Abstract
Agrocybe aegerita, a bark mulch- and wood-colonizing basidiomycete, was found to produce a peroxidase (AaP) that oxidizes aryl alcohols, such as veratryl and benzyl alcohols, into the corresponding aldehydes and then into benzoic acids. The enzyme also catalyzed the oxidation of typical peroxidase substrates, such as 2,6-dimethoxyphenol (DMP) or 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonate) (ABTS). A. aegerita peroxidase production depended on the concentration of organic nitrogen in the medium, and highest enzyme levels were detected in the presence of soybean meal. Two fractions of the enzyme, AaP I and AaP II, which had identical molecular masses (46 kDa) and isoelectric points of 4.6 to 5.4 and 4.9 to 5.6, respectively (corresponding to six different isoforms), were identified after several steps of purification, including anion- and cation-exchange chromatography. The optimum pH for the oxidation of aryl alcohols was found to be around 7, and the enzyme required relatively high concentrations of H2O2 (2 mM) for optimum activity. The apparent Km values for ABTS, DMP, benzyl alcohol, veratryl alcohol, and H2O2 were 37, 298, 1,001, 2,367 and 1,313 μM, respectively. The N-terminal amino acid sequences of the main AaP II spots blotted after two-dimensional gel electrophoresis were almost identical and exhibited almost no homology to the sequences of other peroxidases from basidiomycetes, but they shared the first three amino acids, as well as two additional amino acids, with the heme chloroperoxidase (CPO) from the ascomycete Caldariomyces fumago. This finding is consistent with the fact that AaP halogenates monochlorodimedone, the specific substrate of CPO. The existence of haloperoxidases in basidiomycetous fungi may be of general significance for the natural formation of chlorinated organic compounds in forest soils.
Heme peroxidases are found in plants, fungi, bacteria, and animals and have been grouped on the basis of sequence similarity into two superfamilies; animal peroxidases form one superfamily (18), and plant, fungal, and bacterial peroxidases form another superfamily (45). Peroxidases belonging to the latter superfamily have been proposed to have various functions in the individual organisms, including roles in the polymerization and detoxification of phenolic compounds in secondary metabolism, as well as in the decomposition of wood and humus (7, 32).
Over the last two decades, particular attention has been paid to the peroxidases of white rot fungi, which are involved in the biodegradation of lignin, humic materials, and organopollutants (13, 16, 33). These enzymes are capable of oxidizing recalcitrant aromatic molecules by one-electron abstractions, resulting in the formation of unstable radicals, which tend to disintegrate spontaneously (19). Numerous isozymes of lignin peroxidase (LiP) and manganese peroxidase (MnP), as well hybrid forms of both enzymes (the so-called versatile peroxidases), have been purified and characterized from different basidiomycetes, including Phanerochaete chrysosporium, Phlebia radiata, Trametes versicolor, Pleurotus eryngii, Bjerkandera spp., and Stropharia coronilla (14, 31). In addition to these ligninolytic enzymes, which preferentially oxidize nonphenolic aromatic compounds and/or Mn2+ ions, there have been reports of other peroxidases from basidiomycetous fungi that resemble plant peroxidases and oxidize phenolic and amino aromatic compounds (e.g., peroxidases from Coprinus cinereus and related species and versatile peroxidases) (15, 25, 26). Heme haloperoxidases, which introduce chlorine, bromine, or iodine into organic molecules and also oxidize various aromatic and aliphatic compounds, are other examples of versatile fungal heme proteins; however, they have not been found in basidiomycetes to date, although they have been found in several ascomycetes (6, 12, 27).
In this paper, we describe a novel type of peroxidase from the basidiomycetous fungus Agrocybe aegerita, a common edible mushroom in Mediterranean countries, which oxidizes aryl alcohols and aldehydes at neutral pH and shows haloperoxidase activity.
MATERIALS AND METHODS
Organisms.
A. aegerita (Brig.) Sing. (a synonym of Agrocybe cylindracea; black poplar mushroom) is an agaric fungus that colonizes deciduous wood and bark mulch, preferably stumps of poplar trees. This fungus is found in North America, Europe, and Asia, and it seems to prefer warm or mild climates. A. aegerita is a popular edible mushroom in southern Europe, especially in Italy (Pioppino mushroom), where it is also commercially cultured (37). We performed most experiments with A. aegerita TM A1, which was isolated by G. Gramss from fungal fruiting bodies that developed on a wheat straw pile in Jena, Germany. For purposes of comparison, the following strains obtained from the Deutsche Sammlung für Mikroorganismen, Braunschweig, Germany (DSM), and the Centraalbureau voor Schimmelcultures, Baarn, The Netherlands (CBS), or deposited in the culture collection of the Institute of Microbiology, University of Jena, Jena, Germany (TM strains) were used: A. aegerita TM A1, CBS 358.51, CBS 127.88, TM ae, and TM AAN (= DSM 9613), Agrocybe praecox DSM 3355, Agrocybe vervactii TM N18, and Agrocybe dura A71-2 (= DSM 8353).
Culture conditions.
Fungal stock cultures were maintained on malt extract agar in culture slants and were stored at 4°C in the dark. The fungus was routinely precultured on 2% malt extract agar plates for 2 weeks. The contents of agar plates were homogenized in 40 ml of a sterile NaCl solution (0.9% NaCl), and each mycelial suspension was used to inoculate liquid cultures (2.5 to 5%, vol/vol). The basal liquid medium contained 30 g of soybean meal (Hensel Voll-Soja; Schoeneberger GmbH, Magstadt, Germany) per liter. In addition to soy meal, various concentrations of Bacto Peptone (Difco Laboratories, Detroit, Mich.) were tested. Fungal cultures were agitated in 500-ml flasks containing 200 ml of medium on a rotary shaker (100 rpm) at 24°C in the dark for 14 days. Samples were taken every 1 to 3 days, and the activities of peroxidases and laccase were measured.
The medium used for production of larger amounts of A. aegerita peroxidase (AaP) in a 5-liter stirred-tank bioreactor (Biostat B; Braun Biotech International GmbH, Melsungen, Germany) consisted of 20 g of soybean meal per liter and 5 g of Bacto Peptone per liter and was inoculated with 200 ml of a fungal suspension precultured as described above (fermentation parameters: agitation at 300 rpm and 100% pO2 for dissolved oxygen concentration). The peroxidase and laccase activities, as well as the pH, were determined every 1 to 2 days during a total fermentation period of 11 days.
Chemicals.
2,2′-Azinobis-(3-ethylbenzothiazoline-6-sulfonate) (ABTS), 2,6-dimethoxyphenol (DMP), veratryl alcohol, veratraldehyde, veratric acid, benzyl alcohol, benzaldehyde, and anisyl alcohol, as well as glucose oxidase (from Aspergillus niger), were purchased from Sigma-Aldrich (Steinheim, Germany). Chloroperoxidase (CPO) from Caldariomyces (Leptoxyphium) fumago and 2-chloro-5,5-dimethyl-1,3-cyclohexanedione (monochlorodimedone [MCD]) were obtained from Fluka (Buchs, Switzerland).
Enzyme assays and UV-visible spectra.
The activity of AaP was measured at 310 nm by monitoring the oxidation of veratryl alcohol into veratraldehyde (ɛ310 = 9.3 mM−1 cm−1). Unlike the LiP reaction (40), the reaction was carried out at pH 7 in sodium phosphate/citrate buffer (McIlvaine buffer) and was started by addition of 2 mM H2O2. AaP activities with benzyl alcohol and benzaldehyde were measured under identical conditions by monitoring either the formation (ɛ280 = 1.4 cm−1 mM−1) or the conversion (ɛ300 = 0.6 cm−1 mM−1) of benzaldehyde. Oxidation of DMP was determined by using the same assay mixture and an ɛ569 of 49.6 cm−1 mM−1 (44); for ABTS oxidation (ɛ420 = 36 cm−1 mM−1) the pH of the buffer was adjusted to 5. Laccase activity was also determined with ABTS, but H2O2 was omitted from the reaction mixture (9).
Haloperoxidase activity was measured quantitatively by monitoring the chlorination or bromination of MCD (ɛ290 = 20.1 cm−1 mM−1) in potassium phosphate buffer (pH 2.75) (12).
For comparison, commercial CPO from the ascomycete C. (L.) fumago (Fluka) was tested by using conditions identical to those used in the AaP assays. All enzyme activities were expressed in units (micromoles of product formed per minute or micromoles of substrate converted per minute).
UV-visible spectra of resting, oxidized, and reduced AaP were recorded in 10 mM sodium phosphate buffer (pH 7) in the wavelength range from 200 to 700 nm by using a Lambda 2 spectrophotometer (Perkin-Elmer, Fremont, Calif.). Oxidation and reduction of AaP (3.3 μM) were achieved by addition of H2O2 (15 or 100 mM) and a small crystal of sodium dithionite, respectively.
Enzyme purification and characterization.
Culture fluid (3.9 liters) from the liquid fermentation was frozen (−20°C) and then thawed to precipitate the free extracellular glucan that was produced by the fungus under the culture conditions used. Precipitated material was removed by centrifugation and subsequent filtration through glass fiber filters. Filtrates were concentrated 60-fold by ultrafiltration by using a tangential-flow cassette (Omega open channel; molecular mass cutoff, 10 kDa; Pall-Filtron, Dreieich, Germany) and a 250-ml filter chamber unit equipped with a 10-kDa-cutoff polysulfone membrane (Amicon, Beverly, Mass.).
Crude enzyme was purified further by several steps of ion-exchange chromatography by using Q Sepharose, SP Sepharose, and Mono S columns, as well as an ÄKTA fast protein liquid chromatography (FPLC) system (Amersham Biosciences, Freiburg, Germany). Separation was carried out by using sodium acetate (10 mM, pH 4.25 to 6.0) as the solvent and an increasing sodium chloride gradient (0 to 0.3 or 0.6 M) for protein elution (see Fig. 2).
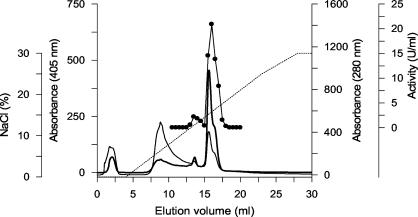
FIG. 2.
FPLC elution profile of peroxidases from A. aegerita TM A1 on a Mono S column. Absorption at 405 nm (thick line) and 280 nm (thin line), AaP activity (•), and the NaCl gradient (dotted line) were determined. AaP activity was measured with veratryl alcohol as the substrate.
Molecular weights of proteins were determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (Hoefer SE 600 Ruby; Amersham Biosciences) as described by Laemmli (21). After electrophoresis, the gels were stained, and protein bands were visualized with AgCl. A low-molecular-weight protein calibration kit (MBI Fermentas, St. Leon Roth, Germany) was used as the standard. Two-dimensional (2-D) electrophoresis (SDS-PAGE followed by isoelectric focusing) was performed by using a Multiphor II electrophoresis system (Amersham Biosciences) and DryStrip gels (pH 3 to 11), which were cut to obtain a final pH range from 3 to 7; staining of 2-D gels was carried out with guaiacol or Coomassie blue.
For N-terminal amino acid analysis, purified AaP was separated by 2-D electrophoresis, and the spots of the AaP II fraction (spots a and b) stained with Coomassie blue (see Fig. 3B) were transferred by electroblotting with a Trans Blot cell (Bio-Rad, Munich, Germany) onto a polyvinylidene difluoride membrane (ProBlott; Perkin-Elmer, Applied Biosystems, Foster City, Calif.). N-terminal amino acid sequences were determined by Sequence Laboratories GmbH, Göttingen, Germany.
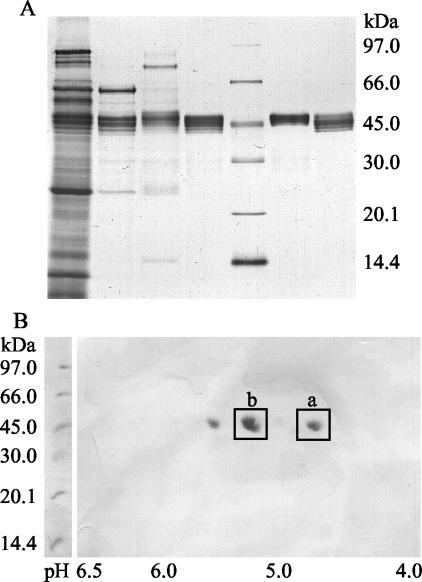
FIG. 3.
Electrophoretic characterization of purified AaP. (A) SDS-PAGE after each purification step. From left to right the lanes contained crude extract, AaP fractions after Q Sepharose separation, AaP I after SP Sepharose separation, AaP I after Mono S separation, protein standards, AaP II after SP Sepharose separation, and AaP II after Mono S separation. (B) 2-D plot of purified AaP II after isoelectric focusing and SDS-PAGE. Spots b (basic form) and a (acidic form) were plotted and used for determination of N-terminal amino acid sequences.
The apparent Michaelis-Menten constants (Km) and catalytic constants (kcat) of purified AaP II were determined for benzyl alcohol, veratryl alcohol, ABTS, DMP, and H2O2 (the expression apparent Km is used because peroxidases do not operate by classic Michaelis-Menten kinetics). Lineweaver-Burk plots were prepared from the initial rates obtained with various substrate concentrations while the concentration of the second substrate was kept constant.
Substrate oxidation experiments.
The following substrates (200 μM) were treated with purified AaP (1 U [= 12 μg = 0.26 μM]) in stirred 3-ml vials containing 1 ml of a reaction solution: veratryl alcohol, anisyl alcohol, benzyl alcohol, and ethanol. Each reaction mixture contained sodium phosphate/citrate buffer (pH 7), 15 mM glucose, and 0.2 U of glucose oxidase (Sigma-Aldrich). The reaction mixtures were incubated at 25°C, and the alcohol and aldehyde concentrations were determined after 20 and 30 min. High-performance liquid chromatography analyses were performed by using a model HP 1090 liquid chromatograph (Agilent, Waldbronn, Germany) equipped with a Merck LiChrospher 5-μm RP-18 reversed-phase column (4.6 by 125 mm). A mixture of acetonitrile and 0.05% acetic acid (40:60, vol/vol) was used as the solvent at a flow rate of 1 ml min−1 under isocratic conditions. Eluted substances were detected in the wavelength range from 190 to 550 nm and were identified by using authentic standards.
RESULTS
Peroxidase production.
A. aegerita produced appreciable amounts of a peroxidase (AaP) that oxidized veratryl alcohol at pH 7 only in N-rich, complex liquid media that contained soybean meal. All strains of the fungus tested secreted the enzyme, but there were considerable differences in the individual enzyme levels (range, 5 to 1,336 U liter−1) (Table 1). A. aegerita TM A1 proved to be the most active strain and was used for all further studies. The other Agrocybe spp. tested did not show any peroxidase activity in the soybean medium.
TABLE 1.
Peroxidase production by several strains of A. aegerita, as well as three other species of the genus Agrocybe, in soybean meal medium
| Fungal strain | Peroxidase activity (U liter−1)a |
|---|---|
| A. aegerita TM A1 | 1,336 ± 194 |
| A. aegerita TM AAN (= DSM 9613) | 281 ± 40 |
| A. aegerita TM ae | 252 ± 33 |
| A. aegerita CBS 358.51 | 15 ± 4 |
| A. aegerita CBS 127.88 | 5 ± 3 |
| A. dura DSM 8353 | 0 |
| A. praecox DSM 3355 | 0 |
| A. vervactii DSM 8353 | 0 |
Peroxidase activity was determined by the oxidation of veratryl alcohol at pH 7.
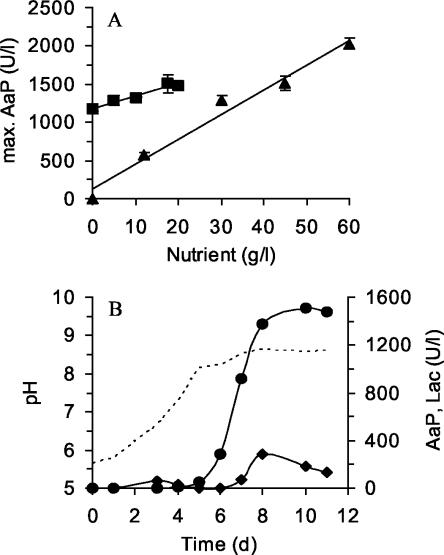
Increasing the amount of soybean meal in the medium (from 12.5 to 60 g liter−1) resulted in an almost linear increase in the AaP activity; in the absence of soybean meal, AaP activity was not detectable (Fig. 1A). The maximum AaP activity (2,021 U liter−1) was found in the presence of 60 g of soybean meal per liter. Bacto Peptone (0 to 20 g liter−1) further stimulated enzyme production; however, the effect was less pronounced, and Bacto Peptone was not required (Fig. 1A).
FIG. 1.
(A) Effect of soybean meal (▴) and Bacto Peptone (▪) on the production of peroxidase by A. aegerita TM A1. The concentration of one component was varied in the presence of a constant amount of the other component (5 g of Bacto Peptone per liter or 20 g of soybean meal per liter). (B) Time course of peroxidase and laccase titers in a 5-liter bioreactor with a soybean meal-Bacto Peptone medium. Peroxidase activity (•) was measured by determining the oxidation of veratryl alcohol into veratraldehyde at pH 7; laccase activity (⧫) was measured with ABTS. The dotted line indicates the time course of pH.
To obtain larger quantities of AaP for the subsequent purification procedure, A. aegerita TM A1 was cultured in a stirred-tank bioreactor. Figure 1B shows the fermentation profile for a 5-liter device. Due to the high viscosity of the growth medium, smaller amounts of soybean meal and Bacto Peptone (20 and 5 g liter−1, respectively) were used. In the course of fermentation, the pH of the medium increased from 5.7 to 8.6. At the beginning of the experiment the amount of dissolved O2 was kept at 100% by constantly bubbling air into the medium (2 liter min−1), but due to the consistency of the soybean slurry, it dropped to 30% after 5 days of fermentation. Production of AaP started on day 6, and the maximum activity (1,550 U liter−1) was reached after 10 days of cultivation. In addition to AaP, the fungus produced laccase, but the level of laccase was noticeably lower (maximum activity, 290 U liter−1). Activities of MnP and LiP were not detectable.
Purification of AaP.
Culture filtrate from the bioreactor was harvested after 10 days and frozen to remove dissolved polysaccharides. This procedure led, after centrifugation, to loss of about 45% of the activity, which can be explained by the partial adsorption of AaP to the precipitated polysaccharides (Table 2). After ultrafiltration, the concentrated crude extract was separated by three steps of ion-exchange chromatography by using FPLC equipment. Figure 2 shows the elution profile for the third purification step with a Mono S column (strong cation exchanger). Two fractions of the enzyme (designated AaP I and AaP II) absorbing at 405 nm (heme) and exhibiting peroxidase activity with veratryl alcohol were distinguished. The final specific activities of both AaP fractions were around 165 U mg−1, but the total activity of AaP II was considerably higher than that of AaP I (244 versus 20 U); the Reinheitszahl values of AaP I and AaP II were 1.65 and 1.85, respectively.
TABLE 2.
Purification of peroxidase from A. aegerita TM A1a
| Purification step | Amt of protein (mg) | Total activity (U) | Sp act (U mg−1) | Yield (%) | Purifi- cation (fold) |
|---|---|---|---|---|---|
| Culture liquid | 465 | 1,380 | 3 | 100 | 1 |
| Ultrafiltration (10 kDa) | 123 | 756 | 6 | 55 | 2 |
| Q Sepharose | 8.2 | 531 | 64 | 38 | 22 |
| SP Sepharose (AaP I) | 0.46 | 43 | 92 | 3 | 31 |
| SP Sepharose (AaP II) | 2.73 | 299 | 110 | 22 | 37 |
| Mono S (AaP I) | 0.12 | 20 | 167 | 1 | 56 |
| Mono S (AaP II) | 1.48 | 244 | 165 | 18 | 55 |
Enzyme activities are based on the oxidation of veratryl alcohol at pH 7.
Characterization of AaP.
SDS-PAGE revealed the same molecular mass (46 kDa) for both AaP fractions (Fig. 3A). 2-D electrophoresis revealed that there was little difference in the isoelectric points. The two AaP fractions each split into three spots; the pIs for AaP I were 4.9, 5.2, and 5.4, and the pIs for AaP II were 4.9, 5.3, and 5.7, indicating the presence of six isoforms of the enzyme (Fig. 3B). The most intense spots of AaP II (spots b and a) were blotted and used for determination of N-terminal amino acid sequences.
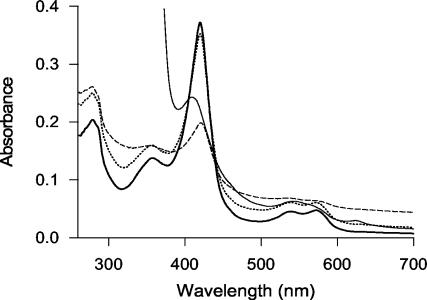
The UV-visible spectrum of the resting enzyme (AaP II after purification) had a characteristic absorption peak at 420 nm (Soret band), which gave the purified enzyme its characteristic reddish color, and two absorption maxima at 540 and 572 nm (Fig. 4). Addition of catalytic amounts of H2O2 (0.2 to 15 mM) led to only slight changes in the spectrum, and the maxima did not shift (Fig. 4); adding excess H2O2 (100 mM) resulted in the immediate formation of gas bubbles and in a drastic decrease in the absorption intensity, but again no shift of the absorption maxima was observed. Addition of sodium dithionite to the resting enzyme caused a shift in the peak from 420 to 409 nm, and in the longer-wavelength range, the peak at 572 nm became a shoulder and a new maximum appeared at 625 nm (Fig. 4).
FIG. 4.
Spectral characteristics of AaP (3.3 μM), including the resting enzyme (thick line), the enzyme oxidized by 15 mM H2O2 (dotted line) or 100 mM H2O2 (dashed line), and the reduced enzyme (after addition of a sodium dithionite crystal) (thin line).
The N-terminal peptide sequencing showed that the spots cut out after 2-D electrophoresis belonging to the AaP II fraction were almost identical, apart from a few uncertain amino acid positions that probably resulted from obstacles during the sequencing procedure (Fig. 5). The levels of sequence identity to other peroxidases from basidiomycetes were very low (≤7%), but the first three amino acids, as well as two additional amino acids (positions 7 and 9), were identical to amino acids at the N terminus of CPO from the ascomycete C. (L.) fumago (35% sequence identity).
FIG. 5.
N-terminal sequence alignment of two AaP II isoforms and several fungal peroxidases, including A. aegerita peroxidase (AaP II spot a and AaP II spot b after 2-D electrophoresis), CPO from C. fumago (CP), C. cinereus peroxidase (CiP), LiP H8 b from P. chrysosporium (PcLiP), MnP from A. bisporus (AbMnP), and versatile peroxidase from P. eryngii (PeVP).
Oxidation of different substrates.
Purified AaP II was used to examine the substrate spectrum of the enzyme. When a combined assay mixture in which glucose oxidase continuously generated H2O2 was used, AaP was able to oxidize several aryl alcohols and aldehydes; ethanol was not subject to AaP attack (Table 3). High-performance liquid chromatography analysis showed that benzyl and veratryl alcohols were first converted into benzaldehyde and veratraldehyde and then into benzoic and veratric acids, respectively. The latter finding was confirmed by the direct conversion of benzaldehyde and veratraldehyde into the corresponding benzoic acids. Anisyl alcohol was also oxidized into the corresponding aldehyde; however, the acid product could not be identified (due to the lack of authentic anisic acid). The following trends were observed for the oxidation of aryl alcohols and aldehydes by AaP: anisyl alcohol > benzyl alcohol ≫ veratryl alcohol and benzaldehyde > anisaldehyde ≫ veratraldehyde.
TABLE 3.
Oxidation of several alcohols and aldehydes by purified AaPa
| Substrate | Amt of substrate converted (μM) | Identified product(s) |
|---|---|---|
| Anisyl alcohol | 157 | Anisaldehyde |
| Anisaldehyde | 44 | NDb |
| Benzyl alcohol | 144 | Benzaldehyde, benzoic acid |
| Benzaldehyde | 62 | Benzoic acid |
| Veratryl alcohol | 92 | Veratraldehyde, veratric acid |
| Veratraldehyde | 11 | Veratric acid |
| Ethanol | 0 |
Each reaction mixture (1 ml) consisted of sodium phosphate/citrate buffer (pH 7), 200 μM substrate (alcohols), 15 mM glucose, 0.2 U of glucose oxidase, and 1 U of AaP. The reaction time was 20 min with continuous stirring at 25°C. The substrate concentration was determined by high-performance liquid chromatography or gas chromatography-mass spectrometry (ethanol). Oxidation of intermediately formed aldehydes was calculated by the decrease in the course (20 min) of the enzymatic reaction. The data are means for three parallel experiments; the standard deviation was <5% in all cases.
ND, not determined.
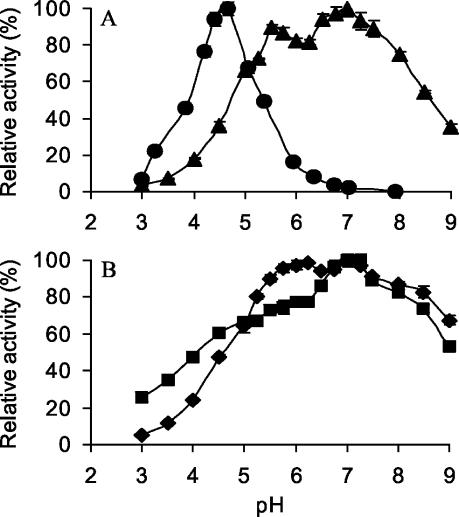
Besides aryl alcohols and aldehydes, AaP was found to oxidize typical peroxidase substrates, such as ABTS and DMP. Table 4 shows the apparent Km and kcat values determined photometrically, as well as those of veratryl alcohol, benzyl alcohol, and H2O2. The pH optima of AaP for the oxidation of ABTS, benzyl alcohol, and DMP differed noticeably. Whereas the curves for benzyl alcohol, veratryl alcohol, and DMP oxidation showed broad, almost neutral pH optima (pH 6.8 to 7.2), ABTS oxidation occurred in a narrower pH range (pH 2 to 6.5), with an acidic maximum around pH 4.7 (Fig. 6). Interestingly, AaP seems to have two pH optima for the oxidation of benzyl alcohol and veratryl alcohol (pH 6.2 and 7.2 and pH 5.5 and 7, respectively). AaP was found to be active between pH 2.5 and 9 and at the latter pH and in the presence of some substrates lost only 30 to 70% of its activity. The enzyme required relatively large amounts of peroxide; thus, in the presence of 5 mM veratryl alcohol, optimal activity was observed with 2 mM H2O2, but the enzyme still exhibited 35% of the maximum activity with 10 mM H2O2 (data not shown).
TABLE 4.
Apparent Michaelis-Menten constants (Km) and catalytic constants (kcat) of AaP II for veratryl alcohol, benzyl alcohol, ABTS, and DMPa
| Substrate | pH | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Veratryl alcohol | 7 | 2,367 | 85 | 3.58 × 104 |
| Benzyl alcohol | 7 | 1,001 | 269 | 2.69 × 105 |
| ABTS | 4.5 | 37 | 283 | 7.67 × 106 |
| DMP | 7 | 298 | 108 | 3.61 × 105 |
| H2O2 | 7 | 1,313 | 367 | 2.79 × 105 |
Reactions were performed in sodium phosphate/citrate buffer (pH 7 or 4.5) in the presence of 2 mM H2O2. The Km for H2O2 was determined in the presence of 5 mM benzyl alcohol. The values are means for three parallel experiments, and the standard deviations were <5%.
FIG. 6.
Effect of pH on the oxidation of ABTS (0.3 mM) (•) and veratryl alcohol (5 mM) (▴) (A) and on the oxidation of DMP (1 mM) (▪) and benzyl alcohol (⧫) (B) by AaP II. Reactions were performed in sodium phosphate/citrate buffer in the presence of 2 mM H2O2 at 25°C. The data are means for three parallel experiments, and the error bars indicate standard deviations.
Comparison of AaP and CPO.
The halogenating activity of AaP was proved by monitoring the chlorination or bromination of MCD. For comparison, authentic heme CPO from C. fumago was tested under identical conditions. We found that AaP is in fact capable of chlorinating and brominating MCD, although the specific halogenating activities (at least at pH 2.75) were considerably lower than those of CPO (21- and 14-fold lower for chloride and bromide, respectively) (Table 5). On the other hand, the activity of AaP with ABTS at pH 5 was 250 times higher than that of CPO, and the activities of AaP with DMP and benzyl alcohol at neutral pH were 10 and 50 times higher than those of CPO. Benzaldehyde, which was moderately oxidized by AaP, was not a substrate for CPO (even highly concentrated CPO was not capable of oxidizing benzaldehyde [Table 5]).
TABLE 5.
Oxidation of different substrates and halogenation of MCD by purified AaP and CPO from C. fumagoa
| Substrate | Sp act of AaP (U mg−1) | Sp act of CPO (U mg−1) |
|---|---|---|
| ABTS | 295.7 | 1.2 |
| DMP | 99.6 | 1.9 |
| Benzyl alcohol | 234.2 | 18.1 |
| Benzaldehyde | 236.7 | 0 |
| MCD (Cl−) | 71.8 | 1,537 |
| MCD (Br−) | 354.3 | 2,859 |
Most reactions were carried out in sodium citrate/phosphate buffer (pH 7); the exceptions were chlorination and bromination of MCD and oxidation of ABTS, which were carried out in phosphate buffer at pH 2.75 and 5.0, respectively. The experiments were carried out in triplicate, and the standard deviations were <5%.
DISCUSSION
A. aegerita produces, in the presence of soybean meal, a peroxidase (AaP) that oxidizes aryl alcohols into the corresponding aldehydes and then into the corresponding benzoic acids. This enzyme also catalyzes the conversion of typical peroxidase substrates, such as ABTS or DMP, and halogenates MCD. The optimum pH for nonhalogenating AaP activities is around 7 (except for ABTS), and the enzyme requires relatively large amounts of H2O2. Two AaP fractions (AaP I and AaP II), which split further into six isoforms, can be distinguished on the basis of the results obtained for the FPLC elution profiles and 2-D electrophoresis gels. The N-terminal amino acid sequence of the major AaP fraction (AaP II) shows almost no homology to the sequences of other peroxidases from basidiomycetes, but it shares the first three amino acids and two additional amino acids with the sequence of heme CPO from the ascomycete C. fumago.
CPOs are known from bacteria and ascomycetous fungi and are among the most versatile oxidoreductases (7). Among the fungal enzymes, two types, vanadium and heme-thiol peroxidases, are distinguished (5, 43). Both types are capable of chlorinating and brominating organic substrates, but they also catalyze various other oxidation reactions (7, 8, 38). So far, both fungal vanadium and heme-thiol CPOs have been found only in ascomycetes and related deuteromycetes (2, 12, 24), although their presence in basidiomycetes was proposed some years ago (6) and very recently there have been indications that there is a putative CPO gene in Agaricus bisporus (white button mushroom) (41). Brominating activities have been described for LiP and MnP, but these enzymes do not chlorinate the assay substrate MCD (10, 36). On the other hand, there is extensive information concerning absorbable organic halogens formed by basidiomycetes as secondary metabolites (6, 11, 42). Most of the more than 80 halogenated metabolites identified from basidiomycetes to date are chlorinated, and they are produced by fungi belonging to 68 genera and 20 different families (6). Taxonomic hot spots are found in the order Agaricales, particularly in the families Strophariaceae and Tricholomataceae (6, 42), which are closely related to the Bolbitiaceae, which is the family that A. aegerita belongs to.
Despite the fact that AaP shares the first three amino acids of the N terminus with Caldariomyces CPO and halogenates MCD, the former enzyme differs from the known heme CPOs in its pH behavior, substrate spectrum, and specificity, as well as in the UV-visible spectrum. Thus, the spectrum of the resting enzyme shows the Soret maximum at 420 nm and additional absorption maxima at 359, 540, and 572 nm. Interestingly, these maxima correspond with those reported for CPO compound II (358, 438, 542, and 571 nm) rather than with those of the native enzyme (403, 515, 542, and 650 nm) (7, 12). AaP reduced by sodium dithionite showed more similarity to the corresponding form of CPO. It had a Soret maximum at 409 nm, a shoulder in the 460-nm region, and two peaks around 550 and 640 nm; only the shoulder at 420 nm was lacking in the reduced form of AaP.
The substrate spectrum of AaP comprises phenolic compounds, ABTS, aryl alcohols, and aldehydes, as well as halogenides (Cl−, Br−). CPO is also able to oxidize most of these substrates; however, the specific activities (except those for halogenides) are lower, and aldehydes are apparently not CPO substrates. CPO-catalyzed oxidation of aryl alcohols into the corresponding aldehydes has been described for the Caldariomyces enzyme (1). Similar to AaP, this CPO converted benzyl and anisyl alcohols into the corresponding aldehydes; however, the specific constants reported for CPO are considerably lower than those of AaP. The CPO reaction with aryl alcohols has been proposed to occur via an attack on the benzylic carbon, resulting in the formation of the benzylic radical and cation (1); a similar mechanism is also conceivable for AaP. Furthermore, it was found that CPO possesses prochiral selectivity in the oxidation of aryl alkanols that makes the enzyme interesting for applications in chemical syntheses (1, 30). This is also true for AaP, especially when its broader substrate spectrum and higher pH optima are taken into consideration.
AaP seems to act not only as a peroxidase but also as a catalase, which was shown by the intensive formation of gas bubbles (very probably O2) after addition of larger amounts of H2O2 in the absence of a second available substrate. CPO from Caldariomyces also has strong catalase activity, and there is a whole class of so-called catalase-peroxidases that are found in both prokaryotes and eukaryotes (8, 39).
CPO production by C. fumago was carried out at a larger scale in complex media comprising fructose and Phytone (papaic digest of soybean meal) in airlift fermentors (3), and the enzyme can now be produced as a recombinant protein (e.g., in A. niger) (4). Culturing A. aegerita in a heavily stirred bioreactor was possible, and the enzyme production was only slightly reduced due to shear stress. For AaP production, soybean meal was found to be the key nutrient, and the concentration of the meal correlated with the AaP activity. Complex N-rich media were also found to promote fungal growth and peroxidase production in other basidiomycetes. Thus, mixtures of Bacto Peptone, yeast extract, and sugars enabled the production of large amounts of versatile peroxidase (a hybrid of MnP and LiP) by Bjerkandera sp. strain BOS55 and P. eryngii (23, 25). On the other hand, nitrogen was shown to repress the production of ligninolytic peroxidases in other fungi (for example, P. chrysosporium or P. radiata) (20, 22). A medium quite similar to that used in our study, consisting of soybean meal, Bacto Peptone, and maltose, was successfully used to produce laccases and peroxidases in a stirred bioreactor with dung- and grassland-dwelling agaric basidiomycetes (Panaeolus spp., Coprinus friseii) (15).
Two isoenzymes of Caldariomyces CPO, isoenzymes A and B, were reported to have molecular masses of 40 and 46 kDa; later work showed that there are multiple forms that differ only in the carbohydrate content, which varies between 20 and 30% (17, 34). We also found two fractions of AaP based on the FPLC elution profiles, and there were indications of multiple forms of the enzyme (at least six isoforms) that became visible in the SDS-PAGE and 2-D gel electrophoresis plots.
At the moment, it is difficult to speculate about the physiological role of AaP. According to previously published reports, involvement of the enzyme in the production of halogenated metabolites that act as antibiotics or as part of the ligninolytic system can be proposed (6). This possibility is particularly interesting because A. aegerita apparently lacks MnP and LiP (32) but colonizes wood and causes, according to some authors, a moderate white rot (35, 46). Moreover, the ability of Caldariomyces CPO to chlorinate and cleave recalcitrant lignin structures has been reported very recently (29), suggesting that this type of enzyme could be involved in the biodegradation of lignocellulosic materials.
Hence, future studies on AaP should focus on two questions. (i) What is the physiological function of AaP, and is the enzyme somehow involved in lignin biodegradation? (ii) Do other basidiomycetes, particularly those secreting large amounts of absorbable organic halogens, produce AaP-like peroxidases? If so, what ecological and environmental significance does this type of halogenation of organic compounds, for example, have for the recently reported formation of stable chlorinated hydrocarbons in weathering plant material (28)? In this context, the substrate spectrum of AaP should be studied more in detail and compared with the substrate spectra of CPO and other peroxidases. The first attempts in this direction are currently being undertaken in our laboratories.
Acknowledgments
This work was supported financially by the German Environmental Foundation (DBU) (grant 13663 to R.U., J.N., and K.S.), the Academy of Finland (grant 52063 to M.H.), and the administration of the International Graduate School of Zittau (R. Konschak, Zittau, Germany).
We thank G. Gramss (Jena, Germany) for providing the fungal strain A. aegerita TM A1, R. Upadhyay (Solan, India) for useful preinvestigations, and I. Schwabe (Jena, Germany) and U. Schneider (Zittau, Germany) for excellent technical assistance.
REFERENCES
- 1.Baciocchi, E., M. Fabbrini, O. Lanzalunga, L. Manduchi, and G. Pochetti. 2001. Prochiral selectivity in H2O2-promoted oxidation of arylalcanols catalysed by chloroperoxidase. Eur. J. Biochem. 268:665-672. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Nun, N., S. Shcolnick, and A. M. Mayer. 2002. Presence of a vanadium-dependent haloperoxidase in Botrytis cinerea. FEMS Microbiol. Lett. 217:121-124. [DOI] [PubMed] [Google Scholar]
- 3.Carmichael, R. D., and M. A. Pickard. 1989. Continuous and batch production of chloroperoxidase by mycelial pellets of Caldariomyces fumago in an airlift fermenter. Appl. Environ. Microbiol. 55:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conesa, A., F. van de Velde, F. van Rantwijk, R. A. Sheldon, C. A. M. J. J. van den Hondel, and P. J. Punt. 2001. Expression of the Caldariomyces fumago chloroperoxidase in Aspergillus niger and characterization of the recombinant enzyme. J. Biol. Chem. 276:17635-17640. [DOI] [PubMed] [Google Scholar]
- 5.Dawson, J. H., and M. Sono. 1987. Cytochrome P-450 and chloroperoxidase:thiolate-ligated heme enzymes. Spectroscopic determination of their active site structures and mechanistic implications of thiolate ligation. Chem. Rev. 87:1255-1276. [Google Scholar]
- 6.de Jong, E., and J. A. Field. 1997. Sulfur tuft and turkey tail: biosynthesis and biodegradation of organohalogens by basidiomycetes. Annu. Rev. Microbiol. 51:375-414. [DOI] [PubMed] [Google Scholar]
- 7.Dunford, H. B. 1999. Heme peroxidases. Wiley-VCH, New York, N.Y.
- 8.Dunford, H. B. 2000. Peroxidase-catalyzed halide ion oxidation. Redox Rep. 5:169-171. [DOI] [PubMed] [Google Scholar]
- 9.Eggert, C., U. Temp, and K.-E. L. Eriksson. 1996. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl. Environ. Microbiol. 62:1151-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farhangrazi, Z. S., R. Sinclair, I. Yamazaki, and L. S. Powers. 1997. Haloperoxidase activity of Phanerochaete chrysosporium lignin peroxidases H2 and H8. Biochemistry 31:10763-10768. [DOI] [PubMed] [Google Scholar]
- 11.Field, J. A., F. J. M. Verhagen, and E. de Jong. 1995. Natural organohalogen production by basidiomycetes. Trends Biotechnol. 13:451-456. [Google Scholar]
- 12.Hager, L. P., D. R. Morris, F. S. Brown, and H. Eberwein. 1966. Chloroperoxidase. II. Utilization of halogen anions. J. Biol. Chem. 241:1769-1777. [PubMed] [Google Scholar]
- 13.Hatakka, A. 1994. Lignin-modifying enzymes from selected white-rot fungi: production and role in lignin degradation. FEMS Microbiol. Rev. 13:125-135. [Google Scholar]
- 14.Hatakka, A. 2001. Biodegradation of lignin, p. 129-180. In M. Hofrichter and A. Steinbüchel (ed.), Lignin, humic substances and coal, vol. 1. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 15.Heinzkill, M., L. Bech, T. Halkier, P. Schneider, and T. Anke. 1998. Characterization of laccases and peroxidases from wood-rotting fungi (family Coprinaceae). Appl. Environ. Microbiol. 64:1601-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofrichter, M. 2002. Review: lignin conversion by manganese peroxidase (MnP). Enzyme Microb. Technol. 30:454-466. [Google Scholar]
- 17.Kenigsberg, P., G. H. Fang, and L. P. Hager. 1987. Post-translational modifications of chloroperoxidase from Caldariomyces fumago. Arch. Biochem. Biophys. 254:409-415. [DOI] [PubMed] [Google Scholar]
- 18.Kimura, S., and M. Ikeda-Saito. 1988. Human myeloperoxidase and thyroid peroxidase, two enzymes with separate and distinct physiological functions, are evolutionarily related members of the same gene family. Proteins 3:113-120. [DOI] [PubMed] [Google Scholar]
- 19.Kirk, T. K., and R. L. Farrell. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465-505. [DOI] [PubMed] [Google Scholar]
- 20.Kirk, T. K., S. Croan, M. Tien, K. E. Murtagh, and R. L. Farrell. 1986. Production of multiple ligninases by Phanerochaete chrysosporium: effect of selected growth conditions and use of a mutant strain. Enzyme Microb. Technol. 8:27-32. [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lundell, T., A. Leonowicz, J. Rogalski, and A. Hatakka. 1990. Formation and action of lignin-modifying enzymes in cultures of Phlebia radiata supplemented with veratric acid. Appl. Environ. Microbiol. 56:2623-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez, M. J., F. J. Ruiz-Dueñas, F. Guillén, and A. T. Martínez. 1996. Purification and catalytic properties of two manganese peroxidase isoenzymes from Pleurotus eryngii. Eur. J. Biochem. 237:424-432. [DOI] [PubMed] [Google Scholar]
- 24.McInnes, A. G., J. A. Walter, J. L. C. Wright, L. C. Vining, N. Ramada, R. Bentley, and E. P. McGovern. 1990. Biosynthesis of mollisin by Mollisin caesia. Can. J. Chem. 68:1-4. [Google Scholar]
- 25.Moreira, M. T., A. Torrado, G. Feijoo, and J. M. Lema. 2000. Manganese peroxidase production by Bjerkandera sp. BOSS55. 2. Operation in stirred tank reactors. Biotechnol. Lett. 22:663-667. [Google Scholar]
- 26.Morita, Y., H. Yamashita, B. Mikami, H. Iwamoto, S. Aibara, M. Terada, and J. Minami. 1988. Purification, crystallization, and characterization of peroxidase from Coprinus cinereus. J. Biochem. 103:693-699. [DOI] [PubMed] [Google Scholar]
- 27.Morris, D. R., and L. P. Hager. 1966. Chloroperoxidase. I. Isolation and properties of the crystalline glycoprotein. J. Biol. Chem. 241:1763-1768. [PubMed] [Google Scholar]
- 28.Myneni, S. C. B. 2002. Formation of stable chlorinated hydrocarbons in weathering plant material. Science 295:1039-1104. [DOI] [PubMed] [Google Scholar]
- 29.Ortiz-Bermúdez, P., E. Srebotnik, and K. E. Hammel. 2003. Chlorination and cleavage of lignin structures by fungal chloroperoxidase. Appl. Environ. Microbiol. 69:5015-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pickard, M. A., T. A. Kadima, and R. D. Carmicheal. 1991. Chloroperoxidase, a peroxidase with potential. J. Ind. Microbiol. 7:1123-1131. [Google Scholar]
- 31.Piontek, K., T. Glumoff, and K. Winterhalter. 1993. Low pH crystal structure of glycosylated lignin peroxidase from Phanerochaete chrysosporium at 2.5 Å resolution. FEBS Lett. 315:119-124. [DOI] [PubMed] [Google Scholar]
- 32.Poulos, T. L. 1993. Peroxidases. Curr. Opin. Biotechnol. 4:484-489. [DOI] [PubMed] [Google Scholar]
- 33.Sack, U., T. M. Heinze, J. Deck, R. Martens, F. Zadrazil, C. E. Cerniglia, and W. Fritsche. 1997. Comparison of phenanthrene and pyrene degradation by different wood-decaying fungi. Appl. Environ. Microbiol. 63:3919-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sae, A. S. W., and B. A. Cunningham. 1979. Isolation and properties of chloroperoxidase isozymes. Phytochemistry 18:1785-1787. [Google Scholar]
- 35.Sapparat, M. C. N., A. M. M. Bucsinszky, H. A. Tournier, M. N. Cabello, and A. M. Arambarri. 2000. Extracellular ABTS-oxidizing activity of autochthonous fungal strains from Argentina in solid medium. Rev. Iberoam. Micol. 17:64-68. [PubMed] [Google Scholar]
- 36.Sheng, D., and M. H. Gold. 1997. Haloperoxidase activity of manganese peroxidase from Phanerochaete chrysosporium. Arch. Biochem. Biophys. 345:126-134. [DOI] [PubMed] [Google Scholar]
- 37.Stamets, P. 2000. Growing gourmet and medicinal mushrooms, 3rd ed. Ten Speed Press, Berkeley, Calif.
- 38.ten Brink, H. B., H. L. Dekker, H. E. Schoemaker, and R Wever. 2000. Oxidation reactions catalyzed by vanadium chloroperoxidase from Curvularia inaequalis. J. Inorg. Biochem. 80:91-98. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, J. A., D. R. Morris, and L. P. Hager. 1970. Chloroperoxidase. VII. Classical peroxidatic, catalytic and halogenating forms of the enzyme. J. Biol. Chem. 245:3129-3134. [PubMed] [Google Scholar]
- 40.Tien, M., and T. K. Kirk. 1988. Lignin peroxidase of Phanerochaete chrysosporium. Methods Enzymol. 161:238-249. [Google Scholar]
- 41.van Pée, K.-H., and S. Zehner. 2003. Enzymology and molecular genetics of biological halogenation, p. 171-199. In The handbook of environmental chemistry, vol. 3, part P. Springer-Verlag, Berlin, Germany.
- 42.Verhagen, F. J. M., H. J. Swarts, J. B. P. A. Wijnberg, and J. A. Field. 1998. Organohalogen production is a ubiquitous capacity among basidiomycetes. Chemosphere 37:2091-2104. [Google Scholar]
- 43.Vilter, H. 1995. Vanadium-dependent haloperoxidases. Met. Ions Biol. Syst. 31:325-362. [PubMed] [Google Scholar]
- 44.Wariishi, H., K. Valli, F. S. Brown, and M. H. Gold. 1992. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. J. Biol. Chem. 267:23688-23695. [PubMed] [Google Scholar]
- 45.Welinder, K. G. 1992. Superfamily of plant, fungal and bacterial peroxidases. Curr. Opin. Struct. Biol. 2:388-393. [Google Scholar]
- 46.Zadrazil, F. 2000. Industrial production of Agrocybe aegerita (Brig.) Sing.: will Agrocybe aegerita become a new culture mushroom. Champignon 412:296-301. (In German.) [Google Scholar]