Abstract
A case-control study involving 24 case farms with at least one recent case of listeriosis and 28 matched control farms with no listeriosis cases was conducted to probe the transmission and ecology of Listeria monocytogenes on farms. A total of 528 fecal, 516 feed, and 1,012 environmental soil and water samples were cultured for L. monocytogenes. While the overall prevalence of L. monocytogenes in cattle case farms (24.4%) was similar to that in control farms (20.2%), small-ruminant (goat and sheep) farms showed a significantly (P < 0.0001) higher prevalence in case farms (32.9%) than in control farms (5.9%). EcoRI ribotyping of clinical (n = 17) and farm (n = 414) isolates differentiated 51 ribotypes. L. monocytogenes ribotypes isolated from clinical cases and fecal samples were more frequent in environmental than in feed samples, indicating that infected animals may contribute to L. monocytogenes dispersal into the farm environment. Ribotype DUP-1038B was significantly (P < 0.05) associated with fecal samples compared with farm environment and animal feedstuff samples. Ribotype DUP-1045A was significantly (P < 0.05) associated with soil compared to feces and with control farms compared to case farms. Our data indicate that (i) the epidemiology and transmission of L. monocytogenes differ between small-ruminant and cattle farms; (ii) cattle contribute to amplification and dispersal of L. monocytogenes into the farm environment, (iii) the bovine farm ecosystem maintains a high prevalence of L. monocytogenes, including subtypes linked to human listeriosis cases and outbreaks, and (iv) L. monocytogenes subtypes may differ in their abilities to infect animals and to survive in farm environments.
Listeria monocytogenes, a facultative intracellular pathogen, is responsible for severe foodborne infections in humans and can also cause invasive disease in many different animal species, including farm ruminants (cattle, sheep, and goats). In 1999, Mead et al. (24) estimated that approximately 2,500 cases of clinical human invasive listeriosis (including 500 deaths) occur each year in the United States. Although several animal-derived L. monocytogenes-contaminated food products, including raw milk, pasteurized milk, chocolate milk, butter, soft cheeses, and processed meat and poultry products, have been implicated as sources of human listeriosis cases and outbreaks (7, 15, 22, 32), L. monocytogenes present in raw materials is effectively inactivated by the time-temperature combinations typical for production of most processed foods (e.g., pasteurization of milk). In general, L. monocytogenes contamination of processed ready-to-eat food products occurs by cross-contamination of the finished product from the food processing plant environment (13). While infected animals and contaminated agricultural environments rarely appear to directly cause human infections, animal-derived food products that are not processed before consumption (e.g., raw milk) and raw foods of plant origin that have been contaminated by manure from infected or shedding animals represent direct links between human infections and L. monocytogenes in farm animals and farm environments. For example, a 1981 outbreak involving 42 human listeriosis cases in Nova Scotia was linked to consumption of coleslaw. This coleslaw had been produced from cabbage harvested from fields fertilized with untreated sheep manure that had been obtained from a farm with a history of ovine listeriosis (36). In rare cases, direct transmission of L. monocytogenes from animals to humans has been observed. In these cases, human symptoms have typically reflected localized cutaneous infections rather than systemic involvement (3).
In ruminants, L. monocytogenes primarily causes encephalitis and uterine infections. Uterine infections are characterized by late-term abortions or septicemia in neonates. The encephalitic form of animal listeriosis is characterized by neurological signs, including circling, excessive salivation, and unilateral facial paralysis (21, 30, 38). In addition, L. monocytogenes can cause eye infections and keratitis in ruminants; these symptoms have been linked to direct inoculation of the eye with L. monocytogenes present in feeds, especially silage (47). L. monocytogenes can be shed in the fecal material of clinically affected animals; however, healthy animals also can be latent L. monocytogenes carriers (28). Multiple studies have shown that up to 50% of fecal samples collected from animals with no clinical symptoms of listeriosis (including cattle, sheep, goats, pigs and poultry) may contain L. monocytogenes (25, 44). Although most animal listeriosis appears to be caused by ingestion of silage contaminated with high levels of L. monocytogenes, not all cases are feedborne (48). While it has been hypothesized that, as ecological systems, livestock farms may function as a natural reservoir for L. monocytogenes and, ultimately, as a primary source of food processing plant environment contamination (44), our understanding of the transmission of L. monocytogenes in the farm ecosystem is limited. For example, it is not clear whether infected animals contribute to the transmission of L. monocytogenes or whether farm animals represent “dead-end” hosts. Many authors agree that a likely scenario for L. monocytogenes transmission on farms includes initial contamination of crops and soil by wildlife, birds, or manure used to fertilize fields. While farm animals may be directly exposed to L. monocytogenes in soil and crops through grazing, L. monocytogenes numbers acquired through this route are likely to be too small to cause infection. On the other hand, improperly fermented silage (pH > 5.0 to 5.5) that was contaminated initially by soil and crops can allow subsequent amplification of L. monocytogenes to high numbers; therefore, silage feeding appears to represent a common route of infection for farm animals (38).
While L. monocytogenes has been isolated from numerous host species and different environmental sources, knowledge of the transmission dynamics and ecology of L. monocytogenes in the primary food production system is limited (23). To gain a more comprehensive understanding of the transmission dynamics and ecology of L. monocytogenes at the preharvest food system level, a case-control study of listeriosis in production ruminants (cattle, sheep, and goats) was conducted on 24 case and 28 control farms. A total of 431 L. monocytogenes isolates were collected from clinical ruminant listeriosis cases, fecal samples of asymptomatic ruminants, animal feedstuffs, and the farm environment (soil and water) and characterized by DNA subtyping. The specific objectives of this study were to (i) determine the prevalence of L. monocytogenes in healthy ruminants, animal feeds, and the farm environment; (ii) gain a better understanding of L. monocytogenes transmission at the farm level to help with the development of compartmental transmission models; and (iii) elucidate the ecology of L. monocytogenes subtypes found in ruminant farms and their relationships to human and animal disease.
MATERIALS AND METHODS
Study population.
Ruminant (cattle, goat, and sheep) listeriosis case farms enrolled in this study were identified either by the New York State Animal Health Diagnostic Laboratory at Cornell University, Ithaca, N.Y. (through positive L. monocytogenes culture results from specimens submitted by field veterinarians) or by veterinary diagnoses reported to project collaborators. Clinical listeriosis cases were defined as cases meeting one or more of the following criteria: (i) isolation of L. monocytogenes from specific organs obtained from ruminants by necropsy; (ii) gross pathology or histopathology findings indicating a L. monocytogenes infection; or (iii) veterinary diagnosis of clinical listeriosis symptoms. The study population consisted of 52 ruminant farms primarily located in New York state, including 24 case and 28 control farms. Bovine (dairy cattle, n = 15; beef cattle, n = 1) and goat (n = 4) case farms were pair matched with a single control farm, while each sheep (n = 4) case farm was matched with two control farms. Control farms were matched by the case farm species, breed, herd size, location, and overall management system (i.e., freestall, tiestall, etc.). Sample collection took place from June 2001 to June 2003.
Sample collection.
Approximately 40 samples were collected from each farm, including 10 fecal, 10 feedstuff, 10 soil, and 10 water samples. Farm management groups (i.e., milking, dry cow, etc.) were identified, and the number of samples collected in each category (fecal, soil, feedstuff, and water) was distributed approximately evenly across management groups. Each sample was collected into a sterile Whirl-Pak bag (Nasco, Fort Atkinson, Wis.) by the use of clean gloves or sampling utensils. Rectal fecal grab samples were collected from randomly selected animals in each management group. Soil samples were collected from diverse locations, including grazing pastures, crop fields, and the farmyard. Feedstuff samples collected included any ration component with the potential to support microbial growth (i.e., silage, haylage, high-moisture corn), the total mixed ration, and plant material from grazing pastures. Water samples were collected from all sources of water available to animals, including group water troughs and individual water buckets in barns as well as ponds and streams in pastures. All samples were stored in clean coolers with ice packs for transit to the laboratory. Samples were processed within 24 h of collection.
Bacteriological analysis.
For fecal materials and other solid samples, 10- and 25-g samples, respectively, were aseptically transferred into sterile stomacher closure bags (Seward Ltd., London, United Kingdom) and Listeria enrichment broth (Difco, Detroit, Mich.) was added to achieve a 1:10 dilution. For water samples, approximately 500 ml was filtered through a 0.45-μm-pore-size filter unit (Nalgene, Rochester, N.Y.). The filter was aseptically transferred to a stomacher bag, and 100 ml of Listeria enrichment broth was added. All enrichments were homogenized by stomaching (Stomacher 400, Seward) at normal speed for 1 min. Alternatively, when samples were known to contain sharp elements that might puncture bags during automatic stomaching (e.g., corn in feces), they were homogenized manually for 1 min until solid matter was completely suspended in the enrichment solution. Enrichments were incubated at 30°C, and 50-μl aliquots of the enrichments were plated onto Oxford medium (Difco) after 24 and 48 h of incubation. Oxford plates were incubated for 48 h at 30°C, and up to four well-isolated colonies with black halos and typical Listeria colony morphology were substreaked for isolation onto L. monocytogenes plating medium (LMPM; Biosynth Biochemica & Synthetica, Naperville, Ill.). LMPM plates were incubated at 35°C for 48 h. L. monocytogenes and L. ivanovii show characteristic colony morphology and a turquoise-blue color on LMPM, due to bacterial hydrolysis of a colorimetric phospholipase substrate, while other Listeria spp. will form white colonies (31). Phospholipase-positive Listeria spp. identified on LMPM were screened using a PCR assay targeting the hemolysin gene (hly), which is not found in phospholipase-positive Listeria spp. other than L. monocytogenes (27). L. monocytogenes isolates were preserved at −80°C in 15% glycerol.
Ribotyping.
A single L. monocytogenes isolate from each L. monocytogenes-positive sample (n = 414) and each clinical isolate (n = 17) was subtyped by automated EcoRI ribotyping using a Riboprinter (DuPont Qualicon, Wilmington, Del.) as described previously (4). Images were acquired with a charge-coupled device camera and processed using the Riboprinter's custom software. This software normalizes fragment pattern data for band intensity and relative band position (4). The Riboprinter generated DuPont identification numbers (IDs) (e.g., DUP-1039) for the majority of the isolates analyzed. When the Riboprinter was unable to assign a DuPont ID (i.e., for a new pattern with <0.85 similarity to existing patterns in the DuPont database), we assigned a unique type designation based on the “ribogroup” that had been assigned by the instrument (e.g., ribogroup 116-233-S-5). All DuPont IDs were confirmed by visual inspection. When an assigned DuPont ID included more than one distinct ribotype pattern (e.g., patterns differing by a single weak band), each pattern was designated with an additional alphabetized letter (e.g., DUP-1039A and DUP-1039B). Ribotype patterns for isolates in this study are available for comparison through Pathogen Tracker (www.pathogentracker.net). Ribotypes were also used to assign isolates to one of three previously described L. monocytogenes lineages (49).
Data analysis and statistical methods.
All statistical analyses were conducted with Statistical Analysis Systems software (SAS Institute, Cary, N.C.). The significance level for all statistical tests was P < 0.05. The distribution of L. monocytogenes-positive samples among the different sample categories (fecal, soil, feed, and water) between case and control farms was analyzed using a χ2 test of independence. The χ2 test of independence was also used to compare the prevalence of L. monocytogenes in bovine farms with that in small-ruminant farms for all sample categories. Fisher's exact test was used to analyze comparisons in which two or more of the expected values in a two-by-two table were less than 5.
Simpson's index of diversity (SID) was calculated as previously described (17) to assess the suitability of ribotyping for differentiation of L. monocytogenes isolates and to describe the diversity of L. monocytogenes subtypes from different sample populations. Specifically, SID was calculated for isolates from all farms, all case farms, and all control farms pooled as well as individually for each herd with five or more L. monocytogenes-positive samples. The Wilcoxon-Mann-Whitney exact test was performed using the one-way nonparametric procedure to compare L. monocytogenes molecular subtype diversity of case farms to that of control farms and of bovine farms to that of small-ruminant farms. The Wilcoxon-Mann-Whitney test was selected for its ability to test the distribution of ordinal-scaled responses in independently sampled populations and for its sensitivity to differences in location of sample populations (39).
L. monocytogenes molecular subtypes were stratified by sample category (i.e., fecal, feed, soil, and water) and by farm system (i.e., case and control) to calculate the distribution frequency for each ribotype and lineage. Clinical isolate subtypes were not included in analysis of ribotype distribution frequency. Only ribotypes with a frequency higher than 5 were analyzed individually; all ribotypes with an observed frequency lower than 5 were combined into a subtype category termed “other.” McNemar's test was employed to assess the association of specific molecular subtypes (ribotype and lineage) for all possible pairwise comparisons between sample categories (e.g., fecal versus soil). Univariate logistic regression correcting for within-farm correlations of multiple isolates by the general estimating equation approach was used to examine the relationship of subtypes to farm system (case versus control) and host species (cattle versus small ruminant). A variable linking each matched case and control farm pair was forced into each regression model to account for the matched-pair study design.
RESULTS
Prevalence of L. monocytogenes on all farms.
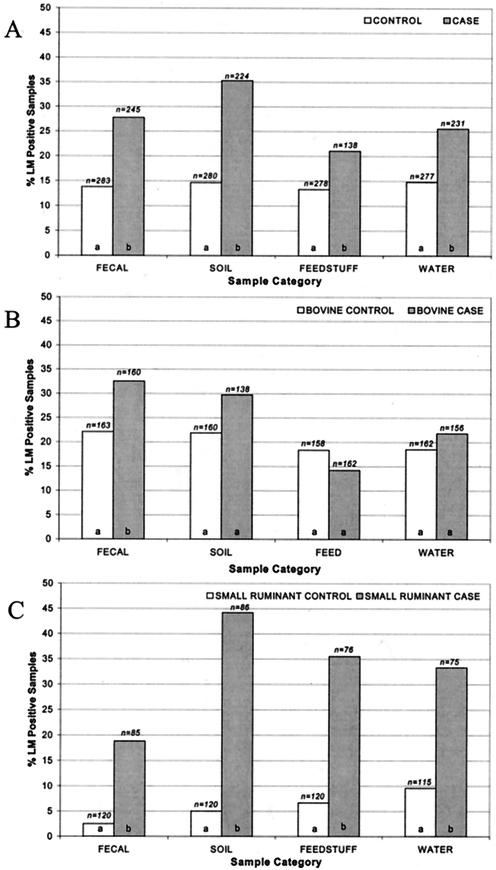
Of the 24 case farms enrolled in this study, 16 had case animals with encephalitis, 4 had case animals with abortions, 3 had cows with clinical mastitis, and 1 had an animal with keratitis. A total of 17 clinical L. monocytogenes isolates from 16 case farms were available (for 1 farm, 2 clinical isolate subtypes were obtained from two different animals); 8 case farms were identified by veterinarian diagnosis of listeriosis, and no clinical isolates were available for these farms. Overall, 2,056 samples, including 528 fecal, 504 soil, 516 feedstuff, and 508 water samples, were collected from 24 case and 28 control farms. A total of 414 samples (107 fecal, 120 soil, 87 feedstuff, and 100 water) were positive for L. monocytogenes, yielding an overall prevalence of 20.1%. Case farms showed a significantly (P < 0.0001) higher overall prevalence of L. monocytogenes (27.3%) than control farms (14.1%). Specifically, case farms showed a significantly (P < 0.0001) higher prevalence of L. monocytogenes in fecal and soil samples (27.8 and 35.3%, respectively) than control farms (13.8 and 14.6%, respectively). L. monocytogenes was more prevalent (P < 0.05) in animal feed samples from case farms (21.0%) than in those from with control farms (13.3%). Case farms, furthermore, had a significantly (P < 0.005) higher prevalence of L. monocytogenes in water samples (25.5%) than control farms (14.8%) (Fig. 1A).
FIG. 1.
Prevalence of L. monocytogenes (LM) in fecal, soil, feedstuff, and water samples collected on all farms (A), cattle farms (B), and small-ruminant farms (goat and sheep farm results were pooled) (C). Different letters indicate statistically different levels of L. monocytogenes in comparisons of prevalence of L. monocytogenes in each sample category of case and control farms at the P < 0.05 level. The total number (n) of samples collected in each sample category is indicated above each bar.
Prevalence of L. monocytogenes on bovine farms.
Data were stratified by species to further analyze the ecology of L. monocytogenes. A total of 1,259 samples, including 323 fecal, 298 soil, 320 feedstuff, and 318 water samples, were collected from bovine case and control farms. L. monocytogenes was found significantly (P < 0.05) more frequently in the feces of healthy cattle collected on case farms than in fecal samples collected on control farms. No significant (P > 0.05) differences were observed between bovine case and control farms in terms of the prevalence of L. monocytogenes in animal feeds and the farm environment (soil and water). L. monocytogenes was isolated from bovine control farm feedstuff samples at a higher rate (18.4%) than it was isolated from bovine case farm samples (14.2%), although the difference was not statistically significant (P = 0.3) (Fig. 1B).
Prevalence of L. monocytogenes on small-ruminant farms.
Because the prevalence of L. monocytogenes in goat farms was statistically similar to that in sheep farms (P > 0.05) but different from the prevalence in cattle farms (P < 0.05), goat and sheep farm data were pooled and termed small-ruminant farm results. A total of 797 samples (205 fecal, 206 soil, 196 feedstuff, and 190 water) were collected from small-ruminant farms. Small-ruminant case farms showed a significantly (P < 0.0001) higher prevalence of L. monocytogenes than control farms for all sample categories (animal feces, feed, soil, and water) (Fig. 1C).
Prevalence of L. monocytogenes in bovine versus small-ruminant farms.
Overall, 22.2% of samples collected on bovine farms and 16.8% of samples collected on small-ruminant farms tested positive for L. monocytogenes. The prevalence of L. monocytogenes in soil, feedstuff, and water samples from cattle farms was statistically similar (P > 0.05) to that for the corresponding samples from small-ruminant farms. Also, on the basis of pooled data from case and control farms, L. monocytogenes was found more frequently in fecal samples from healthy cattle than in samples collected from healthy small ruminants (P < 0.0001). When bovine control farms were compared with small-ruminant control farms, bovine control farms showed a higher (P < 0.05) prevalence of L. monocytogenes in fecal, feed, soil, and water samples. On the other hand, small-ruminant case farms showed a higher prevalence of L. monocytogenes in feed (P < 0.001), soil (P < 0.05), and water (P = 0.06) samples than did bovine case farms.
Transmission of L. monocytogenes.
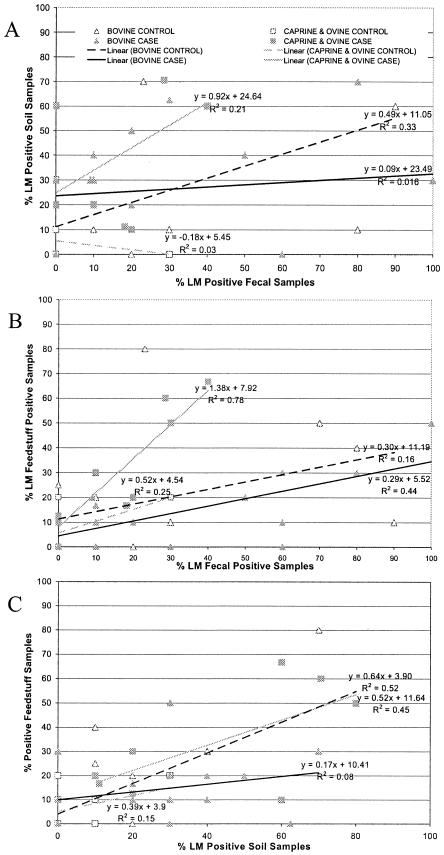
To assess the relationship between the prevalence of L. monocytogenes-positive samples in one sample category (e.g., animal feed) and the prevalence of positive-testing samples in all other sample categories, herd-level prevalence scatter plots were constructed and regression analyses were performed (Fig. 2). While scatter plots were constructed for all six possible relationships of sample categories, R2 values were low for many regression analyses and only the three regression analyses (soil versus fecal, feed versus fecal, and soil versus feed) that yielded high R2 values and are of particular relevance to transmission of L. monocytogenes are reported here (Fig. 2).
FIG. 2.
Scatter plots of percentages of L. monocytogenes (LM)-positive fecal samples versus L. monocytogenes-positive soil samples (A), L. monocytogenes-positive fecal samples versus L. monocytogenes-positive feed samples (B), and L. monocytogenes-positive soil samples versus feed samples (C). Triangles represent bovine farms, while squares indicate small-ruminant farms (goat and sheep farm results were pooled). Control farms are depicted by open symbols, while case farm symbols are shaded. Regression equations and R2 values determined by regression lines for each farm system in each scatter plot comparison are indicated.
The scatter plot of L. monocytogenes prevalence in fecal samples versus that in soil samples (Fig. 2A) shows that the slope of the regression equation for all farms was less than 1.0, indicating that L. monocytogenes was more prevalent in animal feces than in soil. Under the assumption that L. monocytogenes transmission is most likely to proceed from highly contaminated compartments to less-contaminated compartments, these data indicate that L. monocytogenes is transmitted from animal feces to soil. The regression line for small-ruminant case farms showed a slope close to 1.0 (Fig. 2A), possibly indicating that farm environment, particularly soil, contamination by L. monocytogenes plays a more important role in the incidence of listeriosis in small ruminants than in cattle.
The scatter plot of L. monocytogenes prevalence in fecal samples versus that in feed samples (Fig. 2B) further shows that there is a clear difference between the transmission characteristics of L. monocytogenes in small-ruminant and cattle case farms. The regression line for small-ruminant case farms shows a slope of more than 1.0, indicating that L. monocytogenes was more prevalent in feed samples than in fecal samples, consistent with a transmission model that proceeds from feed to animals. On bovine farms, L. monocytogenes was more prevalent in animal feces than in feed (slope < 1.0). These data are consistent with exposure of cattle to L. monocytogenes via contaminated feeds followed by intrahost amplification to high levels and subsequent fecal shedding. The scatter plot of L. monocytogenes prevalence in soil samples versus that in animal feeds (Fig. 2C) indicates that in all farms, L. monocytogenes was more prevalent in soil samples than in animal feeds (slope < 1.0). Thus, farm soil appears to be an important source of ruminant feedstuff contamination by L. monocytogenes.
Molecular subtypes of L. monocytogenes on farms.
All 17 clinical and 414 fecal, feed, and environmental L. monocytogenes isolates were characterized by EcoRI ribotyping. A total of 51 unique EcoRI ribotypes were differentiated. The frequencies of L. monocytogenes subtypes observed at a level of more than 5 among all sample categories and farm systems are summarized in Table 1. Ribotypes DUP-1039C (n = 61), DUP-1042B (n = 60), DUP-1039E (n = 39), and DUP-1045A (n = 24) were the most commonly observed L. monocytogenes molecular subtypes. A variety of L. monocytogenes ribotypes, including DUP-1038B, DUP-1042B, and DUP-1044A, which have been shown to be associated with human listeriosis cases and outbreaks (19, 34) were commonly present in ruminant farms.
TABLE 1.
Distribution of L. monocytogenes ribotypes among case and control farms
| Subtype (lineage) | No. of L. monocytogenes isolates
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case farms
|
Control farms
|
|||||||||
| Fecal | Feed | Soil | Water | Total (%) | Fecal | Feed | Soil | Water | Total (%) | |
| DUP-1023A (II) | 2 | 0 | 4 | 2 | 8 (3.1) | 0 | 1 | 2 | 1 | 4 (2.5) |
| DUP-1023B (II) | 1 | 1 | 0 | 1 | 3 (1.2) | 0 | 2 | 0 | 0 | 2 (1.3) |
| DUP-1030B (II) | 0 | 1 | 2 | 4 | 7 (2.7) | 2 | 0 | 2 | 0 | 4 (2.5) |
| DUP-1038B (I) | 6 | 2 | 1 | 1 | 10 (3.9) | 1 | 0 | 0 | 2 | 3 (1.9) |
| DUP-1039A (II) | 1 | 2 | 3 | 2 | 8 (3.1) | 1 | 3 | 1 | 3 | 8 (5.1) |
| DUP-1039C (II) | 9 | 4 | 15 | 11 | 39 (15.2) | 11 | 4 | 5 | 2 | 22 (13.9) |
| DUP-1039E (II) | 7 | 3 | 2 | 3 | 15 (5.9) | 5 | 5 | 4 | 10 | 24 (15.2) |
| DUP-1042B (I) | 13 | 9 | 18 | 5 | 45 (17.6) | 0 | 4 | 4 | 7 | 15 (9.5) |
| DUP-1042C (I) | 4 | 1 | 2 | 1 | 8 (3.1) | 0 | 0 | 0 | 0 | 0 |
| DUP-1044A (I) | 1 | 0 | 0 | 0 | 1 (0.4) | 5 | 2 | 1 | 0 | 8 (5.1) |
| DUP-1044B (I) | 1 | 4 | 3 | 0 | 8 (3.1) | 0 | 0 | 0 | 1 | 1 (0.6) |
| DUP-1045A (II) | 0 | 1 | 3 | 2 | 6 (2.3) | 3 | 5 | 6 | 4 | 18 (11.4) |
| DUP-1045B (II) | 5 | 0 | 1 | 4 | 10 (3.9) | 0 | 0 | 1 | 0 | 1 (0.6) |
| DUP-1045D (II) | 1 | 6 | 1 | 1 | 9 (3.5) | 1 | 1 | 1 | 2 | 5 (3.2) |
| DUP-1045E (II) | 2 | 3 | 4 | 6 | 15 (5.9) | 0 | 0 | 0 | 0 | 0 |
| DUP-1052A (I) | 0 | 3 | 0 | 0 | 3 (1.2) | 0 | 2 | 2 | 0 | 4 (2.5) |
| DUP-1054A (II) | 1 | 1 | 1 | 1 | 4 (1.6) | 1 | 0 | 0 | 0 | 1 (0.6) |
| DUP-1062D (II) | 2 | 0 | 2 | 0 | 4 (1.6) | 0 | 2 | 0 | 2 | 4 (2.5) |
| DUP-1062E (II) | 2 | 1 | 0 | 3 | 6 (2.3) | 1 | 1 | 1 | 1 | 4 (2.5) |
| DUP-1062F (II) | 0 | 0 | 1 | 0 | 1 (0.4) | 1 | 0 | 4 | 0 | 5 (3.2) |
| 110-239-S2 | 2 | 2 | 3 | 0 | 7 (2.7) | 0 | 0 | 1 | 0 | 1 (0.6) |
| Other ribotype | 8 | 6 | 13 | 12 | 39 (15.2) | 7 | 5 | 6 | 6 | 24 (15.2) |
| Lineage I | 32 | 24 | 34 | 12 | 102 (39.8) | 9 | 9 | 10 | 10 | 38 (24.1) |
| Lineage II | 35 | 26 | 45 | 47 | 153 (59.8) | 30 | 28 | 29 | 30 | 117 (74.1) |
| Lineage III | 1 | 0 | 0 | 0 | 1 (0.4) | 0 | 0 | 2 | 1 | 3 (1.9) |
| Total | 68 | 50 | 79 | 59 | 256 | 39 | 37 | 41 | 41 | 158 |
Diversity of L. monocytogenes on farms.
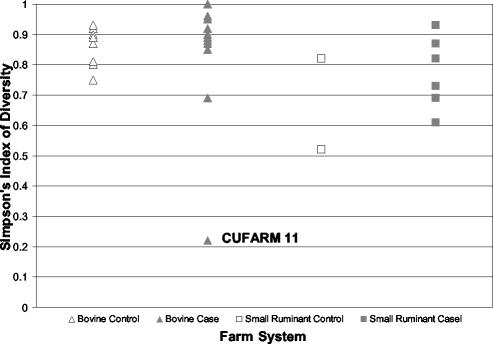
SID quantifies the ability of a molecular subtyping method to discriminate among isolates that are truly different and measures the diversity of subtypes within independent populations. Values for SID range between 0 and 1, with a value of 1 indicating the most diverse population. The SID value for all farms was 0.94, with a variance of 3.0 × 10−5, while the SID values for case and control farms were 0.93 (variance, 6.6 × 10−5) and 0.93 (variance, 7.8 × 10−5), respectively. SID values for individual farms are shown in Fig. 3. The mean SID values for all control farms (n = 12) and for all case farms (n = 19) were identical (SID = 0.82). The mean SID for bovine farms (n = 23; SID = 0.84) was significantly (P < 0.05; Wilcoxon-Mann-Whitney test) larger than the mean SID for small-ruminant farms (n = 8; SID = 0.75). Not all pairwise comparisons were performed due to the small number (n = 2) of small-ruminant control farms with more than five L. monocytogenes isolates.
FIG. 3.
Scatter plot of SID for individual farms with more than four L. monocytogenes-positive samples. Triangles represent cattle farms, while squares indicate small-ruminant farms (goat and sheep farm results were pooled). Control farms are depicted by open symbols, while case farm symbols are shaded. CUFARM 11, results from a bovine case farm that showed an extremely low SID (0.22); eight of the nine isolates from this farm were identical with respect to ribotype (DUP-1039E).
Analysis of L. monocytogenes ribotypes.
Ribotype data were also used to further probe L. monocytogenes transmission in farms. We specifically analyzed the relationship between L. monocytogenes isolates from animals with clinical disease and L. monocytogenes isolates recovered from the respective case farm environments (Table 2). Clinical L. monocytogenes isolates were available for 16 of the 24 case farms in this study. Ribotype DUP-1039C was the predominant clinical L. monocytogenes strain and was isolated from animals with clinical listeriosis on eight farms from which clinical isolates were available. Overall, the same L. monocytogenes ribotype isolated from clinical cases was also identified in feces and/or environmental (feed, soil, or water) samples in 10 of the 16 respective farms. Specifically, the clinical L. monocytogenes ribotype was also detected in fecal samples from healthy animals on six farms and in soil samples on four farms as well as in feed and water samples on two farms each. Organs from multiple animals with clinical listeriosis were available from one farm (CUFARM 8). Two different ribotypes were detected among the clinical L. monocytogenes isolates on this farm; eight clinical isolates were ribotype DUP-1045E, and one was ribotype DUP-1039C. Both clinical ribotypes were recovered in soil samples collected on this farm (Table 2). A case-control comparison showed that a ribotype matching the clinical ribotype was significantly more likely to be isolated from the respective case farm than from the matched control farm (P < 0.05; χ2 test of independence). This indicates that there is a significant association between the presence of a given subtype from a clinical case animal and the presence of this L. monocytogenes subtype in the farm environment, animal feed, or fecal samples from healthy animals on the farm.
TABLE 2.
Recovery of clinical L. monocytogenes subtypes in samples from respective case and matched control farms
| Farm | Animal species | Clinical subtype (lineage) | No. of isolates with matching ribotype recovered on case farm from:
|
Isolates with matching ribotype recovered from samples collected on the matched control farm | |||
|---|---|---|---|---|---|---|---|
| Feces | Soil | Feed | Water | ||||
| 1 | Bovine | DUP-1039C (II) | 1 | 1 | 0 | 0 | No |
| 3 | Bovine | DUP-1038B (I) | 1 | 0 | 0 | 0 | No |
| 7 | Small ruminant | DUP-1042B (I) | 0 | 0 | 0 | 0 | Yes |
| 8 | Small ruminant | DUP-1039C (II) | 0 | 1 | 0 | 0 | No |
| 8 | Small ruminant | DUP-1045E (II) | 2 | 4 | 4 | 3 | No |
| 11 | Bovine | DUP-1039C (II) | 0 | 0 | 0 | 0 | Yes |
| 12 | Bovine | DUP-1039C (II) | 0 | 2 | 0 | 0 | No |
| 13 | Bovine | DUP-1038B (I) | 0 | 0 | 0 | 0 | No |
| 20 | Bovine | DUP-1030A (II) | 1 | 0 | 0 | 0 | No |
| 21 | Bovine | DUP-1045B (II) | 1 | 0 | 0 | 0 | No |
| 22 | Small ruminant | DUP-1039C (II) | 0 | 0 | 1 | 0 | Yes |
| 23 | Bovine | DUP-1030B (II) | 0 | 0 | 0 | 0 | No |
| 30 | Bovine | DUP-1039C (II) | 0 | 2 | 0 | 1 | No |
| 36 | Bovine | DUP-1042B (I) | 0 | 0 | 0 | 0 | No |
| 39 | Bovine | DUP-1039C (II) | 0 | 0 | 0 | 0 | No |
| 43 | Bovine | DUP-1039C (II) | 1 | 0 | 0 | 0 | Yes |
| 46 | Small ruminant | DUP-1045D (II) | 0 | 0 | 0 | 0 | No |
Recovery of fecal ribotypes in animal feed and the farm environment.
To further elucidate the transmission of L. monocytogenes, we also analyzed the relationship between L. monocytogenes ribotypes isolated from fecal samples on both case and control farms and ribotypes isolated from animal feeds and the farm environment of each respective L. monocytogenes fecal-sample-positive herd (Table 3). Overall, 28 different ribotypes were identified among the 107 fecal L. monocytogenes isolates obtained from 30 herds that had at least one animal with a L. monocytogenes-positive fecal sample. Ribotypes identical to those observed in fecal samples were also found in soil, feed, and water samples collected from the same farm on 16, 13, and 16 occasions, respectively. This analysis indicated that ruminant fecal shedding of L. monocytogenes contributes to the dispersal of L. monocytogenes fecal subtypes into the farm environment.
TABLE 3.
Recovery of fecal L. monocytogenes subtypes in environmental samples
| Subtype (lineage) | No. of farms with ribotype isolated from fecal sample | No. of farms where subtype was also recovered from:
|
||
|---|---|---|---|---|
| Soil | Feed | Water | ||
| DUP-1023A (II) | 2 | 1 | 0 | 0 |
| DUP-1023B (II) | 1 | 0 | 0 | 0 |
| DUP-1025A (I) | 2 | 1 | 0 | 0 |
| DUP-1030B (II) | 2 | 0 | 0 | 0 |
| DUP-1038B (I) | 7 | 1 | 0 | 1 |
| DUP-1039A (II) | 2 | 0 | 1 | 1 |
| DUP-1039C (II) | 12 | 4 | 2 | 5 |
| DUP-1039E (II) | 4 | 0 | 1 | 3 |
| DUP-1042A (I) | 1 | 0 | 0 | 0 |
| DUP-1042B (I) | 8 | 4 | 4 | 2 |
| DUP-1042C (I) | 3 | 1 | 0 | 1 |
| DUP-1044A (I) | 2 | 0 | 0 | 0 |
| DUP-1044B (I) | 1 | 0 | 0 | 0 |
| DUP-1045A (II) | 2 | 2 | 1 | 0 |
| DUP-1045B (II) | 3 | 0 | 0 | 1 |
| DUP-1045D (II) | 2 | 0 | 1 | 0 |
| DUP-1045E (II) | 1 | 1 | 1 | 1 |
| DUP-1051C (I) | 1 | 0 | 1 | 1 |
| DUP-1054A (II) | 2 | 0 | 0 | 0 |
| DUP-1062B (II) | 1 | 0 | 0 | 0 |
| DUP-1062C (II) | 1 | 0 | 0 | 0 |
| DUP-1062E (II) | 2 | 0 | 0 | 0 |
| DUP-1062F (II) | 1 | 0 | 0 | 0 |
| DUP-16635B (I) | 2 | 0 | 0 | 0 |
| 116-110-S2 (III) | 1 | 0 | 0 | 0 |
| 116-239-S2 (I) | 1 | 1 | 1 | 0 |
| 116-890-S3 (II) | 1 | 0 | 0 | 0 |
| 116-915-S2 (?)b | 1 | 0 | 0 | 0 |
| Total no. of positive-testing herds | 30a | 16 | 13 | 16 |
Total number of farms with L. monocytogenes-positive fecal samples.
?, lineage not known.
Statistical analysis of ribotype distribution.
The distribution of L. monocytogenes subtypes (both ribotypes and lineages) between case and control farms was evaluated to determine whether specific subtypes were more or less likely to be associated with animal listeriosis. Associations between specific L. monocytogenes subtypes and different sample categories (fecal, soil, feed, and water) as well as associations between subtypes and bovine or small-ruminant host farms were also evaluated. After within-herd correlation corrections of L. monocytogenes subtypes were implemented, ribotype DUP-1045A was significantly (P < 0.05) associated with control farms. While several ribotypes (e.g., DUP-1038B, DUP-1042B, DUP-1042C, DUP-1045B, and DUP-1045E; Table 1) were more common in case than in control farms, these strains were clustered in a single or few herds and no specific ribotypes were significantly (P > 0.05) associated with case farms after within-herd correlation corrections were implemented. Ribotype DUP-1039E was detected in both bovine and small-ruminant farms but was significantly (P < 0.01) more common in bovine farms than in small-ruminant farms. McNemar's test indicated that ribotype DUP-1038B was more commonly isolated from ruminant feces than from animal feed (P < 0.05), farm soil (P < 0.05), or water (P = 0.06). Additionally, L. monocytogenes subtype DUP-1045A was more commonly isolated from farm soil (P < 0.05) than from ruminant feces. Before corrections for clustering of ribotypes within a herd were implemented, lineage I isolates were significantly associated with case farms (P < 0.001) and lineage II isolates were associated with control farms (P < 0.01). However, due to the high level of correlation of subtypes in farms, these associations were not significant after correction using the general estimating equation approach.
DISCUSSION
Using a case-control study design, we have collected comprehensive datasets on the prevalence and subtype composition of L. monocytogenes in ruminant feces, animal feeds, and the farm environment. Our data suggest that (i) the epidemiology and transmission characteristics of L. monocytogenes differ between small-ruminant and cattle farms; (ii) cattle contribute to amplification and dispersal of L. monocytogenes into the farm environment; (iii) the ruminant, and particularly the bovine, farm ecosystem maintains a high prevalence of L. monocytogenes, including subtypes linked to human listeriosis cases and outbreaks, and may thus constitute a significant natural reservoir for L. monocytogenes; and (iv) L. monocytogenes subtypes may differ in their abilities to infect animals and to survive in farm environments. Our findings provide insight into L. monocytogenes transmission in ruminant farming systems. Ultimately, work of this nature will allow better control of listeriosis in animals and the reduction of the incidence of foodborne listeriosis in humans by providing information on potential reservoirs and sources of L. monocytogenes that contribute to introduction of this pathogen into the human food supply.
L. monocytogenes ecology and transmission characteristics differ between bovines and small ruminants.
Our data clearly indicate that the epidemiology and ecology characteristics of L. monocytogenes differ between bovine and small-ruminant farms. A significantly higher prevalence of L. monocytogenes-positive samples was found in bovine farms without listeriosis cases than in small-ruminant farms without listeriosis cases. These findings are consistent with previous preliminary observations regarding the epidemiology of listeriosis among bovines and small ruminants (21, 30, 44). For example, in 1999, Wesley (44) noted that L. monocytogenes fecal shedding was more frequent in cattle (33%) than in sheep (8%), comparable with our data. Also, while listeriosis has been reported as primarily sporadic in cattle, high morbidity rates have been reported among sheep flocks and goat herds (30). We also found that the diversity of L. monocytogenes populations on bovine farms was greater than that observed in small-ruminant farms. This may indicate that small-ruminant farms are characterized by a single or a few subtypes that cause disease in small ruminants and that bovine case and control farms maintain highly diverse L. monocytogenes populations, with animals frequently exposed to multiple L. monocytogenes subtypes.
Our feed prevalence data indicate that small ruminants on farms without listeriosis appear less likely to be exposed to L. monocytogenes than cattle, which appear to be frequently exposed to L. monocytogenes through contaminated feeds and consequently may be immune or less susceptible to infection. Our feed prevalence data also suggest that while exposure to L. monocytogenes through contaminated feeds appears to be sufficient to cause listeriosis in small ruminants, clinical listeriosis in cattle may require additional contributing factors (since L. monocytogenes prevalence does not differ between bovine case and control farms). For example, the combination of exposure to particularly virulent L. monocytogenes subtypes and predisposing host factors, such as postpartum immune deficiency or pregnancy, may be required for clinical listeriosis in cattle (2, 26).
Cattle contribute to amplification and dispersal of L. monocytogenes into the farm environment.
While many studies have indicated that poor-quality silage is commonly contaminated with L. monocytogenes (6, 8, 16, 40, 51) and may represent the predominant source of listeriosis in ruminants (33, 41, 42, 50), little is known about the actual transmission dynamics of L. monocytogenes in production agriculture systems. For example, it is not known whether, and to what extent, infection of animals is necessary for L. monocytogenes dispersal into the environment or whether ruminants and possibly other mammals represent dead-end hosts that do not contribute to the survival and ecological success of this pathogen. Overall, our data indicate that L. monocytogenes subtypes found in clinically affected animals, as well as those subtypes isolated from fecal samples, were also markedly associated with isolation from environmental sources on the respective farms. While our data show a clear association between the presence of L. monocytogenes in clinical and fecal samples and its presence in environmental samples, the nature of a cross-sectional study makes it difficult to draw conclusions on directionality of transmission. We thus used herd-level prevalence scatter plots and regression analyses to allow a preliminary assessment of the relationship between L. monocytogenes prevalence values in different sample categories. This analysis indicated that bovine case and control farms showed similar L. monocytogenes transmission dynamics. Although small-ruminant control farms showed regression patterns similar to the bovine farm patterns, we do not consider these patterns reliable enough to be included in this discussion due to the low R2 values (R2 ≤0.25; Fig. 2). Assuming that transmission is most likely to proceed from highly contaminated compartments to less-contaminated compartments, it appears that animals on bovine farms are the source of environmental contamination, since L. monocytogenes is more prevalent in fecal samples than in soil samples. Similarly, on bovine farms, L. monocytogenes prevalence was higher in fecal samples than in feed samples. These data are consistent with the hypothesis that animals exposed to L. monocytogenes through contaminated silage amplify the pathogen, which leads to higher prevalence in feces than in feed. In 1996, Fenlon and coworkers (11) also found that soil samples from pastures of silage-fed ruminants are more likely to harbor L. monocytogenes compared to other pastures, further supporting the idea that L. monocytogenes strains ingested through silage are amplified and reintroduced into the farm environment by cattle.
In all farm systems, we found evidence that the prevalence of L. monocytogenes in soil was higher than that seen in feed, indicating that soil may serve as a source of animal feed contamination by L. monocytogenes. These findings are consistent with other reports (10) and are further supported by our molecular subtype data, which showed that the same L. monocytogenes molecular subtype was observed in both feed and soil samples on 7 of 26 farms with positive soil and feedstuff sample results; 12 of the 19 farms with no matching soil and feed subtypes had a single L. monocytogenes-positive soil or feed sample, thus permitting no meaningful analyses for these farms. On the other hand, on small-ruminant case farms, L. monocytogenes prevalence in feeds was higher than in fecal samples. These data support our hypothesis that the epidemiology characteristics of listeriosis differ between bovine and small-ruminant farms and may indicate that small ruminants are less likely to appreciably amplify ingested L. monocytogenes. We conclude that bovine hosts amplify L. monocytogenes ingested through feeds and thus represent a critical factor in maintaining high levels of L. monocytogenes contamination on bovine farms.
Ruminant farm ecosystems maintain a high prevalence of L. monocytogenes, including subtypes linked to human listeriosis cases and outbreaks.
While L. monocytogenes is generally considered ubiquitous, limited data on the prevalence of this pathogen in different environments have been available so far. While 20.1% of the 2,056 farm samples collected were positive for L. monocytogenes, the prevalence of L. monocytogenes among 1,805 soil, water, and other environmental samples from urban and pristine environments (i.e., locations with little direct animal and human contact, such as state parks, national forests, etc.), also collected by our group during the same time period and in the same region (New York state), was considerably lower (1.3 and 7.3% for pristine and urban environments, respectively) (35; B. D. Sauders and M. Wiedmann, unpublished data). Similarly, other smaller-scale studies have also shown L. monocytogenes to be quite prevalent in farm environments (5, 10). For example, Garcia and coworkers (12) detected L. monocytogenes in 7.8% of water-trough samples, 1.2% of feed samples, 11.4% of bedding samples, 8.3% of farmyard soil samples, and 2.7% of ewe fecal samples in a study with 15 sheep farms. Our finding that fecal samples from healthy cattle frequently contain L. monocytogenes is consistent with other reports that L. monocytogenes prevalence ranged from 3.1 to 51.0% in fecal samples from healthy cattle (18, 37). The high prevalence of L. monocytogenes in samples collected on ruminant farms, particularly in contrast with the low prevalence in samples collected in other environments, suggests that ruminant farms may represent an important natural reservoir for L. monocytogenes.
Molecular subtype data for all 431 L. monocytogenes isolates collected in this study were used to further probe the importance of the L. monocytogenes populations on ruminant farms as reservoirs for human infections. Interestingly, many of the L. monocytogenes EcoRI ribotypes isolated during this study have also been isolated from human clinical listeriosis cases and outbreaks that have occurred in the same state as the farms enrolled in our study (27, 34; additional data available at www.pathogentracker.net). For example, ribotype DUP-1042B (the second most commonly isolated ribotype in this study) has been linked to two human listeriosis clusters that occurred in New York state in 1998 and 1999 (34) as well as to outbreaks in Boston (United States; 1979) and Massachusetts (United States; 1983) (19). Similarly, ribotypes DUP-1038B and DUP-1044A, which have also been linked to multiple human listeriosis outbreaks (34, 19), were also isolated on multiple farms. While ribotyping provides a highly standardized subtyping method for L. monocytogenes (46), which is well suited for population-based studies, our ribotype data do not show that individual farms have served as direct or indirect sources of specific human listeriosis. Rather, our data indicate that the L. monocytogenes subtype populations found on farms overlap with those responsible for human listeriosis cases. While most raw foods originating from ruminant farms undergo heat treatments that effectively inactivate L. monocytogenes, farms may serve as a source of L. monocytogenes strains that are inadvertently introduced into food-processing environments and subsequently transferred onto foods through postprocessing contamination (1, 45). For example, subtype data for a total of 475 Listeria isolates from different dairy processing facilities and farms showed that 8 L. monocytogenes and 12 Listeria species ribotypes were found in both dairy-processing and farm environments, supporting the potential of on-farm sources to contribute to processing plant contamination (1).
L. monocytogenes subtypes may differ in their abilities to infect animals and survive in farm environments.
While previous studies using molecular subtyping and phenotypic characterization of human, food, and animal isolates have led to the hypothesis that L. monocytogenes subtypes differ in their virulence and transmission characteristics (20, 46), due to the rare nature of listeriosis, few groups have utilized case-control study data to robustly test this hypothesis. Our data support the idea that L. monocytogenes subtypes can differ in their transmission characteristics. For example, while ribotype DUP-1038B was significantly associated with fecal samples compared to environmental samples and was responsible for clinical listeriosis cases on two farms, ribotype DUP-1045A was not observed among the clinical isolates and was associated with control farms and environmental samples. Interestingly, ribotype DUP-1038B has been identified previously as a L. monocytogenes clonal group with high-level virulence potential; this ribotype has been linked to multiple human listeriosis outbreaks (e.g., in Los Angeles in 1985) (19), while ribotype DUP-1045A has been isolated only rarely among human clinical isolates (<0.5% of a collection of more than 600 human clinical isolates [M. Wiedmann, B. D. Sauders, and E. D. Fortes, unpublished data]). Ribotype DUP-1039C, the most common subtype isolated in this study, accounted for 8 of the 17 clinical L. monocytogenes isolates and was also found in fecal samples from 12 herds and in a variety of environmental samples. Our data thus suggest that some L. monocytogenes subtypes (e.g., DUP-1038B) may be adapted to infect mammalian hosts, some (DUP-1045A) may be adapted to environmental survival, and others (e.g., DUP-1039C) may be equally adapted to ruminant hosts and environmental survival and, thus, may be particularly successful at surviving by establishing high population densities in multiple niches.
Conclusions.
Our data support the hypothesis that ruminant, and particularly bovine, farms represent a reservoir for human L. monocytogenes infections. A reservoir may be described as a single or multiple epidemiologically related populations (e.g., ruminants) and/or environments (e.g., animal feed and farm environment) in which a specific pathogen is sustained; additionally, the pathogen is transmitted from the proposed reservoir to cause infections in a population of interest (e.g., humans) (14). In support of this hypothesis, we have shown that maintenance and on-farm transmission of L. monocytogenes appears to rely on ingestion of L. monocytogenes-contaminated feeds and amplification of L. monocytogenes by bovine hosts both with or without clinical disease, followed by fecal dispersal into the farm environment. We have further shown that L. monocytogenes subtypes found on bovine farms have also been associated with human listeriosis cases and outbreaks. A high prevalence of L. monocytogenes on farms provides additional support for the role of ruminant farms as reservoirs for this foodborne pathogen.
While farms may serve as direct or indirect sources of L. monocytogenes strains that are introduced into the human food chain (1, 45), they also represent ecosystems that could facilitate the emergence of new and more virulent L. monocytogenes subtypes in a high-transmission-frequency environment. While cattle have been identified as potentially important reservoirs for Escherichia coli O157:H7 (29) and certain Salmonella serotypes (9, 43), their role as a reservoir for L. monocytogenes is likely to be different and more complex, since, in contrast with Salmonella and E. coli O157:H7 infections, raw animal-derived food products are rarely a direct source of human listeriosis. As additional data on the transmission and ecology of L. monocytogenes along the farm-to-table continuum become available, we will gain a more complete understanding of the natural history of this pathogen which will facilitate rational and effective control strategies to reduce both human and animal listeriosis cases.
Acknowledgments
This work was supported by National Institutes of Health grant award R01GM63259.
We are very grateful for the willingness of the farmers to participate in this study. We thank Frank Welcome, Daryl Nydam, and Mary Smith for their assistance in identifying farms to be enrolled in this study. We are also indebted to Katy Windham for expert technical assistance in the molecular subtyping of our isolates.
REFERENCES
- 1.Arimi, S. M., T. J. Pritchard, and C. W. Donnelly. 1997. Diversity of Listeria ribotypes recovered from dairy cattle, silage and dairy processing environments. J. Food. Prot. 60:811-816. [DOI] [PubMed] [Google Scholar]
- 2.Beatz, A. L., and I. V. Wesley. 1995. Detection of anti-listeriolysin O in dairy cattle experimentally infected with Listeria monocytogenes. J. Vet. Diagn. Investig. 7:82-86. [DOI] [PubMed] [Google Scholar]
- 3.Blenden, D. C., E. H. Kampelmacher, and M. J. Torres-Anjel. 1987. Listeriosis. J. Am. Vet. Med. Assoc. 191:1546-1551. [PubMed] [Google Scholar]
- 4.Bruce, J. 1996. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 50:77-81. [Google Scholar]
- 5.Cooper, J., and R. D. Walker. 1998. Listeriosis. Vet. Clin. N. Am. Food Anim. Pract. 14:113-125. [DOI] [PubMed] [Google Scholar]
- 6.Crump, J. A., P. M. Griffin, and F. J. Angulo. 2002. Bacterial contamination of animal feed and its relationship to human foodborne illness. Clin. Infect. Dis. 35:859-865. [DOI] [PubMed] [Google Scholar]
- 7.Dalton, C. B. C. C. Austin, J. Sobel, P. S. Hayes, W. F. Bibb, L. M. Graves, B. Swaminathan, M. E. Proctor, and P. M. Griffin. 1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336:100-105. [DOI] [PubMed] [Google Scholar]
- 8.Driehuis, F., and S. J. W. H. Oude Elferink. 2000. The impact of the quality of silage on animal health and food safety: a review. Vet. Q. 22:212-216. [DOI] [PubMed] [Google Scholar]
- 9.Eperigin, H. E., and K. V. Nagaraja. 1998. Salmonella. Vet. Clin. N. Am. Food Anim. Pract. 14:17-29. [PubMed] [Google Scholar]
- 10.Fenlon, D. R. 1999. Listeria monocytogenes in the natural environment, p. 21-38. In E. T. Ryser and E. H. Marth (ed.), Listeria listeriosis and food safety, 2nd ed. Marcel Decker, Inc., New York, N.Y.
- 11.Fenlon, D. R., J. Wilson, and W. Donachie. 1996. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J. Appl. Bacteriol. 81:641-650. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, E., M. de Paz, J. L. Rodriguez, P. Gaya, M. Medina, and M. Nunez. 1996. Exogenous sources of Listeria contamination in raw ewe's milk. J. Food Prot. 59:950-954. [DOI] [PubMed] [Google Scholar]
- 13.Gravani, R. 1999. Incidence and control of Listeria in food-processing facilities, p. 657-709. In E. T. Ryser and E. H. Marth (ed.), Listeria listeriosis and food safety, 2nd ed. Marcel Decker, Inc., New York, N.Y.
- 14.Haydon, D. S. Cleaveland, L. H. Taylor, and M. K. Laurenson. 2002. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg. Infect. Dis. 8:1468-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Headrick, M. L., and L. Tollefson. 1998. Food borne disease summary by food commodity. Vet. Clin. N. Am. Food Anim. Pract. 14:91-99. [DOI] [PubMed] [Google Scholar]
- 16.Hinton, M. H. 2000. Infections and intoxications associated with animal feed and forage which may present a hazard to human health. Vet. J. 159:121-128. [DOI] [PubMed] [Google Scholar]
- 17.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of discrimination ability of typing systems: an application of Simpson's Index of Diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Husu, J. R. 1990. Epidemiological studies on the occurrence of Listeria monocytogenes in the feces of dairy cattle. J. Vet. Med. Ser. B 37:276-282. [DOI] [PubMed] [Google Scholar]
- 19.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 20.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 21.Low, J. C., and W. Donachie. 1997. A review of Listeria monocytogenes and listeriosis. Vet. J. 153:9-29. [DOI] [PubMed] [Google Scholar]
- 22.Maijala, R., O. Lyytikainen, T. Autio, T. Aalto, L. Haavisto, and T. Honknen-Buzalski. 2001. Exposure of Listeria monocytogenes within an epidemic caused by butter in Finland. Int. J. Food Microbiol. 70:97-109. [DOI] [PubMed] [Google Scholar]
- 23.McLaughlin, J. 1997. Animal and human listeriosis: a shared problem? Vet. J. 153:3-5. [DOI] [PubMed] [Google Scholar]
- 24.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCraig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng, J., and M. P. Doyle. 1997. Emerging issues in microbiological food safety. Annu. Rev. Nutr. 17:255-275. [DOI] [PubMed] [Google Scholar]
- 26.Miettinen, A., and J. Husu. 1991. Antibodies to listeriolysin O reflect the acquired resistance to Listeria monocytogenes in experimentally infected goats. FEMS Microbiol. Lett. 77:181-186. [DOI] [PubMed] [Google Scholar]
- 27.Norton, D. M., M. A. McCamey, K. L. Gall, J. M. Scarlett, K. J. Boor, and M. Wiedmann. 2001. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl. Environ. Microbiol. 67:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pell, A. N. 1997. Manure and microbes: public and animal health problem? J. Dairy Sci. 80:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen, M. A., and T. A. Casey. 2001. Environmental and food safety aspects of Escherichia coli O157:H7 infections in cattle. Crit. Rev. Microbiol. 27:57-73. [DOI] [PubMed] [Google Scholar]
- 30.Rebhun, W. C. 1995. Diseases of dairy cattle, p. 410-413. Williams & Wilkins, Media, Pa.
- 31.Restaino, L., E. W. Frampton, R. M. Irbe, G. Schabert, and H. Spitz. 1999. Isolation and detection of L. monocytogenes using fluorogenic and chromogenic substrates for phosphatidylinositol-specific phospholipase C. J. Food Prot. 62:244-251. [DOI] [PubMed] [Google Scholar]
- 32.Ryser, E. T. 1999. Foodborne listeriosis, p. 39-73. In E. T. Ryser and E. H. Marth (ed.), Listeria listeriosis and food safety, 2nd ed. Marcel Decker, Inc., New York, N.Y.
- 33.Ryser, E. T., S. M. Arimi, and C. W. Donnelly. 1997. Effects of pH on distribution of Listeria ribotypes in corn, hay and grass silage. Appl. Environ. Microbiol. 63:3695-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauders, B. D., E. D. Fortes, D. L. Morse, N. Dumas, J. A. Kiehlbauch, Y. H. Schukken, J. R. Hibbs, and M. Wiedmann. 2003. Molecular subtyping to detect human listeriosis clusters. Emerg. Infect. Dis. 6:672-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauders, B. D., K. G. Evans, K. Nightingale, and M. Wiedmann. 2002. Listeria monocytogenes from pristine, urban and agricultural sources. International Conference on Emerging Infectious Diseases, Atlanta, Ga.
- 36.Schlech, W. F. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 37.Skovgaard, N., and C. A. Morgen. 1988. Detection of Listeria spp. in faeces from animals, in feeds, and in raw foods or animal origin. Int. J. Food Microbiol. 6:229-242. [DOI] [PubMed] [Google Scholar]
- 38.Smith, M. C., and D. M. Sherman. 1994. Goat medicine, p. 141-144. Lea & Febiger, Malvern, Pa.
- 39.Stokes, M. E., C. S. Davis, and G. G. Koch. 2000. Categorical data analysis using the SAS system, 2nd ed. SAS Institute Inc., Cary, N.C.
- 40.Ueno, H., K. Yokota, T. Arai, Y. Muramatsu, H. Taniyama, T. Iida, and C. Morita. 1996. The prevalence of Listeria monocytogenes in the environment of dairy farms. Microbiol. Immunol. 40:121-124. [DOI] [PubMed] [Google Scholar]
- 41.Vazquez-Boland, J. A., L. Dominguez, M. Blanco, J. Rocourt, J. F. Fernandez-Garayzabal, C. B. Gutierrez, R. I. Tascon, and E. F. Rodriguez-Ferri. 1992. Epidemiologic investigation of a silage-associated epizootic of ovine listeriosis encephalitis, using a new Listeria-selective enumeration medium and phage typing. Am. J. Vet. Res. 53:368-371. [PubMed] [Google Scholar]
- 42.Vela, A. I., J. F. Fernandez-Garayzabal, J. A. Vazquez, M. V. Latre, M. M. Blanco, M. A. Moreno, L. de la Fuente, J. Marco, C. Franco, A. Cepeda, A. A. Rodriguez Moure, G. Suarez, and L. Dominguez. 2001. Molecular typing by pulsed-field gel electrophoresis of Spanish animal and human Listeria monocytogenes isolates. Appl. Environ. Microbiol. 67:5840-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veling, J., H. Wilpshaar, K. Frankena, C. Bartels, and H. W. Barkena. 2002. Risk factors for clinical Salmonella enterica subsp. enterica serovar Typhimurium infection on Dutch dairy farms. Prev. Vet. Med. 54:157-168. [DOI] [PubMed] [Google Scholar]
- 44.Wesley, I. V. 1999. Listeriosis in animals, p. 39-73. In E. T. Ryser and E. H. Marth (ed.), Listeria listeriosis and food safety, 2nd ed. Marcel Decker, Inc., New York, N.Y.
- 45.Wiedmann, M. 2003. ADSA Foundation Scholar Award—an integrated science-based approach to dairy food safety. Listeria monocytogenes as a model system. J. Dairy Sci. 86:1865-1875. [DOI] [PubMed] [Google Scholar]
- 46.Wiedmann, M. 2002. Subtyping of bacterial foodborne pathogens. Nutr. Rev. 60:201-208. [DOI] [PubMed] [Google Scholar]
- 47.Wiedmann, M., and K. E. Evans. 2002. Infectious diseases of dairy animals: Listeriosis, p. 777-782. In J. W. Fuquay and P. F. Fox (ed.), Encyclopedia of dairy science. Academic Press, London, United Kingdom.
- 48.Wiedmann, M., S. Mobini, J. R. Cole, Jr., C. K. Watson, G. Jeffers, and K. J. Boor. 1999. Molecular investigation of a listeriosis outbreak in goats caused by an unusual strain of Listeria monocytogenes. J. Am. Vet. Med. Assoc. 215:369-371. [PubMed] [Google Scholar]
- 49.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiedmann, M., J. Czajka, N. Bsat, M. Bodis, M. C. Smith, T. J. Divers, and C. A. Batt. 1994. Diagnosis and epidemiological association of Listeria monocytogenes strains in two outbreaks of listerial encephalitis in ruminants. J. Clin. Microbiol. 32:991-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida, T., Y. Kato, M. Sato, and K. Hirai. 1998. Sources and routes of contamination of raw milk with Listeria monocytogenes and its control. J. Vet. Med. Sci. 60:1165-1168. [DOI] [PubMed] [Google Scholar]