Abstract
Mesentericin Y105 is a 37-residue bacteriocin produced by Leuconostoc mesenteroides Y105 that displays antagonistic activity against gram-positive bacteria such as Enterococcus faecalis and Listeria monocytogenes. It is closely related to leucocin A, an antimicrobial peptide containing β-sheet and α-helical structures. To analyze structure-function relationships and the mode of action of this bacteriocin, we generated a collection of mesentericin derivatives. Mutations were obtained mostly by PCR random mutagenesis, and the peptides were produced by an original system of heterologous expression recently described (D. Morisset and J. Frère, Biochimie 84:569-576, 2002). Ten derivatives were obtained displaying modifications at eight different positions in the mesentericin Y105 sequence. Purified peptides were incorporated into lysophosphatidylcholine micelles and analyzed by circular dichroism. The α-helical contents of these peptides were compared and related to their respective bactericidal activities. Moreover, studies of the intrinsic fluorescence of tryptophan residues naturally occurring at positions 18 and 37 revealed information about insertion of the peptides in micelles. A model for the mode of action of mesentericin Y105 and related bacteriocins is proposed.
Bacteriocins are proteinaceous compounds produced by many lactic acid bacteria. They generally display a narrow spectrum of antibacterial activity against species closely related to the producing strain. They have been classified on the basis of their biochemical characteristics (29, 36). Class I peptides are the lantibiotics, which are small posttranslationally modified peptides that contain unusual amino acid derivatives such as lanthionine. The thermoresistant class II bacteriocins are further subdivided into three classes, namely, IIa (anti-Listeria peptides), IIb (two-component peptides), and IIc (sec-dependent bacteriocins). Class III bacteriocins are thermosensitive proteins. The fourth class, composed of complex molecules, has been disputed (45) and is currently disregarded. Among the class IIa bacteriocins, also known as pediocin-like bacteriocins, at least 24 anti-Listeria peptides were described previously (1, 3, 4, 9, 10, 16, 17, 20, 24, 26, 30-33, 39, 40, 46, 51, 53, 54, 56, 57, 60). These compounds share a YGNGVxCxxxxC consensus and a conserved disulfide bond within the N-terminal part of the peptide; they tend to differ mainly in their C-terminal domains (15). It is commonly accepted that these peptides exert bactericidal activity by forming pores in target cell membranes (9). Class IIa bacteriocins are mostly secreted from their producing cell by dedicated ATP-binding cassette (ABC) transporters and their accessory proteins, with concomitant cleavage of their N-terminal leader sequence (25, 29). However, some of these bacteriocins are exported by the general secretory pathway (sec) with cleavage of their N-terminal signal peptide (10, 32).
Mesentericin Y105 (MesY105) is a 37-amino-acid class IIa bacteriocin produced by Leuconostoc mesenteroides Y105 (26). Its sequence differs from that of leucocin A, isolated from Leuconostoc gelidum, by only two residues (A22F and I26V) (24), resulting in an enhanced bactericidal activity for MesY105 compared to that of leucocin A (21).
The three-dimensional structure, in an artificial membrane, of MesY105 remains unknown. Nevertheless, a circular dichroism (CD) study has shown that this bacteriocin is unstructured in aqueous solution and becomes structured in the anisotropic solvent trifluoroethanol (21). Moreover, structure predictions for pediocin PA-1/AcH, another class IIa bacteriocin, showed a β-sheet conformation in the N-terminal portion and an α-helix conformation in the C terminus of the peptide (8, 59). Finally, leucocin A (22), carnobacteriocin B2 (58), and sakacin P (55) structures were analyzed using nuclear magnetic resonance (NMR) spectroscopy with trifluoroethanol and phospholipid micelles as membrane-mimicking environments. All bacteriocins share a well-defined amphipathic α-helix (residues 19 to 39 for carnobacteriocin B2, residues 17 to 31 for leucocin A, and residues 18 to 33 for sakacin P) followed by an unstructured C-terminal tail. This amphipathic helix could interact with phospholipids and then disrupt the bacterial membrane (15, 42). Surprisingly, the N terminus of carnobacteriocin B2 appears highly disordered (58) whereas the corresponding regions in leucocin A and sakacin P form a well-defined antiparallel β-sheet (22, 55) despite sharing a high level of sequence similarity for the first 17 residues.
In attempts to elucidate the structure-function relationships of class II bacteriocins, many studies analyzed either the biological activity (5, 11, 18, 19, 21, 34, 41, 47) or the in vitro binding to phospholipid vesicles (7) of bacteriocin derivatives. It is generally observed that mutations are deleterious for bactericidal activity, but in some cases, they reinforce the antagonist potency (18, 34, 41). All these “enhancing” substitutions involve an increase in the net positive charge, H12K, T20K, and 44K in sakacin P (34). Binding of pediocin PA-1, studied by tryptophan fluorescence (7), showed that the K11L-H12L derivative bound weakly to the lipid vesicles. These studies thus suggest that electrostatic interactions, in the N-terminal half of bacteriocins, govern the initial peptide binding to the target cell and influence the target cell specificity. It has also been determined that target cell specificity is dependent on both the C-terminal region (17) and the C-terminal disulfide bridge, which contributes to the widening of the antimicrobial spectrum as well as to higher potency at elevated temperatures (18).
Here, we present the results of tryptophan fluorescence and CD analyses of MesY105 derivatives, obtained by randomized mutagenesis or chemical synthesis, investigating the role of specific amino acids in the binding to phospholipid vesicles, in an attempt to determine the structure and mode of action of bacteriocins.
MATERIALS AND METHODS
Strains, vectors, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α was grown in Luria-Bertani broth (50) with vigorous shaking at 37°C. Lactic acid bacteria were grown in de Man-Rogosa-Sharpe (MRS; Difco Laboratories) medium at 30°C. The Listeria ivanovii Li4(pVS2) (2) indicator strain was propagated in brain heart infusion (Difco Laboratories) at 37°C. Agar plates contained 1.5% (wt/vol) agar. Soft agar was made with 1% (wt/vol) agar. Bacteriocin-producing strains were propagated in MRS broth at 30°C. Antibiotics (Sigma-Aldrich) were added as selective agents when appropriate: erythromycin (150 μg/ml) and ampicillin (50 μg/ml) for E. coli and erythromycin (10 μg/ml) and chloramphenicol (10 μg/ml) for L. mesenteroides and L. ivanovii.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Escherichia coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 (rK−mK+) deoR thi-1 supE44 λ−gyrA96 relA1 | Invitrogen |
| Leuconostoc mesenteroides subsp. mesenteroides Y105 | Wild strain | 26 |
| Leuconostoc mesenteroides subsp. dextranicum DSM20484 | Wild strain | DSMa |
| Listeria ivanovii Li4(pVS2) | Indicator strain | 2 |
| pGEM-T easy | Cloning vector; bla | Promega |
| pMK4 | Shuttle vector E. coli-gram-positive bacteria; bla cat | 52 |
| pFBYC04 | mesYI mesCDE erm | 6 |
| pDMJF01 | mesI mesCDE erm | 43 |
| pGEM-T easy:YI | pGEM-T easy derivative; mesYI | This work |
| pDMJF:YI | pMK4 derivative; mesYI | 43 |
DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen.
Molecular techniques.
Plasmid DNA was isolated from L. mesenteroides as described by Hechard et al. (27). Restriction and modification enzymes from various sources were used as recommended by the suppliers. Transformation of L. mesenteroides by electroporation was done by the method of Raya et al. (48) with a Gene Pulser apparatus (Bio-Rad; 0.2-cm cuvettes; settings, 25 μF, 2.5 kV, and 200 Ω). Other DNA manipulations were carried out according to standard procedures (50). PCR primers were purchased from Invitrogen Corporation. Nucleotide sequences were obtained with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer) and analyzed with the ABI Prism 310 genetic analyzer (Perkin-Elmer).
Mutagenesis procedure and plasmid construction.
Base substitutions were introduced as described by Miller et al. (41) with modifications as follows. Mutagenesis reaction mixtures contained 50 U of Taq DNA polymerase (Promega) per ml; 2 ng of pFBYC04 (6) template per ml; 5 mM MgCl2; 1 mM MnCl2; 1 mM (each) dATP, dCTP, and dTTP; 30 μM dGTP; and 0.6 μM (each) primer. A 30-cycle protocol consisting of 90 s of strand denaturation at 96°C, 60 s of primer annealing at 50°C, and 60 s of primer extension at 72°C was used, on a Mastercycler personal system (Eppendorf), to amplify the mesYI codon encoding pre-MesY105 and its immunity protein MesI, with tot13 (5′-CACATAACACTCATTATTCT-3′) and tot14 (5′-CGGAATTCATCATGTAGTTTCAT-3′) primers. Amplified DNA fragments were introduced into the pGEM-T easy vector (Promega) and sequenced. Then, the mutated mesYI operons were purified and transferred into the pMK4 vector (52), leading to pDMJF:YI plasmids. These plasmids were individually transformed in L. mesenteroides DSM20484(pDMJF01) (43), and transformants were plated on MRS agar containing chloramphenicol and erythromycin.
Solid-phase synthesis of MesY105 tryptophan substitution derivatives.
[Phe18] MesY105, [Phe37] MesY105, and [1-36] MesY105, also named W18F, W37F, and Mes36 peptides, respectively, were prepared by stepwise solid-phase synthesis with 9-fluorenylmethoxy carbonyl polyamide active ester chemistry on a 433A peptide synthesizer (Applied Biosystems) as described by Fleury et al. (21).
Bacteriocin activity assays.
Antimicrobial activity in cultures was determined and quantification of bacteriocins was performed using the well diffusion method as described by Fleury et al. (21). Supernatants of overnight cultures obtained by centrifugation at 12,000 × g for 10 min at 4°C were adjusted to pH 6.0 with 1 N NaOH, heated for 30 min at 65°C, and stored at −20°C until used. Fifty-microliter aliquots of supernatants containing MesY105 or its derivatives were used to fill wells (6-mm diameter) cut in cooled soft agar plates seeded with L. ivanovii Li4(pVS2) (2) (2% [vol/vol] of overnight cultures). After 2 h at 4°C, plates were incubated at 37°C for growth, and 24 h later, the diameters of the growth inhibition zones were measured. For quantification of bacteriocin activity, 50 μl of serial dilutions in MRS broth was added to each well. The MIC was defined as the concentration of bacteriocin showing a 1-mm zone of inhibition around the well. The bacteriocin activity values presented are the results of at least three independent measurements.
Bacteriocin purification.
Wild-type and mutant bacteriocins were purified by applying the bacterial culture directly on a cation exchanger followed by reverse-phase chromatography according to the three-step method of Guyonnet et al. (23). The purity level of peptides was more than 95%, according to the high-pressure liquid chromatography elution profile of the purified extracts (data not shown). Molecular masses of the peptides were determined by mass spectrometry with a Perkin-Elmer Sciex API 165 mass spectrometer equipped with an ion spray source.
The concentration of purified bacteriocins was determined by measuring UV absorption at 280 nm, which was converted to protein concentration with molecular extinction coefficients, calculated from the contributions of individual amino acid residues.
Computer analysis.
Multiple alignments of class IIa bacteriocins were made using the Needleman-Wunsch algorithm (44) and the Blosum62 substitution matrix from the PileUp software (University of Wisconsin Genetics Computer Group package) (13). The mean hydrophobic moment of the putative helix from native MesY105 and derivatives was calculated using the normalized amino acid hydrophobicity scale and the calculation method of Eisenberg et al. (14). Amphipathy profiles and helical wheel representations were drawn using the hydrophobicity scale of Kyte and Doolittle (37). Personal computer tools were designed for the graphic representation of both the hydrophobic moment and amphipathy. The assigning of secondary structure of the leucocin A structures was made with the Promotif tool of the Protein Data Bank server (http://www.rcsb.org/pdb/). A MesY105 model was obtained by the fully automated protein structure homology-modeling SWISS-MODEL server, accessible via the ExPASy web server (http://swissmodel.expasy.org/).
CD analysis.
Purified bacteriocins were added to an acetonitrile-water solution (50%, vol/vol) or a dodecyl-lysophosphatidylcholine (LPC) preparation (molar weight [MW], 495.6; concentration varying from 250 to 300 mM) buffered at pH 5.5 with 2-(N-morpholino)ethanesulfonic acid (MES buffer; Sigma) (10 mM). All the lipid concentrations used were far above the critical micellar concentration of 7 μM (corresponding to a lipid/peptide molar ratio [Ri] around 1). The peptide concentration was fixed at 25 mg/ml (approximately 6.5 μM). Spectra were recorded between 190 and 250 nm, at 25°C, with an ISA Jobin Yvon CD6 CD spectrophotometer (Jobin Yvon) calibrated with epiandrosterone. Data were collected using a quartz cell with 1- and 3-mm path lengths for aqueous and LPC solutions, respectively. All the spectra were recorded at least fivefold with a bandwidth of 1 nm, a scan speed of 1 nm/s, and a time constant of 1s. In all cases, baseline scans of acetonitrile-water solution were subtracted from the experimental readings. Results are recorded in units of absorbance of variability between right circular and left circular polarized light (ΔA). Then, spectra were converted in molar absorbance units per residue [Δɛ/n (square centimeters per mole)] with the following equation: Δɛ = ΔA · MW/C · n · l, where MW is the MW of the peptide (grams per mole), n is the residue number in the peptide, C is the peptide concentration (grams per liter), and l is the path length.
Estimation of protein helical content for each bacteriocin was done using the following formula: Hα = 100 · (θ222nm − 3,000)/−39,000 (θ = 3,298 Δɛ), where Hα is the α-helical content and θ is the molar ellipticity per residue (degrees · square centimeter · decimole−1).
Measurements of binding of MesY105 and derivatives.
The binding to lipids of MesY105 and its derivatives was tested by monitoring changes in the fluorescence of tryptophan residues, when the peptides were exposed to LPC micelles. Tryptophan fluorescence was measured with a spectrofluorometer (Fluoromax; Spex Industries). Emission spectra from 300 to 400 nm, with an increment of 1 nm, were recorded upon excitation at 295 nm. LPC from a stock solution (5 g/liter, i.e., 10 mM) was added in a stepwise manner to a 6.5 μM MesY105 solution in 50% (vol/vol) acetonitrile-water, until a lipid/peptide molar ratio (Ri) of 40 was reached. The peptide-lipid mixture was continuously stirred in cuvettes, and the fluorescence emission spectra were recorded after each lipid addition. For each lipid concentration, equilibrium was rapidly reached since emission spectra were the same after 1 and 3 min of incubation. The baseline of fluorescence measured for the acetonitrile-water solution in the absence of peptide was subtracted from each fluorescence spectrum.
The tryptophan emission spectrum was analyzed to determine the maximum emission wavelength (λmax) and the fluorescence intensity (F). The intensity value was obtained by integrating the emission curve and was corrected for dilution effects. Fluorescence titration curves relate blueshift (Δλmax = λmax0 − λmax, in nanometers), or relative intensity variation (F − F0/F0), to the lipid/peptide concentration ratio (Ri) (the subscript zero denotes the value obtained without lipid).
RESULTS
Production and purification of MesY105 derivatives.
Our heterologous expression system was used to produce and secrete MesY105 derivatives (43) (Fig. 1). Mutations were introduced into the operon mesYI encoding MesY105 and MesI by a PCR random mutagenesis procedure derived from the work of Miller et al. (41). The conditions used generate about one substitution per 100 bp of DNA. The DNA region of the mesYI operon was sequenced from 47 independent pGEM-T easy recombinant plasmids. Only 12 displayed a single codon replacement in the mesY gene. These 12 modified mesYI operons were introduced independently into pMK4 (52), an E. coli-lactic acid bacterium shuttle vector, leading to the pDMJF:YI plasmids, and then transferred into L. mesenteroides DSM20484(pDMJF01) (43). The pDMJF01 plasmid expresses the MesY105 dedicated transport system, allowing the secretion of the bacteriocin if the mesY gene is expressed from a second plasmid (43). The L. mesenteroides DSM20484 transformants harboring the two plasmids, pDMJF01 and pDMJF:YI, were grown at 30°C, and bacteriocins were purified from the culture supernatant as described previously (23).
FIG. 1.
Overview of MesY105 mutants characterized in the present work. The W18F, W37F, and Mes36 peptides were obtained by solid-phase chemical synthesis. The other peptides were obtained by PCR mutagenesis and then expressed and secreted by L. mesenteroides DSM20484.
Bacteriocin gene modifications involving a cysteine codon (Y3C, C14Y/S, R28C, and W37C) lead to an absence of peptide production. We assume that these substitutions could be responsible for an incorrect maturation activity or an altered secretion. In contrast, the seven other variants were produced and the resulting high-pressure liquid chromatography retention times varied from 22 to 25 min, depending on the substitution. The mass spectrometry analyses of these bacteriocin variants showed that all of them had an observed mass 2 Da lower than that of the corresponding theoretical reduced molecule, suggesting that all cysteine residues were in the disulfide form as in MesY105 (data not shown).
Computer analysis.
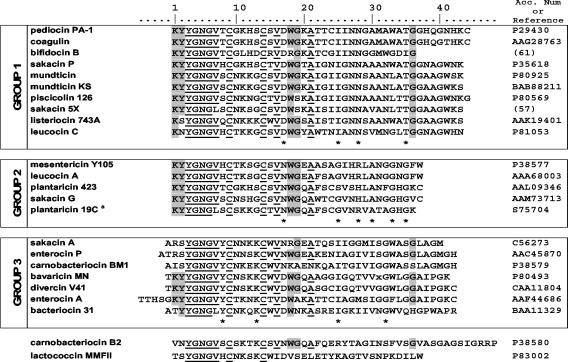
The amino acid sequences of the 24 class IIa characterized bacteriocins are shown in Fig. 2, highlighting their relatively well conserved residues. This figure shows a grouping of the bacteriocins, in three main groups, on the basis of the dendrogram produced by the PileUp software. Pediocin PA-1 is representative of the first group composed of 43- and 44-amino-acid bacteriocins (except bifidocin B) with the characteristic residues Asp17, Ile25, Asn27-Asn28, and Thr35. The second group, corresponding to mesentericin-like bacteriocins, is composed of shorter peptides (36 to 37 amino acids) with Asn17, Gly25, Arg28 or His28, Ala30, Gly33, and Gly35 as characteristic residues. The third group is formed by peptides with elongated N termini. The residues Tyr8, Lys13, Ile25, and Gly32 are typical of this group. Two bacteriocins, carnobacteriocin B2 and lactococcin MMFII, appeared atypical and could not be classified in any group.
FIG. 2.
Multiple sequence alignment of known class IIa bacteriocins. Residues conserved ≥90% are underlined. Residues conserved ≥65% are highlighted. Characteristic residues of each group are underlined with an asterisk. Only a partial sequence is shown for plantaricin 19C, indicated by a superscript “a.”
This subgrouping is in agreement with previously published works (15, 17, 19, 42), which were also based on sequence similarities in the C-terminal portion of the peptide. Nevertheless, in the work of Fimland et al. (19) (i) lactococcin MMFII and carnobacteriocin B2 are not atypical peptides and (ii) the first group includes enterocin A and divercin V41. So, all three groups proposed herein are better defined, and the few typical positions (Fig. 2) are sufficient to classify a new class IIa bacteriocin in one of these groups.
The five group 2 bacteriocins can be sorted into two subgroups based on the number of disulfide bridges (one or two). MesY105 and leucocin A have one disulfide bridge and have highly similar sequences, as they differ by only two residues. The MesY105 variants G13E, S15F, W18F, and A21V affect conserved residues within group 2, while the derivatives H8Y/L, A24P, and W37F concern specific positions of the group 2 bacteriocins with one disulfide bridge. In the peptide R28H, the mutation involves one of the residues that characterize the group 2 peptides.
Solution structures of leucocin A (22), analyzed by NMR spectroscopy in membrane-mimicking environments, indicated that the region encompassing residues 2 to 31 adopts a well-defined tertiary structure, including a three-stranded antiparallel β-sheet (residues 2 to 16) followed by an amphiphilic helix (residues 17 to 31). Analysis of the leucocin A Protein Data Bank structure files 1cw6, 3leu, and 2leu by Promotif shows a residue 19 to 29 helix for the two former sequences and a residue 18 to 29 helix for the latter. Due to the slight discrepancies in secondary structure allocations, we used an α-helical segment spanning residues 18 to 30. The same helix segment was attributed to the putative MesY105 structure because (i) there was high sequence similarity between MesY105 and leucocin A (94% identity), (ii) predictions of MesY105 secondary structure suggested that the peptide could be configured as an amphipathic helix spanning residues 17 to 31 (37), and (iii) automatic molecular modeling with the use of 2leu and 1cw6 as templates provided a satisfactory MesY105 model preserving the α-helical structure (C-alpha root mean square values: 2leu-1cw6 = 3.60 Å, MesY105-1cw6 = 3.52 Å, and MesY105-2leu = 0.41 Å).
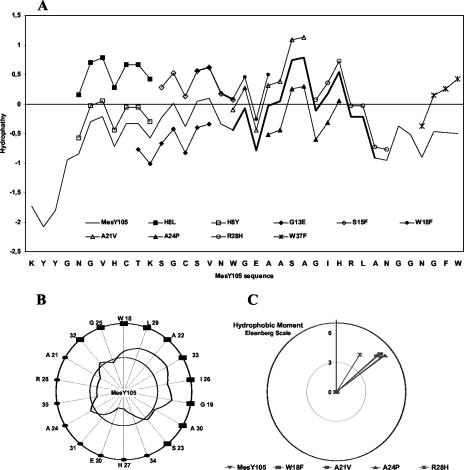
Hydropathy profiles of MesY105 and its derivatives calculated according to the method of Kyte and Doolitle (37) are presented in Fig. 3A. Both the N terminus and the C-terminal tails appear to be hydrophilic, and the H8L, W18F, S15F, and W37F substitutions seem to make the peptides more hydrophobic. In contrast, the helical segment of the peptide is more hydrophobic. The helical wheel projection (Fig. 3B) clearly shows the amphipathic character of the α-helix segment, with one nonpolar half-surface and one polar half-surface. For the variants of the helix segment, no dramatic changes in the hydrophobic moment (Fig. 3C) were observed, except for the R28H peptide, which showed a 20° change in orientation in the lipid-water interface plan (angle value); also, a significant decrease in its amphipathic character was revealed by the hydrophobic moment value (4.46 compared to 5.64 for the native bacteriocin).
FIG. 3.
Hydrophobicity analysis of MesY105 and derivative peptides. (A) Hydropathy profiles drawn, with use of the Kyte and Doolittle table, with a seven-residue mobile window. Negative and positive values are for hydrophilic and hydrophobic regions, respectively. The MesY105 profile is shown as a continuous line without any symbol (the helix segment is extra thick). Only changes in the hydropathy profile of mutated peptides are shown. (B) Helical wheel representation of the putative MesY105 helix. The curve shows the hydropathy for a three-residue window moving on the wheel surface and with use of the Kyte and Doolittle scale. The inner circle is for the hydropathy zero. Full squares indicate the hydrophobic helix surface. (C) Hydrophobic moment value and angle for an ideal MesY105 W18-A30 α-helix. Calculations, with use of the hydrophobicity scale of Eisenberg et al. (14), were made for MesY105 and the variants mutated in the W18-A30 segment. The y axis shows the moment value and is used as the reference for the moment angle (origin = helix center; direction = first residue in the helix, i.e., W18).
CD analysis in aqueous buffer and LPC micelles.
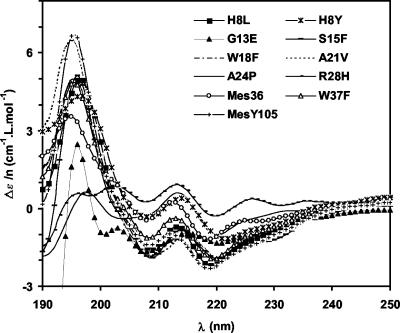
In acetonitrile-MES-buffered water solutions, the CD spectra of MesY105 and its derivatives displayed an important negative value of molar absorbance at 200 nm that is characteristic of a highly unordered structure. Upon addition of dodecyl-LPC micelles and except for R28H, most of these spectra presented characteristics of a helicoid structure with negative maxima between 208 and 222 nm and positive maxima between 195 and 197 nm (Fig. 4).
FIG. 4.
CD spectra of MesY105 and its derivatives in the presence of LPC micelles. Δɛ/n is the variation of molar absorbance per residue.
The CD spectra were also analyzed to estimate the quantitative distribution of secondary structure in aqueous and micellar solutions (Table 2). Analysis shows that, in aqueous solution, a large part of the peptides was unstructured (45 to 77%), depending on their sequence. Generally, upon binding to micelles, the percentage of unordered structure decreased, except for G13E, A24P, and above all R28H, whereas the helical structure increased, except for R28H. In LPC micelles, the helix content of the R28H, Mes36, and A24P derivatives greatly differed, since it was only 9, 14, and 15%, respectively, compared to 24% for MesY105.
TABLE 2.
Ellipticity, tryptophan fluorescence emission shift, and activities of MesY105 and its derivativesa
| Peptide | % by CDb
|
Fluorescence emission shift (Δλmaxc [nm]) | Biological activity (MICd [nM]) | |||
|---|---|---|---|---|---|---|
| Aqueous buffer
|
C13-LPC micelles (Ri = 40)
|
|||||
| Unstructured | Hα | Unstructured | Hα | |||
| MesY105 | 68 | 7 | 26 | 24 | 9 | 1.6 |
| H8Y | 50 | 10 | 59 | 17 | 11 | 31 |
| H8L | 64 | 9 | 31 | 22 | 10 | 83 |
| G13E | 45 | 13 | 46 | 19 | 6 | 130 |
| S15F | 58 | 8 | 37 | 23 | 10 | 4.1 |
| W18F | 76 | 5 | 27 | 24 | 9 | 21 |
| A21V | 61 | 6 | 37 | 24 | 13 | 73 |
| A24P | 58 | 9 | 60 | 15 | 4 | 9,700 |
| R28H | 66 | 8 | 73 | 9 | 1 | 20,000 |
| W37F | 77 | 4 | 37 | 21 | 14 | 47 |
| Mes36 | 76 | 6 | 52 | 14 | 6 | 27,000 |
Standard deviations are less than 5%.
Percentage of amino acid residues in unstructured or α-helix state.
Change in the maximum fluorescence emission wavelength observed upon addition of LPC micelles.
MIC data were obtained with at least three independent activity assays against L. ivanovii Li4(pVS2) (2).
Analysis of the antagonistic activity of the MesY105 derivative peptides.
The antimicrobial activity of the purified bacteriocins was determined against the indicator strain L. ivanovii Li4(pVS2) (2). Each derivative exhibits lower antagonistic activity than that of MesY105 (Table 2). Three groups of mutants can be distinguished according to their MICs. The first one is composed of peptides A24P, R28H, and Mes36, whose MICs are dramatically high (from 9.7 to 27 μM) compared to that of MesY105 (1.6 nM). The second group (W18F, H8Y, W37F, A21V, H8L, and G13E) consists of peptides with low activity (MICs from 20 to 130 nM). Only S15F has a moderately reduced activity (Table 2).
MesY105 and Mes36 MICs (1.6 nM and 27 μM, respectively) are quite different from the previously published values (34 nM and 6 μM, respectively) (21). Such differences could be related to the use of two different indicator strains for the MIC determination, i.e., L. ivanovii BUG496 in the previous study (21) and L. ivanovii Li4(pVS2) in this work.
Binding of MesY105 and derivatives to lipid micelles.
Binding of MesY105 derivatives was monitored by determining the changes in the tryptophan fluorescence parameters, F and λmax, when LPC was added to a peptide solution in order to form micelles.
When LPC was omitted, the emission spectrum of MesY105 (25 mg/ml, MES buffer, pH 5.5) displays a maximum emission wavelength of 356 nm. The two peptides, W37F and Mes36, devoid of tryptophan at position 37, present a maximum emission wavelength of 355 nm (data not shown). Interestingly, W18F, which lacks the tryptophan at position 18, displayed a maximum emission wavelength of 361 nm (data not shown). These results show that, in the W18F variant, tryptophan 37 was probably fully exposed to water as revealed by the important shift in λmax while, in the Mes36 and W37F peptides, Trp18 was less exposed to water. Moreover, in MesY105, it seems that both tryptophan residues were in an aqueous environment but not fully exposed to water. One hypothesis would be that, in the native bacteriocin, both tryptophans were partially shielded from the solvent.
Upon addition of the lipid micelles, the emission intensity of MesY105 slightly increased and the λmax shifted to a lower wavelength (blueshift) (Table 2). This decrease in emission wavelength and increase in emission intensity are characteristic of the translocation of tryptophan(s) from a hydrophilic and polar to a hydrophobic and nonpolar solvent (38). This result means that MesY105 bound to the lipid micelles and that the tryptophan residue(s) integrated into the hydrophobic phase of the lipid bilayer. The W18F peptide behaved similarly to MesY105 upon the addition of lipids (Table 2), with a lower fluorescence intensity because only one tryptophan residue was present (data not shown). In contrast, Mes36 had a slightly weaker blueshift (compared to MesY105) and a very weak increase in fluorescence intensity. The W37F peptide shows a significant blueshift (14 nm) but a moderate increase in emission intensity. These results suggest that both tryptophans have different locations in the membrane.
The binding of the other MesY105 derivatives to LPC micelles was also monitored (Table 2). The R28H, A24P, and G13E peptides seemed to bind weakly to the micelles compared to MesY105, as they displayed a very slight blueshift in tryptophan fluorescence. The H8Y, H8L, and S15F peptides bound to the micelles in a way similar to that of MesY105, and in contrast, A21V penetrated deeper in the micelles (Δλmax = 13 instead of 9 nm for MesY105).
DISCUSSION
Ten MesY105 derivatives were produced, purified, and then characterized for their bactericidal activity, secondary structure, and membrane binding to phospholipid micelles. The substitutions occurred along the bacteriocin amino acid sequence and included partially or well-conserved (Gly13, Trp18, Ala21, and Trp37) as well as nonconserved (His8, Ser15, Ala24, and Arg28) amino acids among class IIa bacteriocins (Fig. 2).
In an aqueous environment, MesY105 and its derivative peptides were mainly in an unordered structure (more than 45%), as shown previously for class IIa bacteriocins (21, 22, 58, 59). In contrast, in LPC micelles, folding of MesY105 was clearly observed by CD (Table 2); this phenomenon was previously detected by NMR studies of leucocin A in dodecylphosphocholine micelles (22).
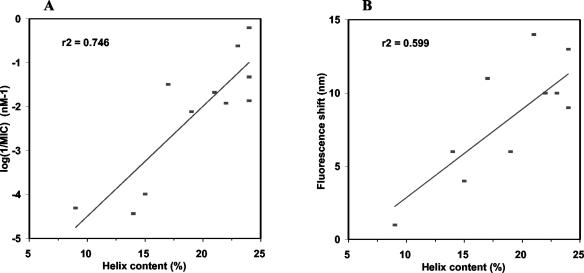
All the derivatives displayed a weaker bactericidal activity than that of MesY105, and most of them harbored modifications in their helix content wherever the mutations were localized. Two weakly defined linear relationships are observed between the helix content and either the activity expressed by log(1/MIC) (Fig. 5A) or the fluorescence emission shift (Fig. 5B). The former suggests that the length of the α-helix would be one of the main factors leading to biological activity, while the latter would be correlated with the interactions of the lipid micelles with Trp18 localized in the putative helix segment since Trp18 (W37F shift, 14 nm) would be more deeply buried in the hydrophobic environment of the lipid chains than would Trp37 (W18F shift, 9 nm) (Table 2).
FIG. 5.
Relation between the helix content and the biological activity (A) or the fluorescence emission shift (B). The antilisterial activity is expressed as the inverse of the MIC.
The A24P substitution led to one of the most altered activities (Table 2). Since proline is well known as a helix breaker, a deep change in helix content was expected and evidenced by the CD data (Table 2). This structural change leading to a very weak binding to the micelles (Table 2) would be sufficient to explain the biological response of the A24P derivative.
Despite the fact that serine 15 is quite conserved in bacteriocin groups 1 and 2 (10 peptides among 15) (Fig. 2), the S15F mutation resulted in the least pronounced effect on the biological activity (Table 2). Such a limited effect from replacing a polar residue with a hydrophobic and aromatic residue was unexpected, but in most of the sequences of group 3 bacteriocins, tryptophan, another aromatic amino acid, is present at the same position (Fig. 2).
Among the substitutions localized in the N-terminal moiety, G13E exhibits the most deleterious effect on the activity. Its helix content would be sufficient to account for both its low biological activity and the emission shift in fluorescence. The G13E substitution introduces a negative charge in the positive segment of the N-terminal half of the bacteriocins that is proposed to be involved in the primary binding of the peptide to the membrane (8, 34).
The mutations H8L and H8Y, which completely eliminate the positive charge at position 8, were less deleterious than the G13E mutation, which introduces a negative charge. As previously suggested (34), it appears that a positive charge at position 8 is not required for optimal biological activity. This is consistent with the fact that only 5 of the 24 bacteriocins (Fig. 2) display a positive charge at position 8. However, all peptides possess an N-terminal eighth amino acid with polar properties. The activity of H8Y, which is double that of H8L, demonstrates the requirement of a polar residue at this position.
Four mutations are localized in the amphipathic helix, which is considered a major element in target selectivity and antagonistic activity. Indeed, this helix is supposed to penetrate the membrane and promote leakage of the cytosolic material, either by forming channels through the membrane or by locally disrupting the bilayer (15, 17, 19, 42).
Substitution R28H-induced dramatic changes in LPC micelle interaction as well as in peptide folding (Table 2) could probably account for the pronounced reduction in the biological activity. However, the effects of this conservative mutation were rather surprising, since similar conservative mutations (i.e., H8K and H12K) slightly affected the sakacin P activity (34). Moreover, a modification of Arg28 was expected to have an impact on activity. At this position, Arg or His residues are present only in group 2 bacteriocins; furthermore, Arg is present solely in peptides with one disulfide bridge while His is observed in those with two bridges. The R28H mutation induced a decrease in the helix hydrophobic moment value, a probable 20° rotation of the helix in the membrane plane (Fig. 3C), and, above all, the loss of the long hydrophobic arginine side chain. Indeed, Arg is known as a membrane-anchoring and positioning residue in several transmembrane α-helical peptides (12, 35, 49). In MesY105, Arg28 could exhibit a similar behavior referred to as “snorkeling,” which involves both electrostatic and hydrophobic interactions between arginine and phospholipids.
Both the A21V and W18F peptides, mutated at highly conserved positions, were among the most strongly active derivatives. These derivatives fully conserved their helical structure and their ability to bind to lipid micelles (Table 2). This result was expected for the first derivative because the A21V mutation is conservative, whereas the W18F derivative lost the hydroxyl group, but this substitution maintained the local bulk and the aromatic property. In these peptides, the hydrophobicity of the N-terminal half of the helix is increased, so that the two helix moieties have similar hydrophobicities (Fig. 3). Thus, the A21V and W18F helices would be oriented parallel to the membrane plane and the Trp18 or Phe18 residue would sink deeper into the lipidic phase, respectively, as suggested for the A21V peptide by the increase in the fluorescence shift (Table 2). Such an orientation would be less favorable for the subsequent formation of transmembrane channels, suggesting that the helix extent is not sufficient per se to explain the antagonist activity. Moreover, the comparison of the MesY105, W18F, and A21V peptides indicates that an increased N-terminal helix hydrophobicity results in a reduction in biological activity, as long as aromaticity is preserved. This is in accordance with the suggestion that at position 18 (and also 41) in sakacin P, aromaticity is more important for bactericidal activity than is hydrophobicity (19).
A tryptophan is conserved at position 41 or 37 of the group 1 or group 2 peptides, respectively, among the bacteriocins with one disulfide bridge. In MesY105, the W37F substitution resulted in a greater reduction in bactericidal activity in comparison to that for W18F (Table 2) as observed with the W37F and W18F sakacin P derivatives (19). The W37F peptide was not significantly affected in its helical structure, in contrast to Mes36, which had a very weak α-helical content and also exhibited a dramatic fall in antagonistic activity (Table 2). This confirmed that Trp37 would be not essential for helix promotion but would play a critical role in helix stabilization in the membrane.
Fimland and coworkers (19) proposed a hairpin structural model for all the group 1 bacteriocins. This model was confirmed by NMR studies on sakacin P (55) and was also suggested for group 2 bacteriocins (19). The results presented herein are in general agreement with such a hairpin-like structure stabilized by Trp18 and either a C-terminal disulfide bridge or a C-terminal Trp. This suggests that some modifications are required to adapt this model to group 2 bacteriocins. like MesY105. The oblique orientation of the helix at the hydrophobic-hydrophilic membrane interface at an angle of 30 to 50° (19) would allow positioning of both Trp18 and Trp37 in this interface. Trp18 is more deeply buried than Trp37 in the apolar phase of the membrane as suggested by its predicted position in the hydrophobic face of the helix and according to its observed higher fluorescence shift. The location of the C-terminal Trp37 could be stabilized by hydrophobic interactions between Phe36 and the membrane core. Moreover, positioning the C-terminal end of the helix would involve the Arg28 residue by a snorkeling process.
In conclusion, the present work shows that all the studied residues in MesY105 are important for bacteriocin activity but that their contribution varies in terms of magnitude. Trp37 and Arg28 are the most important, presumably because they contribute to stabilizing the helical portion in a position crucial for antimicrobial activity. All of the mutations localized in the N-terminal half of the peptide decrease the helix content to various extents. Further structural and biological studies are required to understand the antibacterial activity of MesY105, and other class IIa peptides, since the mode of action on target bacterial cells may require an initial specific interaction of the bacteriocin with a proteinaceous docking molecule (28).
Acknowledgments
We sincerely thank Lars Axelsson for generously providing L. ivanovii Li4(pVS2). Manilduth Ramnath is gratefully acknowledged for valuable review of the text.
D.M. is supported by grants from the MENRT. This work is partially supported by Rhodia Food Company.
REFERENCES
- 1.Atrih, A., N. Rekhif, A. J. Moir, A. Lebrihi, and G. Lefebvre. 2001. Mode of action, purification and amino acid sequence of plantaricin C19, an anti-Listeria bacteriocin produced by Lactobacillus plantarum C19. Int. J. Food Microbiol. 68:93-104. [DOI] [PubMed] [Google Scholar]
- 2.Axelsson, L., T. Katla, M. Bjornslett, V. G. Eijsink, and A. Holck. 1998. A system for heterologous expression of bacteriocins in Lactobacillus sake. FEMS Microbiol. Lett. 168:137-143. [DOI] [PubMed] [Google Scholar]
- 3.Aymerich, T., H. Holo, L. S. Havarstein, M. Hugas, M. Garriga, and I. F. Nes. 1996. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl. Environ. Microbiol. 62:1676-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennik, M. H., B. Vanloo, R. Brasseur, L. G. Gorris, and E. J. Smid. 1998. A novel bacteriocin with a YGNGV motif from vegetable-associated Enterococcus mundtii: full characterization and interaction with target organisms. Biochim. Biophys. Acta 1373:47-58. [DOI] [PubMed] [Google Scholar]
- 5.Bhugaloo-Vial, P., J. P. Douliez, D. Moll, X. Dousset, P. Boyaval, and D. Marion. 1999. Delineation of key amino acid side chains and peptide domains for antimicrobial properties of divercin V41, a pediocin-like bacteriocin secreted by Carnobacterium divergens V41. Appl. Environ. Microbiol. 65:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biet, F., J. M. Berjeaud, R. W. Worobo, Y. Cenatiempo, and C. Fremaux. 1998. Heterologous expression of the bacteriocin mesentericin Y105 using the dedicated transport system and the general secretion pathway. Microbiology 144:2845-2854. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., R. D. Ludescher, and T. J. Montville. 1997. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phospholipid vesicles. Appl. Environ. Microbiol. 63:4770-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y., R. Shapira, M. Eisenstein, and T. J. Montville. 1997. Functional characterization of pediocin PA-1 binding to liposomes in the absence of a protein receptor and its relationship to a predicted tertiary structure. Appl. Environ. Microbiol. 63:524-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chikindas, M. L., M. J. Garcia-Garcera, A. J. Driessen, A. M. Ledeboer, J. Nissen-Meyer, I. F. Nes, T. Abee, W. N. Konings, and G. Venema. 1993. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC 1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl. Environ. Microbiol. 59:3577-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cintas, L. M., P. Casaus, L. S. Havarstein, P. E. Hernandez, and I. F. Nes. 1997. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 63:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbier, C., F. Krier, G. Mulliert, B. Vitoux, and A. M. Revol-Junelles. 2001. Biological activities and structural properties of the atypical bacteriocins mesenterocin 52B and leucocin B-TA33a. Appl. Environ. Microbiol. 67:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Planque, M. R., J. A. Kruijtzer, R. M. Liskamp, D. Marsh, D. V. Greathouse, R. E. Koeppe II, B. de Kruijff, and J. A. Killian. 1999. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J. Biol. Chem. 274:20839-20846. [DOI] [PubMed] [Google Scholar]
- 13.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenberg, D., R. M. Weiss, and T. C. Terwilliger. 1984. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA 81:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24:85-106. [DOI] [PubMed] [Google Scholar]
- 16.Ferchichi, M., J. Fr[grave]ere, K. Mabrouk, and M. Manai. 2001. Lactococcin MMFII, a novel class IIa bacteriocin produced by Lactococcus lactis MMFII, isolated from a Tunisian dairy product. FEMS Microbiol. Lett. 205:49-55. [DOI] [PubMed] [Google Scholar]
- 17.Fimland, G., O. R. Blingsmo, K. Sletten, G. Jung, I. F. Nes, and J. Nissen-Meyer. 1996. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl. Environ. Microbiol. 62:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fimland, G., L. Johnsen, L. Axelsson, M. B. Brurberg, I. F. Nes, V. G. Eijsink, and J. Nissen-Meyer. 2000. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J. Bacteriol. 182:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fimland, G., V. G. Eijsink, and J. Nissen-Meyer. 2002. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508-9515. [DOI] [PubMed] [Google Scholar]
- 20.Fimland, G., K. Sletten, and J. Nissen-Meyer. 2002. The complete amino acid sequence of the pediocin-like antimicrobial peptide leucocin C. Biochem. Biophys. Res. Commun. 295:826-827. [DOI] [PubMed] [Google Scholar]
- 21.Fleury, Y., M. A. Dayem, J. J. Montagne, E. Chaboisseau, J. P. Le Caer, P. Nicolas, and A. Delfour. 1996. Covalent structure, synthesis, and structure-function studies of mesentericin Y 105(37), a defensive peptide from gram-positive bacteria Leuconostoc mesenteroides. J. Biol. Chem. 271:14421-14429. [DOI] [PubMed] [Google Scholar]
- 22.Fregeau Gallagher, N. L., M. Sailer, W. P. Niemczura, T. T. Nakashima, M. E. Stiles, and J. C. Vederas. 1997. Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 36:15062-15072. [DOI] [PubMed] [Google Scholar]
- 23.Guyonnet, D., C. Fremaux, Y. Cenatiempo, and J. M. Berjeaud. 2000. Method for rapid purification of class IIa bacteriocins and comparison of their activities. Appl. Environ. Microbiol. 66:1744-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hastings, J. W., M. Sailer, K. Johnson, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1991. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J. Bacteriol. 173:7491-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Havarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 26.Hechard, Y., B. Dérijard, F. Letellier, and Y. Cenatiempo. 1992. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. J. Gen. Microbiol. 138:2725-2731. [DOI] [PubMed] [Google Scholar]
- 27.Hechard, Y., J. M. Berjeaud, and Y. Cenatiempo. 1999. Characterization of the mesB gene and expression of bacteriocins by Leuconostoc mesenteroides Y105. Curr. Microbiol. 39:265-269. [DOI] [PubMed] [Google Scholar]
- 28.Hechard, Y., and H. G. Sahl. 2002. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84:545-557. [DOI] [PubMed] [Google Scholar]
- 29.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jack, R. W., J. Wan, J. Gordon, K. Harmark, B. E. Davidson, A. J. Hillier, R. E. Wettenhall, M. W. Hickey, and M. J. Coventry. 1996. Characterization of the chemical and antimicrobial properties of piscicolin 126, a bacteriocin produced by Carnobacterium piscicola JG126. Appl. Environ. Microbiol. 62:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaiser, A. L., and T. J. Montville. 1996. Purification of the bacteriocin bavaricin MN and characterization of its mode of action against Listeria monocytogenes Scott A cells and lipid vesicles. Appl. Environ. Microbiol. 62:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalmokoff, M. L., S. K. Banerjee, T. Cyr, M. A. Hefford, and T. Gleeson. 2001. Identification of a new plasmid-encoded sec-dependent bacteriocin produced by Listeria innocua 743. Appl. Environ. Microbiol. 67:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamoto, S., J. Shima, R. Sato, T. Eguchi, S. Ohmomo, J. Shibato, N. Horikoshi, K. Takeshita, and T. Sameshima. 2002. Biochemical and genetic characterization of mundticin KS, an antilisterial peptide produced by Enterococcus mundtii NFRI 7393. Appl. Environ. Microbiol. 68:3830-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kazazic, M., J. Nissen-Meyer, and G. Fimland. 2002. Mutational analysis of the role of charged residues in target-cell binding, potency and specificity of the pediocin-like bacteriocin sakacin P. Microbiology 148:2019-2027. [DOI] [PubMed] [Google Scholar]
- 35.Killian, J. A., and G. von Heijne. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429-434. [DOI] [PubMed] [Google Scholar]
- 36.Klaenhammer, T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-85. [DOI] [PubMed] [Google Scholar]
- 37.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydrophobic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 38.Lakowicz, J. R. 1983. Principles of fluorescence spectroscopy, p. 111-150. Plenum Press, New York, N.Y.
- 39.Le Marrec, C., B. Hyronimus, P. Bressollier, B. Verneuil, and M. C. Urdaci. 2000. Biochemical and genetic characterization of coagulin, a new antilisterial bacteriocin in the pediocin family of bacteriocins, produced by Bacillus coagulans I(4). Appl. Environ. Microbiol. 66:5213-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metivier, A., M. F. Pilet, X. Dousset, O. Sorokine, P. Anglade, M. Zagorec, J. C. Piard, D. Marion, Y. Cenatiempo, and C. Fremaux. 1998. Divercin V41, a new bacteriocin with two disulphide bonds produced by Carnobacterium divergens V41: primary structure and genomic organization. Microbiology 144:2837-2844. [DOI] [PubMed] [Google Scholar]
- 41.Miller, K. W., R. Schamber, O. Osmanagaoglu, and B. Ray. 1998. Isolation and characterization of pediocin AcH chimeric protein mutants with altered bactericidal activity. Appl. Environ. Microbiol. 64:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moll, G. N., S. Brul, W. N. Konings, and A. J. Driessen. 2000. Comparison of the membrane interaction and permeabilization by the designed peptide Ac-MB21-NH2 and truncated dermaseptin S3. Biochemistry 39:11907-11912. [DOI] [PubMed] [Google Scholar]
- 43.Morisset, D., and J. Frere. 2002. Heterologous expression of bacteriocins using the mesentericin Y105 dedicated transport system by Leuconostoc mesenteroides. Biochimie 84:569-576. [DOI] [PubMed] [Google Scholar]
- 44.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 45.Nes, I. F., D. B. Diep, L. S. Havarstein, M. B. Brurberg, V. Eijsink, and H. Holo. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek 70:113-128. [DOI] [PubMed] [Google Scholar]
- 46.Quadri, L. E., M. Sailer, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1994. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J. Biol. Chem. 269:12204-12211. [PubMed] [Google Scholar]
- 47.Quadri, L. E., L. Z. Yan, M. E. Stiles, and J. C. Vederas. 1997. Effect of amino acid substitutions on the activity of carnobacteriocin B2. Overproduction of the antimicrobial peptide, its engineered variants, and its precursor in Escherichia coli. J. Biol. Chem. 272:3384-3388. [DOI] [PubMed] [Google Scholar]
- 48.Raya, R. R., C. Fremaux, G. L. De Antoni, and T. R. Klaenhammer. 1992. Site-specific integration of the temperate bacteriophage φadh into the Lactobacillus gasseri chromosome and molecular characterization of the phage (attP) and bacterial (attB) attachment sites. J. Bacteriol. 174:5584-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ridder, A. N., S. Morein, J. G. Stam, A. Kuhn, B. de Kruijff, and J. A. Killian. 2000. Analysis of the role of interfacial tryptophan residues in controlling the topology of membrane proteins. Biochemistry 39:6521-6528. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Simon, L., C. Fremaux, Y. Cenatiempo, and J. M. Berjeaud. 2002. Sakacin G, a new type of antilisterial bacteriocin. Appl. Environ. Microbiol. 68:6416-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan, M. A., R. E. Yasbin, and F. E. Young. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29:21-26. [DOI] [PubMed] [Google Scholar]
- 53.Tichaczek, P. S., R. F. Vogel, and W. P. Hammes. 1993. Cloning and sequencing of curA encoding curvacin A, the bacteriocin produced by Lactobacillus curvatus LTH1174. Arch. Microbiol. 160:279-283. [DOI] [PubMed] [Google Scholar]
- 54.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1996. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J. Bacteriol. 178:3585-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uteng, M., H. H. Hauge, P. R. Markwick, G. Fimland, D. Mantzilas, J. Nissen-Meyer, and C. Muhle-Goll. 2003. Three-dimensional structure in lipid micelles of the pediocin-like antimicrobial peptide sakacin P and a sakacin P variant that is structurally stabilized by an inserted C-terminal disulfide bridge. Biochemistry 42:11417-11426. [DOI] [PubMed] [Google Scholar]
- 56.Van Reenen, C. A., M. L. Chikindas, W. H. Van Zyl, and L. M. Dicks. 2003. Characterization and heterologous expression of a class IIa bacteriocin, plantaricin 423 from Lactobacillus plantarum 423, in Saccharomyces cerevisiae. Int. J. Food Microbiol. 81:29-40. [DOI] [PubMed] [Google Scholar]
- 57.Vaughan, A., V. G. Eijsink, T. F. O'Sullivan, K. O'Hanlon, and D. van Sinderen. 2001. An analysis of bacteriocins produced by lactic acid bacteria isolated from malted barley. J. Appl. Microbiol. 91:131-138. [DOI] [PubMed] [Google Scholar]
- 58.Wang, Y., M. E. Henz, N. L. Gallagher, S. Chai, A. C. Gibbs, L. Z. Yan, M. E. Stiles, D. S. Wishart, and J. C. Vederas. 1999. Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry 38:15438-15447. [DOI] [PubMed] [Google Scholar]
- 59.Watson, R. M., R. W. Woody, R. V. Lewis, D. S. Bohle, A. H. Andreotti, B. Ray, and K. W. Miller. 2001. Conformational changes in pediocin AcH upon vesicle binding and approximation of the membrane-bound structure in detergent micelles. Biochemistry 40:14037-14046. [DOI] [PubMed] [Google Scholar]
- 60.Yildirim, Z., D. K. Winters, and M. G. Johnson. 1999. Purification, amino acid sequence and mode of action of bifidocin B produced by Bifidobacterium bifidum NCFB 1454. J. Appl. Microbiol. 86:45-54. [DOI] [PubMed] [Google Scholar]