Abstract
A membrane-associated 3,5-dichlorophenol reductive dehalogenase was isolated from Desulfitobacterium frappieri PCP-1. The highest dehalogenase activity was observed with the biomass cultured at 22°C, compared to 30 and 37°C, where the cell suspensions were 2.2 and 9.6 times less active, respectively. The reductive dehalogenase was purified 12.7-fold to apparent homogeneity. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a single band with an apparent molecular mass of 57 kDa. Its dechlorinating activity was not inhibited by sulfate and nitrate but was completely inhibited by 2.5 mM sulfite and 10 mM KCN. A mixture of iodopropane and titanium citrate caused a light-reversible inhibition of the dechlorinating activities, suggesting the involvement of a corrinoid cofactor. Several polychlorophenols were dechlorinated at the meta and para positions. The apparent Km for 3,5-dicholorophenol was 49.3 ± 3.1 μM at a methyl viologen concentration of 2 mM. Six internal tryptic peptides were sequenced by mass spectrometry. One open reading frame (ORF) was found in the Desulfitobacterium hafniense genome containing these peptide sequences. This ORF corresponds to a gene coding for a CprA-type reductive dehalogenase. The corresponding ORF (named cprA5) in D. frappieri PCP-1 was cloned and sequenced. The cprA5 gene codes for a 548-amino-acid protein that contains a twin-arginine-type signal for secretion. The gene product has a cobalamin binding site motif and two iron-sulfur binding motifs and shows 66% identity (76 to 77% similarity) with some tetrachloroethene reductive dehalogenases. This is the first CprA-type reductive dehalogenase that can dechlorinate chlorophenols at the meta and para positions.
Several strictly anaerobic bacteria are able to reductively dehalogenate a large variety of chlorinated compounds and use them as terminal electron acceptors (8). Desulfomonile tiedjei DCB-1, Dehalobacter restrictus PER-K23, Sulfurospirillum (formerly Dehalospirillum) multivorans, and many members of the genus Desulfitobacterium have been the most studied for their dechlorinating activity. Three types of reductive dehalogenases have been isolated from dehalorespiring bacteria. The most frequently reported dehalogenases consist of a single polypeptide containing one corrinoid cofactor and two iron-sulfur clusters: tetrachloroethene (PCE) reductive dehalogenases of S. multivorans (18), Desulfitobacterium sp. strain PCE-S (16), and Desulfitobacterium frappieri TCE-1 (27), ortho-chlorophenol reductive dehalogenases of Desulfitobacterium hafniense (5), Desulfitobacterium dehalogenans (28), and Desulfitobacterium chlororespirans (10), and PCE- and trichloroethene (TCE)-reductive dehalogenases of Dehalococcoides ethenogenes (14). Two reductive dehalogenases with one corrinoid cofactor but without an iron-sulfur cluster have also been reported: the ortho-chlorophenol reductive dehalogenase from D. frappieri PCP-1 (3) and the PCE reductive dehalogenase from Clostridium bifermentans DPH-1 (22). These two proteins are different from all the other dehalogenases already described. The third type of dehalogenase is a heme protein consisting of two subunits and was isolated only from Desulfomonile tiedjei DCB-1 (20). Abiotic dehalogenation of several halogenated compounds was also observed from the heat-inactivated PCE dehalogenase of S. multivorans (17) and from bacterial transition metal coenzymes vitamin B12 (Co), coenzyme F430 (Ni), and hematin (Fe) (9, 17).
Genes coding for the first type of reductive dehalogenases have been reported, such as cprA from D. dehalogenans (24, 28), pceA from S. multivorans (19) and Desulfitobacterium sp. strain Y51 (26), and tceA from Dehalococcoides ethenogenes (15). These genes are all closely linked to genes cprB, pcrB, and tceB, respectively, which encode for hydrophobic proteins potentially acting as membrane anchors for the dehalogenases. Villemur et al. (29) isolated genes from D. frappieri PCP-1 that are highly related to cprA and cprB. Furthermore, they also observed several genes coding for putative CprA and PceA in the genomes of D. hafniense DCB-2 and Dehalococcoides ethenogenes. Genes coding for reductive dehalogenases containing corrinoid cofactor but without an iron-sulfur cluster have been isolated: crdA, coding for an enzyme mediating the ortho-chlorophenol reductive dehalogenation in D. frappieri PCP-1 (3), and pceC, coding the PCE reductive dehalogenase in C. bifermentans DPH-1 (22). Both genes and gene products show no similarity with each other and with the first type of reductive dehalogenases.
D. frappieri PCP-1 is a strictly anaerobic bacterium which can dechlorinate pentachlorophenol (PCP) to 3-chlorophenol (3-CP) and different chlorophenols at the ortho, meta, and para positions (2, 6). At least two inducible dehalogenases are involved in D. frappieri PCP-1, one for ortho dechlorination and a second for meta and para dechlorination. In this paper, we report the purification and the characterization of a reductive dehalogenase that catalyzes the meta and para dechlorination of several chlorophenols and the isolation of the corresponding gene.
MATERIALS AND METHODS
Culture conditions and preparation of membrane fraction.
D. frappieri PCP-1 (ATCC 700357) was cultivated anaerobically at 30°C in a serum bottle containing mineral salt medium supplemented with 55 mM pyruvate and 0.1% yeast extract (2, 6). 3,5-Dichlorophenol (3,5-DCP) (60 μM) was added to induce the dechlorinating activity.
The optimum temperature for the 3,5-DCP dehalogenase production was studied with PCP-1 cultures incubated for 48 h at 22, 30, and 37°C. Cells were harvested by centrifugation at 9,000 × g for 20 min and resuspended in fresh medium without yeast extract at an optical density at 600 nm of 0.5. The 3,5-DCP dechlorinating activity of each preparation was evaluated by determination of the 3-CP produced after 3 h of incubation at 37°C. Culture medium without yeast extract does not sustain the growth of strain PCP-1.
The production of 3,5-DCP dehalogenase was performed in a 14-liter bottle containing 9 liters of anaerobic medium. This medium was inoculated with 450 ml of an exponentially growing culture from a 1-liter bottle incubated at 30°C. The 14-liter bottle was incubated at 30°C for 25 h and then at room temperature (22°C) for the next 15 h. Periodically, the pH was adjusted above 7.0 with a sterile saturated solution of NaHCO3. After approximately 15 and 25 h of incubation, 50 μM of 3,5-DCP was added to the culture. Cells were harvested by centrifugation at 9,000 × g for 20 min at 4°C and fractionated by the method previously described (3). The membrane preparation was resuspended in 10 ml of 50 mM phosphate buffer, pH 7.5, containing 1 mM dithiothreitol (DTT) and 20% (vol/vol) glycerol and stored at −80°C. The latter preparation (1.0 to 2.0 ml) was thawed and diluted in 10 ml of 50 mM phosphate buffer, pH 8.0, containing 1 mM DTT, 20% (vol/vol) glycerol, and 0.1% Triton X-100. The preparation was agitated for 45 min at 4°C and centrifuged at 161,000 × g for 90 min at 4°C, and the supernatant (solubilized dehalogenase) was used for characterization and purification.
The work was performed in an anaerobic chamber (Bactron II; Sheldon Manufacturing, Cornelius, Oreg.) under a gas mixture containing 80% N2, 10% H2, and 10% CO2 or in serum bottles capped under this gas mixture. All the solutions were made anoxic in an anaerobic jar by repeated cycles of vacuum and flushing with oxygen-free gas mixture.
Purification of reductive dehalogenase.
A crude solubilized dehalogenase preparation was loaded onto a Protein Pak DEAE-5PW (8.0 by 75 mm) or Mono Q HR (10 mm by 10 cm) column equilibrated in 50 mM phosphate buffer, pH 8.0, containing 1 mM DTT, 0.1% Triton X-100, and 10% glycerol. The dehalogenase was weakly retained by the column, and the activity was eluted without salt gradient. Fractions with 3,5-DCP dehalogenase activity were pooled. The preparation was mixed with a saturated ammonium sulfate solution to reach 0.5 M. The suspension was filtered through a membrane of 0.2-μm pore size and loaded onto a column (1.2 by 8.5 cm) of macroprep methyl hydrophobic interaction chromatography (HIC) support (Bio-Rad Laboratories, Mississauga, Ontario, Canada) previously equilibrated with 50 mM phosphate buffer, pH 7.5, containing 1 mM DTT, 10% glycerol, and 0.5 M ammonium sulfate. The bound proteins were eluted with a 0.5 to 0 M ammonium sulfate gradient in the latter buffer containing 6 mM CHAPS{3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}. The fractions with dehalogenating activity were collected in a serum bottle with oxygen-free gas mixture.
Analytical methods.
Dehalogenation assays were performed under anaerobic conditions in 12-ml serum bottles containing 2.0 ml of the assay mixtures as described by Boyer et al. (3). The assay mixtures contained 100 mM potassium phosphate buffer, pH 7.0, 2 mM titanium citrate, 2 mM methyl viologen, 20% (vol/vol) glycerol, 1 mM chlorophenol, and 50 μl of enzyme preparation. After a 1-h incubation at 37°C, the reaction was stopped by addition of 10 ml of acetonitrile containing 0.33% (vol/vol) acetic acid. The mixture was centrifuged for 5 min at 5,000 × g, and the products of the reaction were determined by high-pressure liquid chromatography analysis (3).
The optimum pH was determined by carrying out standard enzyme assays at different pH values between 6.2 and 8.0 in 100 mM potassium phosphate buffer at 37°C. Initial characterization of the enzyme was carried out with a semipurified preparation obtained after chromatography on a DEAE-5PW column. The oxygen sensitivity assay was performed in 12-ml serum bottles exposed to air or in anoxic capped serum bottles (control). The dechlorinating activities of each bottle were determined from samples taken after 0, 0.5, 1, 2, 3, and 4 h of incubation at 4°C. The experiment was performed in duplicate. Determination of the optimum enzymatic temperature was carried out in a standard enzyme assay mixture (1-h incubation) between 5 and 65°C. The temperature stability of the enzyme in a standard assay mixture was also determined by first incubating for 2 h the enzyme preparations without 3,5-DCP at the different temperatures. The mixtures were then brought to 37°C, and the enzymatic reaction was started by the addition of the substrate. This experiment was performed in triplicate. The Km and Vmax values were determined with a semipurified preparation with a range of substrate concentration varying from 5 to 500 μM. The values were calculated with Sigma Plot 2002 v. 8.0, enzyme kinetic module 1.1.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide) were carried out by the method of Laemmli (11). Protein concentration was determined with a Bio-Rad assay protein kit (Bulletins 1069 and 1123; Bio-Rad Laboratories). Bovine serum albumin was used as the standard. Trypsin treatments of the protein band on SDS-PAGE were carried out with trypsin (sequencing grade modified trypsin; Promega, Madison, Wis.) as described by Shevchenko et al. (23). The tryptic peptides were purified on ZipTip C18 (Millipore Corporation, Bedford, Mass.) and analyzed with a Quattro II triple quadrupole mass spectrometer (Micromass Canada, Pointe-Claire, Quebec, Canada) equipped with a Nanospray and a Z-spray interface. Samples were introduced into the mass spectrometer by using type D Nanospray probe tips (Micromass Canada) in positive mode. The capillary voltage was 1.0 kV and the cone was at 35 V.
Inactivation of dehalogenase with iodopropane.
Light-reversible alkylation of corrinoids by iodopropane was based on the procedure of Brot and Weissbach (4). Semipurified dehalogenase preparations were incubated for 30 min at 37°C in the dark with 2 mM titanium(III) citrate with 0 (control) and 1 mM 1-iodopropane. The vials were placed on ice and exposed to a 300-W slide projector for 20 min. Dehalogenase activities were determined on the different preparations before and after light exposition.
Gene isolation and sequence.
Extraction of total DNA of strain PCP-1 was performed according to the method of Li et al. (13). The rdA1 and rdA2 sequences were taken from the incomplete genome sequence (January 2004) of D. hafniense DCB-2 at the Joint Genome Institute web site (http://www.jgi.doe.gov/). The sequences were on contig 1065, gene 1880 and 1881 (12 November 2003 gene annotations); it was formerly on contig 2977 as described by Villemur et al. (29). This sequence was used to design the primers. PCRs were carried out with 50-μl reaction mixtures containing 10 to 30 ng of total strain PCP-1 DNA, deoxynucleoside triphosphates (200 μM each), Taq DNA polymerase buffer (10 mM Tris-HCl, pH 9.0, 50 mM KCl, 1.5 mM MgCl2), 10 pmol of each oligonucleotide (HafRdA1-G, 5′ TGC GGC ACT TTT CTT GAT CGC 3′; and HafRdA1-D, 5′ GTC TTA AGC AAA TGA CGC AGC 3′), and 2.5 U of Taq DNA polymerase (Pharmacia). This mixture was heated at 80°C for 2 min before the addition of the DNA sample and then brought to 94°C for 5 min and 55°C for 5 min, followed by 30 cycles of 72°C for 2 min, 94°C for 40 s, and 55°C for 1 min and a final elongation step of 10 min at 72°C. The PCR product was cloned in a T-vector according to manufacturer specifications (pGEM T-easy vector; Promega), and both strands were sequenced. Two clones were also sequenced.
Nucleotide sequence accession number.
The sequences for the strands of the PCR product were submitted to GenBank under accession number AY349165.
RESULTS
Production of 3,5-DCP dehalogenase activity.
The incubation temperature of the PCP-1 culture was an important factor for the production of 3,5-DCP dehalogenase activity. The biomass obtained from a culture incubated at 22°C showed approximately 10 times more dehalogenase activity (4,030 ± 1,280 nmol of 3-CP h−1 mg of protein−1) than was observed at 37°C (420 ± 120 nmol of 3-CP h−1 mg of protein−1). The cells cultured at 30°C had an activity of 1,830 ± 590 nmol of 3-CP h−1 mg of protein−1. For an optimal production of the dehalogenase activity, the incubation temperature for the PCP-1 culture was 30°C for the first 25 h to favor the bacterial growth and 22°C for the last 15 h to achieve the optimal synthesis of the 3,5-DCP dehalogenase.
Initial characterization and purification of the dehalogenase.
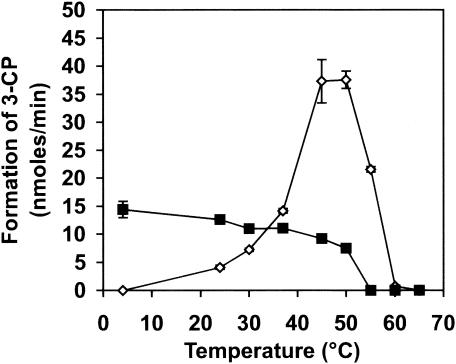
The optimum pH of the 3,5-DCP dehalogenase activity of the crude preparation was observed between 6.8 and 7.0. The dehalogenase was oxygen sensitive and it had a half-life of 110 min upon exposure to air. The optimum temperature of the dechlorinating activity was 50°C (Fig. 1). The enzymatic system was stable under the assay conditions for at least 2 h at temperatures below 50°C. Rapid loss of enzyme activity was observed at higher temperatures. The dechlorinating activity was completely inhibited by 2.5 mM sulfite. Sulfate and nitrate at 75 mM had no effect on the dechlorination. Adding 2.5 and 10 mM KCN to the assay enzyme mixtures showed 75.5 and 100% inhibition, and 5 mM NaN3 showed 13.2% inhibition of a semipurified preparation. Incubation of the dehalogenase preparation with 10 mM EDTA for 1 h had no effect on the dechlorinating activity. In the presence of 1 mM iodopropane and 2 mM titanium citrate, the dehalogenase lost 67% of its initial activity when incubated in the dark. Subsequent exposure to light restored 56.4% of the activity of the control group, suggesting the presence of a cobalamin cofactor.
FIG. 1.
Temperature dependence of a semipurified preparation of 3,5-DCP reductive dehalogenase. ◊, incubation temperature of standard enzyme assay conditions with 3,5-DCP substrate; ▪, thermal stability (complete assay mixtures containing the enzyme preparation but lacking the substrate were incubated at the different temperatures for 2 h). After addition of the 3,5-DCP substrate, the vials were incubated at 37°C for 1 h for the dehalogenation assays.
The substrate specificity of the semipurified dehalogenase toward different chlorophenols is shown in Table 1. The highest activity was observed at the meta position with 3,5-DCP and 2,3,5-trichlorophenol. Dechlorination at the para position was also detected with PCP, 2,3,4,5-tetrachlorophenol, and 3,4,5-trichlorophenol. Dechlorination at the ortho position was observed only with 2,4,6-trichlorophenol, 2,4,5-trichlorophenol, and 2,4-DCP. No dechlorination was detected with 3,4-DCP, 3,6-DCP, 2,5-DCP, 2,3-DCP, 3-CP, 4-CP, 2-CP, and 3-chloro-4-hydroxyphenylacetate. The apparent Km value for 3,5-DCP was 49.3 ± 3.1 μM at a methyl viologen concentration of 2 mM. With the semipurified preparation, the Vmax was 188.1 ± 5.2 nmol of 3-CP min−1 mg of protein−1 for 3,5-DCP.
TABLE 1.
Dechlorinating activity of semipurified preparations of dehalogenase IIa
| Substrate | Product(s) | Position of dechlorination | Mean sp act (nmol min−1 mg of protein−1) |
|---|---|---|---|
| Expt 1 | |||
| PCP | 2,3,5,6-TeCP | para | 73.6 ± 3.1 (0.3)b |
| 3,4,5-TCP | 3,5-DCP | para | 24.4 ± 0.3 (0.09) |
| 3,5-DCP | 3-CP | meta | 272.2 ± 6.0 (1.0) |
| Expt 2 | |||
| 3,4,5-TCP | 3,5-DCP | para | 56.8 ± 0.3 (0.4) |
| 2,4,6-TCP | 2,4-DCP | ortho | 92.6 ± 7.9 (0.6) |
| 2,4-DCP | 4-CP | ortho | 113.2 ± 5.9 (0.7) |
| 3,5-DCP | 3-CP | meta | 159.0 ± 1.7 (1.0) |
| Expt 3 | |||
| 2,3,4,5-TeCP | 2,3,5-TCP | para | 215 ± 1.25 (0.7) |
| +2,5-DCP | meta | 25.8 ± 0.3 (0.08) | |
| 2,4,5-TCP | 3,4-DCP | ortho | 184 ± 6.0 (0.6) |
| 2,3,6-TCP | 2,6-DCP | meta | 143.6 ± 2.8 (0.4) |
| 2,3,5-TCP | 2,5-DCP | meta | 328.6 ± 2.6 (1.0) |
| 2,3-DCP | 2-CP | meta | 160.6 ± 9.3 (0.5) |
| 3,5-DCP | 3-CP | meta | 322.2 ± 7.9 (1.0) |
Preparations made after chromatography on a DEAE-5PW column. TCP, trichlorophenol; TeCP, tetrachlorophenol.
Values in parentheses represent the ratio of activity compared to that with 3,5-DCP as substrate. Chlorophenols not dechlorinated: 3,4-DCP; 2,6-DCP; 2,5-DCP; 2,3-DCP; 3-CP; 4-CP; 2-CP.
The 3,5-DCP dehalogenase was purified by ion-exchange chromatography on DEAE-5PW or Mono Q columns, followed by HIC on a methyl-HIC column. Specific activities of the purified enzyme increased 12.7-fold from the first step for a recovery of 6.8% (Table 2). SDS-PAGE revealed only one band with a molecular mass estimated at 57 kDa (Fig. 2). This band was eluted from the gel, treated with trypsin, and analyzed by mass spectroscopy. The amino acid sequences of six tryptic peptides were determined.
TABLE 2.
Purification scheme for the 3,5-DCP reductive dehalogenase
| Purification step | Total activity (nmol min−1)a | Yield (%) | Total protein (mg)b | Sp act (nmol min−1 mg−1) | Purifi- cation factor |
|---|---|---|---|---|---|
| Solubilized fraction | 152 | 100 | 4.82 | 31.5 | 1 |
| DEAE-5PW | 125.6 | 82.6 | 0.43 | 290.8 | 9.2 |
| Methyl-HIC | 10.4 | 6.8 | 0.026 | 400 | 12.7 |
Amount (nmoles) of 3-CP produced per min at 37°C with 3,5-DCP as substrate.
Total protein was determined with Bio-Rad assay protein using serum albumin as a standard.
FIG. 2.
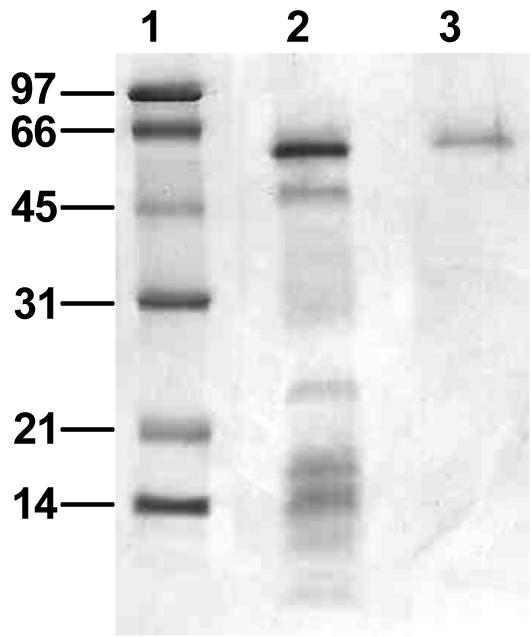
SDS-PAGE with the purified 3,5-DCP reductive dehalogenase. The gel was stained for proteins with Coomassie brilliant blue R-250. Lane 1, molecular size (kDa) markers; lane 2, fraction after ion-exchange chromatography column; lane 3, purified 3,5-DCP reductive dehalogenase after methyl-HIC column.
Gene cloning of the reductive dehalogenase.
The genome sequence available for D. hafniense DCB-2 was screened with the tryptic peptide sequences. Niggemyer et al. (21) showed that strains PCP-1 and DCB-2 share 88.7% of their genome. This allowed us to find one open reading frame (ORF) in the strain DCB-2 genome containing all six tryptic peptide sequences (Fig. 3). This ORF corresponds to the rdA1 gene that was identified previously by Villemur et al. (29) and encodes for a putative reductive dehalogenase. Downstream of rdA1 is another ORF, called rdB1, which has homology with the cprB/pceB gene family coding for anchor proteins of reductive dehalogenases (see Villemur et al. [reference 29] for sequence analyses of both genes).
FIG. 3.
Deduced amino acid sequence of cprA5 from strain PCP-1. Underlined sequences are peptides that were detected by mass spectrometry, and bold and double-underlined sequences are amino acid sequences determined by mass spectrometry. *, amino acid difference between strains DCB-2 and PCP-1 at position 273. Boxed sequences are the cobalamin binding motif (at amino acid 159) and the two iron-sulfur binding motifs. A slash indicates the predicted location of the cleavage of the peptide signal.
From this, we designed and synthesized two oligonucleotides that were used to amplify the two corresponding ORFs from strain PCP-1 genomic DNA. A 2.3-kb fragment was amplified, cloned, and sequenced, and it was compared to the corresponding sequence of D. hafniense DCB-2. This fragment indeed contains the two ORFs, in the same gene arrangement found in the D. hafniense DCB-2 genome. These two ORFs were named cprA5 and cprB5 after the first four cprAB genes identified by Villemur et al. (29). There is one nucleotide difference (C for T) between the DCB-2 and PCP-1 sequences, located at nucleotide 818 in cprA5 which generated an amino acid substitution of a proline in the DCB-2 sequence for a leucine in the PCP-1 sequence at position 273 of CprA5 (Fig. 3). This leucine was revealed by mass spectrometry analysis in one of the tryptic peptides.
cprA5 contains 548 codons. Sequence analysis with the SIGNALP program (http://ca.expasy.org) predicted that the protein contains a peptide signal and the cleavage would occur after amino acid 27. The sequence of this putative peptide signal is indeed highly hydrophobic, as is seen in most signal peptides. This sequence also contains the sequence motif, RRXFXK, found in peptide signals characteristic of the twin-arginine-type secretion pathway. In most cases, this system involves proteins that bind to various cofactors in the cytoplasm and are thus folded before being exported. Such proteins are predominantly involved in respiratory electron transport chains (1, 25), as would be expected for proteins involved in halorespiration.
The predicted mature protein has 521 amino acids with a molecular mass of 58,314 Da, which is around the molecular mass observed by SDS-PAGE. The pI of the predicted mature protein is 5.62. The cprA5 gene product is more related to PCE reductive dehalogenases (PceA) than chloroaromatic reductive dehalogenases (CprA). It has 66% identity (76 to 77% similarity) with pceA gene products from D. frappieri TCE-1 (GenBank accession no. CAD28792), Desulfitobacterium sp. strain Y51 (BAC00915) (26), Desulfitobacterium sp. strain PCE-S (AAO60101), and Dehalobacter restrictus (CAD28790) and 23 to 28% identity (39 to 42% similarity) with two PceAs from S. multivorans (AAC60788, AAG46194) (19) and the tceA gene product of Dehalococcoides ethenogenes (AAF73916) (15). CprA5 has the sequence EYHYNG (Fig. 3) that is related to the motif DXHXXG found in cobalamin binding proteins (7). The aspartic acid replacement by a glutamic acid residue is also listed in the cobalamin-binding conserved domain databases (such as pfam02310.8 or COG5012) for some proteins. Finally, the sequence contains the two iron-sulfur binding motifs characteristic of all CprA/PceA reductive dehalogenases (29).
cprB5 is identical to the rdB1 of D. hafniense DCB-2. It contains 101 codons and codes for a protein of 11,585 Da and a predicted pI of 9.48.
DISCUSSION
In this study we report the production and purification of a new reductive dehalogenase of D. frappieri PCP-1. The incubation at room temperature of the PCP-1 cell culture was important for the production of 3,5-DCP dehalogenase activity. Although its optimum temperature for growth is 38°C (2), incubation at 30 and 37°C resulted in much less dechlorinating activity. This result cannot be imputed to the optimum temperature of the dechlorinating activity, which is 50°C. As the strain PCP-1 originates from soil (12), its natural growth temperature is probably around 20 to 22°C and is more favorable for the synthesis of the CprA5 reductive dehalogenase.
The purified dehalogenase has an apparent molecular mass of 57 kDa by SDS-PAGE. This value is in the same range of molecular mass (47 to 65 kDa) determined for PceA/CprA reductive dehalogenases (5, 10, 14, 16, 18, 28). This value is different from the 3-chlorobenzoate reductive dehalogenase of Desulfomonile tiedjei, which consists of two subunits with an apparent molecular mass of 64 and 37 kDa (20), and from the CrdA and PceC dehalogenases, with 37 and 35 kDa, respectively (3, 22). The presence of a corrinoid cofactor in this enzyme is suggested by the KCN and sulfite inhibition of the dechlorinating activity, the light-reversible inhibition of activity with iodopropane and titanium citrate, and the presence of a cobalamin binding site motif in the deduced amino acid sequence of cprA5 (4, 8, 10). Two iron-sulfur binding motifs (FCXXCXXCXXXCP and CXXCXXXC) are also observed, suggesting the presence of two iron-sulfur clusters in the dehalogenase. These results indicate that this dehalogenase belongs to the group of CprA/PceA reductive dehalogenases. This was confirmed by the 66% identity (76 to 77% similarity) found with some PceA reductive dehalogenases from D. frappieri TCE-1 (GenBank accession no. CAD28792), Desulfitobacterium sp. strain Y51 (BAC00915) (26), Desulfitobacterium sp. PCE-S (AAO60101), and Dehalobacter restrictus (CAD28790).
The cprB5 gene is located 15 nucleotides downstream of cprA5 and codes for a small protein of 11,585 Da. Villemur et al. (29) showed that this gene product has similarities in sequence and hydrophobic profile with all the CprB/PceB family. Because of their hydrophobic features, these proteins would be located in the membrane to anchor the reductive dehalogenases to the membrane (19, 28).
The CprA5 dehalogenase can dechlorinate various chlorophenols at the meta and para positions. The highest dechlorinating activity was observed with 3,5-DCP and not with PCP and highly chlorinated phenols. In contrast, the highest rate of ortho-dechlorination of the CrdA dehalogenase from D. frappieri PCP-1 was with PCP and highly chlorinated phenols (3). The high dechlorinating activity for PCP and highly chlorinated phenols is consistent with the fact that strain PCP-1 was isolated from a PCP-enriched consortium which rapidly ortho-dechlorinates PCP to 3,4,5-trichlorophenol (2). However, the CprA dehalogenases from D. chlororespirans (10) and D. dehalogenans (28) dechlorinate PCP at the ortho position much less rapidly than dichlorophenols and other lesser-chlorinated aromatic compounds. The CprA5 dehalogenase is the first CprA-type dehalogenase that can dechlorinate chlorophenols at the meta and para positions. The differences of specificity observed between these reductive dehalogenases could be dependent on the apoprotein component or the type of corrinoid cofactor found in these dehalogenases, or both. Neumann et al. (17) have already shown that the corrinoid cofactor from heat-inactivated PCE reductive dehalogenase of S. multivorans can abiotically dechlorinate different alkyl halides at a higher rate than the native enzyme. This cofactor is not cyanocobalamin but would be a new type of corrinoid. Future kinetic and structural studies will provide additional insights into the enzyme mechanism and help further attempts to isolate or design enzymes able to dehalogenate other environmentally significant chloroaromatics.
Acknowledgments
We thank Rita Alary for excellent technical assistance.
This study was funded by the Natural Sciences and Engineering Research Council of Canada and by Fonds de Recherche sur la Nature et les Technologies (FQRNT).
REFERENCES
- 1.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard, B., R. Beaudet, R. Villemur, G. McSween, F. Lépine, and J.-G. Bisaillon. 1996. Isolation and characterization of Desulfitobacterium frappieri, sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int. J. Syst. Bacteriol. 46:1010-1015. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, A., R. Pagé-Bélanger, M. Saucier, R. Villemur, F. Lépine, P. Juteau, and R. Beaudet. 2003. Purification, cloning and sequencing of an enzyme mediating the reductive dechlorination of 2,4,6-trichlorophenol from Desulfitobacterium frappieri PCP-1. Biochem. J. 373:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brot, B., and H. Weissbach. 1965. Enzymatic synthesis of methionine: chemical alkylation of the enzyme-bound cobamide. J. Biol. Chem. 240:3064-3070. [PubMed] [Google Scholar]
- 5.Christiansen, N., B. K. Ahring, G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the 3-chloro-4-hydroxy-phenylacetate reductive dehalogenase of Desulfitobacterium hafniense. FEBS Lett. 436:159-162. [DOI] [PubMed] [Google Scholar]
- 6.Dennie, D., I. Gladu, F. Lépine, R. Villemur, J.-G. Bisaillon, and R. Beaudet. 1998. Spectrum of reductive dehalogenation activity of Desulfitobacterium frappieri PCP-1. Appl. Environ. Microbiol. 64:4603-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drennan, C. L., S. Huang, J. T. Drummond, R. G. Matthews, and M. L. Lidwig. 1994. How a protein binds B12: a 3.0 A X-ray structure of B12-binding domains of methionine synthase. Science 266:1669-1674. [DOI] [PubMed] [Google Scholar]
- 8.El Fantroussi, S., H. Naveau, and S. N. Agathos. 1998. Anaerobic dechlorinating bacteria. Biotechnol. Prog. 14:167-188. [DOI] [PubMed] [Google Scholar]
- 9.Gantzer, C. J., and L. P. Wackett. 1991. Reductive dechlorination by bacterial transition-metal coenzymes. Environ. Sci. Technol. 25:715-722. [Google Scholar]
- 10.Krasotkina, J., T. Walters, K. A. Maruya, and S. W. Ragsdale. 2001. Characterization of the B12- and iron-sulfur-containing reductive dehalogenase from Desulfitobacterium chlororespirans. J. Biol. Chem. 276:40991-40997. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Lanthier, M., R. Villemur, F. Lépine, J.-G. Bisaillon, and R. Beaudet. 2001. Geographic distribution of Desulfitobacterium frappieri PCP-1 and Desulfitobacterium spp. in soils from the province of Quebec, Canada. FEMS Microbiol. Ecol. 36:185-191. [DOI] [PubMed] [Google Scholar]
- 13.Li, T., J-G. Bisaillon, R. Villemur, L. Létourneau, K. Bernard, F. Lépine, and R. Beaudet. 1996. Isolation and characterization of a new bacterium carboxylating phenol to benzoic acid under anaerobic conditions. J. Bacteriol. 178:2551-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnuson, J. K., R. V. Stern, J. M. Gosset, S. H. Zinder, and D. R. Burris. 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, E., G. Wohlfarth, and G. Diekert. 1998. Purification and characterization of the tetrachloroethene reductive dehalogenase of strain PCE-S. Arch. Microbiol. 169:497-502. [DOI] [PubMed] [Google Scholar]
- 17.Neumann, A., A. Siebert, T. Trescher, S. Reinhardt, G. Wohlfarth, and G. Diekert. 2002. Tetrachloroethene reductive dehalogenase of Dehalospirillum multivorans: substrate specificity of the native enzyme and its corrinoid cofactor. Arch. Microbiol. 177:420-426. [DOI] [PubMed] [Google Scholar]
- 18.Neumann, A., G. Wohlfarth, and G. Diekert. 1996. Purification and characterization of tertrachloroethene reductive dehalogenase from Dehalospirillum multivorans. J. Biol. Chem. 271:16515-16519. [DOI] [PubMed] [Google Scholar]
- 19.Neumann, A., G. Wohlfarth, and G. Diekert. 1998. Tetrachloroethene dehalogenase from Dehalospirillum multivorans: cloning, sequencing of the encoding genes, and expression of the pceA gene in Escherichia coli. J. Bacteriol. 180:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni, S., J. K. Fredrickson, and L. Xun. 1995. Purification and characterization of a novel 3-chlorobenzoate-reductive dehalogenase from the cytoplasmic membrane of Desulfomonile tiedjei DCB-1. J. Bacteriol. 177:5135-5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niggemyer, A., S. Spring, E. Stackebrandt, and R. F. Rosenzweig. 2001. Isolation and characterization of a novel As(V)-reducing bacterium: implications for arsenic mobilization and the genus Desulfitobacterium. Appl. Environ. Microbiol. 67:5568-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okeke, B. C., Y. C. Chang, M. Hatsu, T. Suzuki, and K. Takamizawa. 2001. Purification, cloning, and sequencing of an enzyme mediating the reductive dechlorination of tetrachloroethylene (PCE) from Clostridium bifermentans DPH-1. Can. J. Microbiol. 47:448-456. [PubMed] [Google Scholar]
- 23.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 24.Smidt, H., M. van Leest, J. van der Oost, and W. M. de Vos. 2000. Transcriptional regulation of the cpr gene cluster in ortho-chlorophenol-respiring Desulfitobacterium dehalogenans. J. Bacteriol. 182:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smidt, H., A. D. Akkermans, J. van der Oost, and W. M. de Vos. 2000. Halorespiring bacteria-molecular characterization and detection. Enzyme Microb. Technol. 27:812-820. [DOI] [PubMed] [Google Scholar]
- 26.Suyama, A., M. Yamashita, S. Yoshino, and K. Furukawa. 2002. Molecular characterization of the PceA reductive dehalogenase of Desulfitobacterium sp. strain Y51. J Bacteriol. 184:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Pas, B., J. Gerritse, W. M. de Vos, G. Schraa, and A. J. M. Stams. 2001. Two distinct enzyme systems are responsible for tetrachloroethene and reductive dehalogenation in Desulfitobacterium strain PCE1. Arch. Microbiol. 176:165-169. [DOI] [PubMed] [Google Scholar]
- 28.Van de Pas, B., H. Smidt, W. R. Hagen, J. van der Oost, G. Schraa, A. J. M. Stams, and W. M. de Vos. 1999. Purification and molecular characterization of ortho-chlorophenol reductive dehalogenase, a key enzyme of halorespiration in Desulfitobacterium dehalogenans. J. Biol. Chem. 274:20287-20292. [DOI] [PubMed] [Google Scholar]
- 29.Villemur, R., M. Saucier, A. Gauthier, and R. Beaudet. 2002. Occurrence of several genes encoding putative reductive dehalogenases in Desulfitobacterium hafniense/frappieri and Dehalococcoides ethenogenes. Can. J. Microbiol. 48:697-706. [DOI] [PubMed] [Google Scholar]