) These are but two obvious physiological functions that enhance the survival of these bacteria when associated with a dinoflagellate cell. It comes as no surprise, therefore, that a preliminary analysis of the genome of Silicibacter sp. strain TM1040 reveals several genetic loci that may serve to enhance the interaction of this bacterium with its dinoflagellate host (unpublished data). These data suggest that TM1040 is a very good model for studying bacterium-dinoflagellate interactions.
Acknowledgments
This work was supported by grants from NOAA ECOHAB (NA86OP0492) and NIH NIEHS (ES9563) to R.B. and an American Society for Microbiology fellowship to P.D.
Footnotes
This is publication 04-651 of the Center of Marine Biotechnology.
REFERENCES
- 1.Adler, J. 1966. Chemotaxis in bacteria. Science 153:708-716. [DOI] [PubMed] [Google Scholar]
- 2.Alavi, M., T. Miller, K. Erlandson, R. Schneider, and R. Belas. 2001. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3:380-396. [DOI] [PubMed] [Google Scholar]
- 3.Alldredge, A. L., and Y. Cohen. 1987. Can microscale chemical patches exist in the sea? Microelectrode study of marine snow, fecal pellets. Science 235:689-691. [DOI] [PubMed] [Google Scholar]
- 4.Armitage, J., and R. Schmitt. 1997. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology 143:3671-3682. [DOI] [PubMed] [Google Scholar]
- 5.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, W. H., J. M. Lang, and R. Mitchell. 1974. Selective stimulation of marine bacteria by algal extracellular products. Limnol. Oceanogr. 19:833-839. [Google Scholar]
- 7.Biegala, I. C., G. Kennaway, E. Alverca, J. Lennon, D. Vaulot, and N. Simon. 2002. Identification of bacteria associated with dinoflagellates (Dinophyceae) Alexandrium spp. using tyramide signal amplification-fluorescent in situ hybridization and confocal microscopy. J. Phycol. 38:404-411. [Google Scholar]
- 8.Blackburn, N., T. Fenchel, and J. Mitchell. 1998. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science 282:2254-2256. [DOI] [PubMed] [Google Scholar]
- 9.Chambers, S. T., C. M. Kunin, D. Miller, and A. Hamada. 1987. Dimethylthetin can substitute for glycine betaine as an osmoprotectant molecule for Escherichia coli. J. Bacteriol. 169:4845-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, J. J. 1982. Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Syst. 13:291-314. [Google Scholar]
- 11.DeLoney-Marino, C. R., A. J. Wolfe, and K. L. Visick. 2003. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl. Environ. Microbiol. 69:7527-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallacher, S., K. J. Flynn, J. M. Franco, E. E. Brueggemann, and H. B. Hines. 1997. Evidence for production of paralytic shellfish toxins by bacteria associated with Alexandrium spp. (Dinophyta) in culture. Appl. Environ. Microbiol. 63:239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giovannoni, S., and M. Rappe. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 14.Gonzalez, J. M., J. S. Covert, W. B. Whitman, J. R. Henriksen, F. Mayer, B. Scharf, R. Schmitt, A. Buchan, J. A. Fuhrman, R. P. Kiene, and M. A. Moran. 2003. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 53:1261-1269. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossart, H. P., L. Riemann, and F. Azam. 2001. Bacterial motility in the sea and its ecological implications. Aquat. Microb. Ecol. 25:247-258. [Google Scholar]
- 17.Hold, G. L., E. A. Smith, M. S. Rappe, E. W. Maas, E. R. B. Moore, C. Stroempl, J. R. Stephen, J. I. Prosser, T. H. Birkbeck, and S. Gallacher. 2001. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol. Ecol. 37:161-173. [Google Scholar]
- 18.Hu, Z., and R. M. Moore. 1996. Kinetics of methyl halide production by reaction of DMSP with halide ion. Mar. Chem. 52:147-155. [Google Scholar]
- 19.Jansen, M., and T. A. Hansen. 1998. Tetrahydrofolate serves as a methyl acceptor in the demethylation of dimethylsulfoniopropionate in cell extracts of sulfate-reducing bacteria. Arch. Microbiol. 169:84-87. [DOI] [PubMed] [Google Scholar]
- 20.Kato, J., A. Ito, T. Nikata, and H. Ohtake. 1992. Phosphate taxis in Pseudomonas aeruginosa. J. Bacteriol. 174:5149-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller, M. D., and W. Korjeff-Bellows. 1996. Physiological aspects of the production of dimethylsulfoniopropionate (DMSP) by marine phytoplankton, p. 131-153. In G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, N.Y.
- 22.Kiene, R. P., L. J. Linn, and J. A. Bruton. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43:209-224. [Google Scholar]
- 23.Miller, T. R., and R. Belas. 2004. Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp. Appl. Environ. Microbiol. 70:3383-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell, J., L. Pearson, and S. Dillon. 1996. Clustering of marine bacteria in seawater enrichments. Appl. Environ. Microbiol. 62:3716-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell, J. G., L. Pearson, A. Bonazinga, S. Dillon, H. Khouri, and R. Paxinos. 1995. Long lag times and high velocities in the motility of natural assemblages of marine bacteria. Appl. Environ. Microbiol. 61:877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palleroni, N. J. 1976. Chamber for bacterial chemotaxis experiments. Appl. Environ. Microbiol. 32:729-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt, R., I. Raska, and F. Mayer. 1974. Plain and complex flagella of Pseudomonas rhodos: analysis of fine structure and composition. J. Bacteriol. 117:844-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiba, T. 1991. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst. Appl. Microbiol. 14:140-145. [Google Scholar]
- 29.Sjoblad, R. D., and R. Mitchell. 1979. Chemotactic responses of Vibrio alginolyticus to algal extracellular products. Can. J. Microbiol. 25:964-967. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, B. F., and P. T. Visscher. 1996. Metabolic pathways involved in DMSP degradation, p. 265-276. In G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, N.Y.
- 31.Wei, X., and W. D. Bauer. 1998. Starvation-induced changes in motility, chemotaxis, and flagellation of Rhizobium meliloti. Appl. Environ. Microbiol. 64:1708-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 86:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmer-Faust, R. K., M. P. de Souza, and D. C. Yoch. 1996. Bacterial chemotaxis and its potential role in marine dimethylsulfide production and biogeochemical sulfur cycling. Limnol. Oceanogr. 41:1330-1334. [Google Scholar]
- 34.Zobell, C. 1946. Marine microbiology: a monograph on hydrobacteriology. Chronica Botanica Company, Waltham, Mass.
- 35.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]