Abstract
Previous studies have demonstrated that metal-reducing microorganisms can effectively promote the precipitation and removal of uranium from contaminated groundwater. Microbial communities were stimulated in the acidic subsurface by pH neutralization and addition of an electron donor to wells. In single-well push-pull tests at a number of treated sites, nitrate, Fe(III), and uranium were extensively reduced and electron donors (glucose, ethanol) were consumed. Examination of sediment chemistry in cores sampled immediately adjacent to treated wells 3.5 months after treatment revealed that sediment pH increased substantially (by 1 to 2 pH units) while nitrate was largely depleted. A large diversity of 16S rRNA gene sequences were retrieved from subsurface sediments, including species from the α, β, δ, and γ subdivisions of the class Proteobacteria, as well as low- and high-G+C gram-positive species. Following in situ biostimulation of microbial communities within contaminated sediments, sequences related to previously cultured metal-reducing δ-Proteobacteria increased from 5% to nearly 40% of the clone libraries. Quantitative PCR revealed that Geobacter-type 16S rRNA gene sequences increased in biostimulated sediments by 1 to 2 orders of magnitude at two of the four sites tested. Evidence from the quantitative PCR analysis corroborated information obtained from 16S rRNA gene clone libraries, indicating that members of the δ-Proteobacteria subdivision, including Anaeromyxobacter dehalogenans-related and Geobacter-related sequences, are important metal-reducing organisms in acidic subsurface sediments. This study provides the first cultivation-independent analysis of the change in metal-reducing microbial communities in subsurface sediments during an in situ bioremediation experiment.
Uranium [U(VI)] is the most common radionuclide contaminant found within the U.S. nuclear weapon complex managed by the U.S. Department of Energy. Nitrate is often a cocontaminant with U(VI) in subsurface sedimentary environments because of the use of nitric acid in the processing of U(VI) waste (39, 45). U(VI) can be microbiologically immobilized from groundwater by its reduction from UO22+ to insoluble U(IV) oxide (28, 31, 33). One of the more promising strategies for the in situ remediation of U(VI) waste involves the “biostimulation” of U(VI) immobilization (39). Biostimulation is defined as addition of nutrients (carbon and other nutrient sources) that serve to increase the number or activity of indigenous microorganisms available for bioremediation activity. Biostimulation can lead to creation of a permeable treatment zone in contaminated aquifers that removes radionuclides from the aqueous phase before they enter sensitive water supplies.
Nitrate serves as a competing and energetically more favorable electron acceptor for metal-reducing bacteria in nitric acid-contaminated sediments (9). Previous studies of microbial U(VI) reduction in sediments cocontaminated with nitrate have indicated that no net U(VI) reduction occurs until nitrate is reduced. Once nitrate is depleted, U(VI) and Fe(III) are reduced concurrently (12, 13, 49). Thus, denitrification intimately affects the fate of U(VI) in anoxic subsurface environments. Subsurface denitrification is believed to be mediated by a group of facultative anaerobes that are phylogenetically and metabolically diverse (35). Many Fe(III)-reducing bacteria (FeRB) and sulfate-reducing bacteria (SRB), such as members of the Geobacteraceae and Shewanellae families, possess the ability to reduce nitrate (9, 31, 43), but few have been observed to carry out complete denitrification.
Optimization of U(VI) bioremediation strategies requires an extensive understanding of metal-reducing microorganisms and the environmental parameters controlling their activity. Most previous work on U(VI)-reducing bacteria has been conducted with pure cultures or enrichments in the laboratory, where it is difficult to reconstruct field conditions. Few studies of known metal reducers capable of U(VI) utilization have been carried out by cultivation-independent techniques with subsurface sediments, and structure-function relationships have not been examined extensively. Thus far, for the FeRB, members of the Geobacteraceae family within the δ-Proteobacteria subdivision were most often detected in abundance in subsurface sediment slurries at neutral pH upon stimulation of concurrent U(VI) and Fe(III) reduction by addition of acetate as an electron donor (18).
Several FeRB and SRB have the ability to reduce U(VI), and some conserve energy for growth from this reaction (6, 32, 57). Not only do these organisms have the ability to enzymatically reduce U(VI), but the products of microbial sulfate and Fe(III) reduction, hydrogen sulfide and Fe(II), can chemically reduce U(VI) (26, 34, 37). In the terrestrial subsurface, oxidized iron is often the most abundant electron acceptor available under anoxic conditions, thus providing a metabolic advantage for FeRB (28). Previous culture studies conducted in our laboratory suggested that upon pH neutralization, members of the δ-Proteobacteria subdivision (Geobacter spp., Anaeromyxobacter spp.) will be important metal reducers in initially acidic subsurface sediments cocontaminated with U(VI) and nitrate (42). Iron(III)-reducing consortia, enriched from the U.S. Department of Energy Natural and Accelerated Bioremediation Research Field Research Center (FRC), where our study was conducted, utilized Fe(III) minerals as the sole electron acceptor (24) and rapidly reduced U(VI) in culture. A most-probable-number (MPN) serial dilution assay for SRB and FeRB revealed that FeRB are far more abundant than SRB in subsurface FRC sediments (L. Petrie, unpublished results). Therefore, the FeRB were the focus of this study, as they represent indigenous microorganisms likely to catalyze the immobilization of U(VI) contamination in situ. Further evidence from preliminary field experiments at the FRC site suggested that subsurface microbial activity was carbon substrate limited (20). Thus, we hypothesized that (i) biostimulation, through the addition of a carbon substrate and pH neutralization, would catalyze the reduction of nitrate, Fe(III), and U(VI) in contaminated FRC sediments and (ii) metal-reducing members of the δ-Proteobacteria subdivision would constitute a substantial proportion of the sedimentary microbial communities after in situ biostimulation. We further proposed that the overall change in microbial community composition during biostimulation must also be understood to place the metal-reducing organisms into the context of competing heterotrophs.
With a combination of push-pull activity tests, solid-phase geochemistry, and cultivation-independent analysis of microbial communities, we examined the in situ change in microbial community composition in subsurface sediments cocontaminated with U(VI) and nitrate during biostimulation. Through quantification of the in situ activity and community composition of metal-reducing microorganisms likely to catalyze U(VI) sorption and precipitation, we provide important inputs for reaction-based biogeochemical models that greatly aid in the development of in situ U(VI) bioremediation strategies.
MATERIALS AND METHODS
Site and sample description.
This study focused on contaminated subsurface sediments collected from the U.S. Department of Energy FRC, located at the Y-12 complex within the Oak Ridge National Laboratory in Oak Ridge, Tenn. It is centered on groundwater plumes that originate from former waste disposal ponds at the Y-12 plant. Contaminated acidic subsurface sediments were sampled from area 1 in the saturated zone of shallow residuum overlying Nolichucky shale, where elevated concentrations of U(VI) and nitrate have been observed. Other contaminants include additional radionuclides (plutonium, technetium), toxic metals (nickel, aluminum, barium, chromium, mercury), chelating agents (EDTA), chlorinated hydrocarbons (trichloroethylene [TCE] and perchloroethylene), polychlorinated biphenyls, and fuel hydrocarbons (toluene, benzene) (http://www.lbl.gov/NABIR/). The “background area” is a pristine site ∼163 ha in size, located in West Bear Creek Valley, approximately 2 km away from the S-3 ponds (http://www.lbl.gov/NABIR/). Subsurface sediments of the background area contain a parent rock mineralogy and sediment characteristics similar to those of contaminated sites of the FRC (http://www.lbl.gov/NABIR/).
Sediment cores were sampled prior to biostimulation, on 30 August 2001, and after biostimulation (following the final push-pull test), on 16 December 2002, with an Acker Drill Co. (TBD-II) Holegator track drill equipped with polyurethane sleeves lining the corer. The ranges of core depths sampled before and after biostimulation experiments were 3.0 to 5.7 and 2.1 to 3.6 m for contaminated area 1 and the background areas, respectively. Cores (0.42 m in diameter, 1.83 m in length) were immediately transferred to a Coy anaerobic chamber adjacent to the field sites, where they were subsampled under aseptic and strictly anoxic conditions with alcohol-rinsed equipment. Intact subsamples were sealed under argon, frozen in liquid nitrogen, and shipped to Florida State University on dry ice via Fed-Ex priority overnight. Before sediment biostimulation, cores FB32, FB33, and FB34 were sampled adjacent to injection wells from the push-pull experiment (42). After biostimulation, cores FB45, FB46, FB47, and FB49 were sampled adjacent to wells corresponding to the same sites where cores FB32 (corresponding to both FB45 and FB46), FB33, and FB34 were sampled, respectively.
Chemical analysis.
The sediment pH was determined by diluting 2 ml of sediment with 2 ml of deionized water. The 1:1 dilution was shaken for 1 h and centrifuged, and then the pH of the supernatant was measured with a calibrated digital pH meter (36). Nitrate was extracted from sediments for 1 h with a 1:1 dilution of sediment sample-deionized water, followed by centrifugation, and the supernatant was analyzed for dissolved nitrate after reduction with V(III) to NO with a chemiluminescence detector (4). Poorly crystalline Fe oxide minerals were quantified by a 1-h 0.5 M HCl extraction (23).
In situ biostimulation.
Push-pull tests for in situ bioremediation activity were conducted as described in a companion paper by Istok et al. (20). Briefly, wells were successively tested five times with approximately 1 to 2 months between treatments. Test solutions consisted of site groundwater that was neutralized with 100 mM sodium bicarbonate, made anoxic by sparging with 80% N2-20% CO2, and amended with either ethanol or glucose to a final concentration of 400 mM. Approximately 200 liters of test solution was injected into each well for each test with a siphon over a period of 1 to 2 days. Following injection, groundwater samples were periodically collected from the same well for up to 1,000 h. Samples were filtered in the field with 0.2-μm-pore-size syringe filters and stored at 4°C until analyzed. Groundwater was analyzed for nitrate by ion chromatography, for U(VI) with a kinetic phosphorescence analyzer (Chemcheck, Richland, Wash.) (5), for Fe(II) by the ferrozine assay (52), for ethanol by gas chromatography with flame ionization detection, and for glucose by the phenol-sulfuric acid method (8). Rates of donor utilization and nitrate and U(VI) reduction were calculated by plotting dilution-adjusted concentrations versus time (see reference 20 for further information).
DNA extraction and 16S rRNA gene analysis.
Microbial community DNA was extracted directly from the sediment by a method adapted from the simultaneous RNA-DNA extraction protocol described previously (19). The DNA was purified twice with a Wizard column (Promega, Madison, Wis.) and resuspended in 50 μl of sterile water. Sterile-water aliquots also extracted by this method served as negative controls. All of the 16S rRNA gene sequences obtained from negative controls were considered laboratory contaminants. Community DNA from uncontaminated sediment was extracted with the Ultra Clean Soil DNA kit (Mo Bio Laboratories, Solana Beach, Calif.) in accordance with the manufacturer's instructions. Immediately after extraction, aliquots (1 μl) of DNA were added to a mixture of PCR reagents for cloning and sequencing. Bacterial 16S rRNA genes were amplified from community DNA with primers 8F (5′-AGA GTT TGA TCM TGG CTC AG-3′) and 1392R (5′-ACG GGC GGT GTG TRC-3′) as previously described (10). The PCR products were cloned with a TOPO TA Cloning kit (Invitrogen, Carlsbad, Calif.) and screened by digestion with restriction enzyme HaeIII (New England Biolabs, Beverly, Mass.) as previously described (11). Clone libraries were generated from three contaminated sediment cores, four biostimulated sediment cores, and one pristine sediment core. Cloned inserts with unique restriction patterns were amplified with M13 primers from whole cells and purified for sequencing with QIAquick PCR purification columns (QIAGEN, Valencia, Calif.). Sequencing was performed with an Applied Biosystems 3100 genetic analyzer with primers G and H (27) at the Florida State University sequencing facility. Sequences were aligned against close relatives from the Ribosomal Database Project with the ARB software package (54) and rRNA secondary-structure diagrams. Dendrograms were constructed with ARB and PAUP* 4.0s (Sinauer Associates, Sunderland, Mass.) with only unambiguously aligned nucleotides. The possibility of chimera formation was analyzed by submitting the sequences to the CHIMERA_CHECK program in the Ribosomal Database Project database (7).
Quantitative PCR analysis.
The relative abundances of 16S rRNA gene sequences closely related to Geobacter sp., Anaeromyxobacter sp., Paenibacillus sp., and Brevibacillus sp. were determined from DNA directly extracted from subsurface sediment by an MPN-PCR technique as previously described (14). Serial 10-fold dilutions of extracted DNA were made in sterile water, and 1-μl aliquots were used as the template in the PCR. Primer sets for Anaeromyxobacter and Paenibacillus-Brevibacillus were designed with the probe design function in the ARB software package (54) (Table 1). Optimum temperature and cycling parameters were determined to be an initial denaturation step of 94°C for 10 min, followed by 35 cycles of 95°C (30 s), 56.5°C (30 s), and 72°C (45 s), with a final extension step of 72°C for 10 min. To test primer specificity, each primer set was compared to the sequences available in the Ribosomal Database Project and GenBank databases with the PROBE_CHECK (Ribosomal Database Project) and BLAST algorithms (1). Clone libraries produced from each primer set were also analyzed to ensure primer specificity. Each primer set produced cloned sequences closely related to their intended target organisms. PCR products were analyzed by gel electrophoresis in 1% agarose gels, stained in an ethidium bromide bath, and visualized by UV transillumination. The highest dilution that yielded a product was noted, and a standard three-tube MPN chart was consulted in order to determine the number of 16S rRNA gene copies per gram of sediment extracted (2). A test with known amounts of template DNA was conducted to verify the accuracy of the 16S rRNA gene abundances determined through MPN-PCR. The accuracy test revealed that the numbers of 16S rRNA gene copies per gram of sediment may be slightly overestimated but are well within the standard error.
TABLE 1.
Primer pairs and amplicon lengths for use in MPN-PCR
| Target organism(s) | Primer name | Primer sequence (5′-3′) | Amplicon length (bp) | Reference |
|---|---|---|---|---|
| Anaeromyxobacter sp. | 60F | CGA GAA AGC CCG CAA GGG T | 401 | This study |
| 461R | ATT CGT CCC TCG CGA CAG T | |||
| Paenibacillus Brevibacillus spp. | 160F | TGA GTA ACA CGT AGG CAA CCT | 174 | This study |
| 334R | TAA TGC GCC GCA GGC CCA T | |||
| Geobacter sp. | 494F | AGG AAG CAC CGG CTA ACT CC | 331 | 18 |
| 825R | TAC CCG CRA CAC CTA GTC T |
Nucleotide sequence accession numbers.
The nucleotide sequences reported here were submitted to the GenBank database under accession numbers AY527742 to AY527816.
RESULTS
In situ push-pull tests for microbial activity.
Microbial activity was quantified by analysis of electron acceptor and donor utilization in groundwater samples collected over regular intervals (Table 2; Fig. 1). Prior to testing, groundwater was aerobic and products of microbial respiration [nitrite, Fe(II)] were not detectable, indicating that microbial activity in the contaminated aquifer was carbon substrate limited (http://www.lbl.gov/NABIR/). Carbon substrate limitation was corroborated by control tests conducted with no added electron donor in which no microbial activity was detected (20).
TABLE 2.
In situ biostimulation data showing effect of electron donor on nitrate and U(VI) reduction rates
| Well | Treat- ment | Avg rate (SD)a
|
||
|---|---|---|---|---|
| Electron donor use (mM/h) | Nitrate reduction (mM/h) | U(VI) reduction (μM/h) | ||
| FB32 | Glucose | 1.135 (1.980) | 0.281 (0.229) | 0.017 (0.017) |
| FB33 | Glucose | 1.116 (1.816) | 0.950 (1.180) | 0.028 (0.011) |
| FB34 | Ethanol | 1.774 (3.271) | 0.915 (1.193) | 0.009 (0.010) |
Values are average rates obtained from five successive push-pull activity tests at each site.
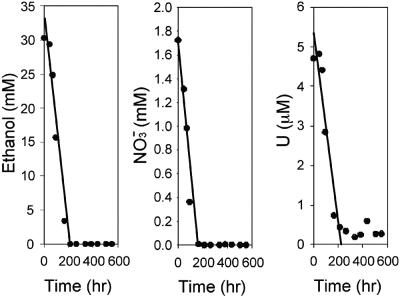
FIG. 1.
Rates of ethanol utilization and nitrate and U(VI) reduction in contaminated well FB34.
Extensive microbial reduction of available electron acceptors [nitrate, Fe(III), U(VI)] and utilization of electron donors (glucose, ethanol) were indicated at all of the sites where in situ tests were conducted at the FRC. A representative plot of activity is shown for well FB34 in Fig. 1, and a summary of the rates obtained from push-pull activity tests at all of the wells is provided in Table 2. Although rates were initially highly variable, substrate utilization was induced upon repeated treatment with an electron donor, such that nearly complete reduction of available nitrate and U(VI) was observed upon completion of the field experiment (Fig. 1). During the later stages of successive push-pull treatment, utilization of electron acceptors and the electron donor occurred simultaneously in a parallel, linear relationship (Fig. 1).
Results from groundwater testing were supported by geochemical analysis of subsurface sediment cores collected before and after biostimulation experiments immediately adjacent to the wells treated by push-pull testing. Sediment pH generally increased by 1 to 2 pH units (the only exception being borehole FB45, with the small decrease in pH possibly due to other simultaneously occurring microbial processes, such as fermentation), and extractable nitrate concentrations decreased by up to 1 to 2 orders of magnitude in treated sediments (Table 3). Additionally, the reduction of amorphous Fe(III) minerals by metal-reducing bacteria was indicated by the accumulation of Fe(II) to between 59 and 88% of the total HCl-extractable Fe (Table 3). Each sediment sample used for DNA extraction in our study came from the same cores as those used for geochemical analysis.
TABLE 3.
Changes in pH, nitrate concentration, and percentage of Fe(II) in total Fe after biostimulation of contaminated FRC sediment
| Core (carbon source added) | Corresponding unstimulated core | pH
|
Nitrate concn (mM)
|
% of Fe(II) in total Fe
|
|||
|---|---|---|---|---|---|---|---|
| Before biostimulation | After biostimulation | Before biostimulation | After biostimulation | Before biostimulation | After biostimulation | ||
| FB45 (glucose) | FB32 | 4.4 | 4.1 | 8.6 | 1.5 | 38.6 | 58.7-85.5 |
| FB46 (glucose) | FB32 | 4.4 | 6.6 | 8.6 | 2.2 | 38.6 | 58.7-85.5 |
| FB47 (glucose) | FB33 | 3.6 | 4.5 | 154.3 | 6.8 | 9.60 | 66.8 |
| FB49 (ethanol) | FB34 | 3.8 | 4.9 | 36.9 | 0.1 | 5.10 | 87.8 |
Clone library analysis.
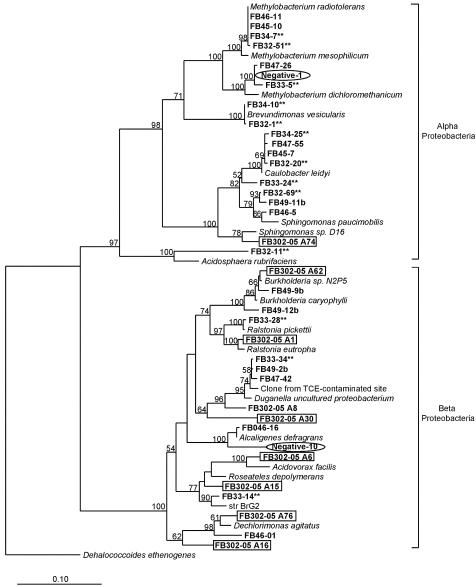
In contaminated FRC sediment before biostimulation, common 16S rRNA gene sequences detected through cloning and sequencing (>10% of the contaminated clone libraries) included species within the α-, β-, and γ-Proteobacteria subdivisions (Fig. 2, 3, and 4). The most abundant (23%) 16S rRNA gene sequences were 98% similar to the Methylobacterium genus within the α-Proteobacteria subdivision, including Methylobacterium radiotolerans, M. dichloromethanicum, and M. mesophilicum. M. dichloromethanicum is able to use dichloromethane as its sole source of carbon and energy (21), and M. mesophilicum is within a group of bacteria capable of degrading EDTA-metal complexes (58). Almost one-fourth of the cloned 16S rRNA genes before biostimulation had 98% sequence identity to organisms cultured from low-nutrient environments, such as Caulobacter leidyi and Brevundimonas vesicularis (22). Also, 18% of retrieved sequences were greater than 95% similar to a cloned 16S rRNA gene sequence within the β-Proteobacteria subdivision initially detected within a TCE-contaminated site undergoing in situ bioremediation treatment (A. B. Carroll and S. H. Zinder, unpublished data). Ten percent of the contaminated clone libraries were composed of 16S rRNA gene sequences related to Pseudomonas anguilliseptica, a γ-Proteobacteria subdivision member previously detected in U(VI) waste piles (55).
FIG. 2.
Phylogenetic tree of α- and β-proteobacterial 16S rRNA gene sequences cloned from contaminated sediment. Asterisks indicate a phylotype detected before biostimulation, while no asterisks indicates a phylotype detected after biostimulation. Circled clones represent negative controls, and boxed clones are from the uncontaminated site.
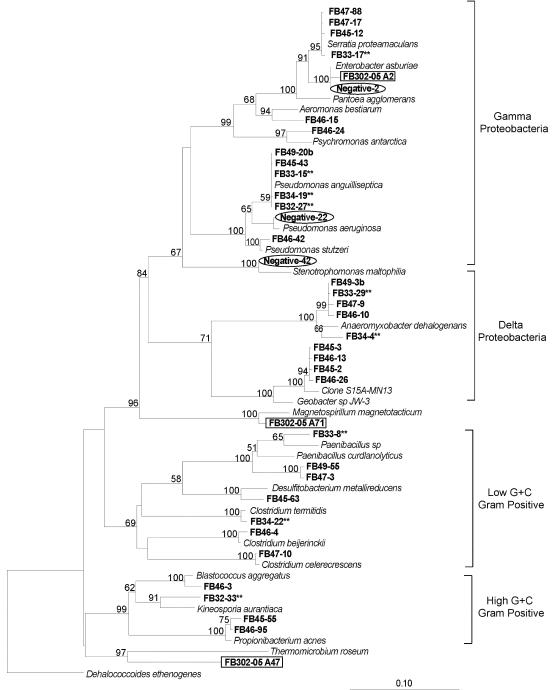
FIG. 3.
Phylogenetic tree of δ- and γ-proteobacterial and gram-positive 16S rRNA gene sequences cloned from contaminated sediment. Asterisks indicate a phylotype detected before biostimulation, while no asterisks indicates a phylotype detected after biostimulation. Circled clones represent negative controls, and boxed clones are from the uncontaminated site.
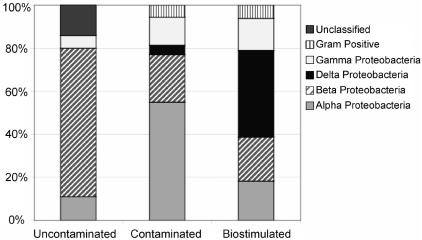
FIG. 4.
Percentages of 16S rRNA gene sequences cloned from contaminated, biostimulated, and uncontaminated sediment.
In the biostimulated sediments, the most common 16S rRNA gene sequences (>10% of the biostimulated clone libraries) detected included species within the α-, β-, δ-, and γ-Proteobacteria subdivisions (Fig. 2, 3, and 4). Forty percent of the sequences retrieved in the biostimulated clone libraries were related to members of the δ-Proteobacteria subdivision. Nearly 25% of the 16S rRNA gene sequences in the biostimulated clone libraries were 96% similar to the δ-Proteobacteria subdivision species Anaeromyxobacter dehalogenans, a known dissimilatory Fe(III)-reducing organism (16). Members of the metal-reducing Geobacteraceae family within the δ-Proteobacteria subdivision were also detected, as several cloned 16S rRNA gene sequences were 93% similar to species in this family. Organisms within the δ-Proteobacteria subdivision represented the greatest increase in frequency of 16S rRNA gene sequences retrieved after biostimulation of FRC sediment, increasing from less than 5% to 37% of the clone libraries. Almost 15% of the biostimulated clone libraries were composed of sequences closely related to polycyclic aromatic hydrocarbon-degrading organisms (38), Burkholderia sp. strain N2P5 and Sphingomonas paucimobilis, within the β- and α-Proteobacteria subdivisions, respectively. Clones closely related to members of the Methylobacterium genus, including M. dichloromethanicum, M. radiotolerans, and M. mesophilicum, were still present (11% of the clone library) after biostimulation experiments but were not as common as in the initial contaminated clone libraries.
To compare sequences from contaminated FRC sediment to sequences from pristine sediment with similar geochemistry, DNA was also extracted from a pristine background site. In the background sediment, a lower diversity of retrieved sequences was observed in comparison with the contaminated and biostimulated clone libraries (Fig. 2, 3, and 4). The most common 16S rRNA gene sequences (>10% of the clone library) were either within the β-Proteobacteria subdivision or were not closely related to any previously identified organisms. Almost 70% of the 16S rRNA gene clones retrieved from the background sediment were within the β-Proteobacteria subdivision. Similar results were obtained in a recent study of a U(VI)-contaminated aquifer in Rifle, Colo., with β-proteobacterial sequences predominating in pristine samples from an upgradient control well (3). Thirty percent of the clones from the background site were 92% similar to an uncultured member of the β-Proteobacteria subdivision most closely related to Duganella zoogloeoides, an aerobic chemoorganotroph isolated from wastewater and activated sludge (44). Almost one-fifth of cloned sequences were greater than 98% similar to the β-Proteobacteria subdivision species Ralstonia eutropha, a well-known facultative chemolithotroph (22). Also within the β-Proteobacteria subdivision, sequences closely related to the species Dechloromonas agitata and Acidovorax facilis were detected in 14% of the clone library.
To ensure that retrieved sequences were not laboratory contaminants, 16S rRNA gene clones generated from sterile-water extractions were used as negative controls (Fig. 2 and 3). About 70% of the cloned sequences in the negative-control library were closely related to members of the γ-Proteobacteria subdivision, in contrast to sediment libraries, where less than 20% of retrieved sequences were related to members of the γ-Proteobacteria subdivision. More than one-third of the negative-control clones were greater than 98% similar to the γ-Proteobacteria subdivision member Enterobacter asburiae, a species recently detected in contaminated agricultural water (25). Sequences related to the γ-proteobacterial species Pseudomonas aeruginosa and Stenotrophomonas maltophilia, two organisms previously identified in a study documenting laboratory contaminants, were also found (56).
MPN-PCR.
Before the in situ addition of carbon substrates (glucose or ethanol) to contaminated FRC sediments, Anaeromyxobacter-related 16S rRNA gene sequences were more abundant than Geobacter- or Paenibacillus-Brevibacillus-related sequences (Table 4). One to 2 orders of magnitude fewer Geobacter-related 16S rRNA gene copies per gram of wet sediment were detected, and in all boreholes but one, Paenibacillus-Brevibacillus-related 16S rRNA genes were the least abundant (Table 4). After biostimulation, the number of Geobacter-related 16S rRNA genes increased by 2 orders of magnitude in two of the four boreholes (one stimulated with glucose and one stimulated with ethanol) (Table 4, borehole sets A and D) and remained relatively stable in the other two (Table 4, borehole sets B and C). In three of the four boreholes (Table 4, borehole sets A, B, and D), the number of Anaeromyxobacter-related and Paenibacillus-Brevibacillus-related 16S rRNA genes decreased by 1 to 2 and 1 to 3 orders of magnitude, respectively. In the fourth borehole (Table 4, borehole set C), the number of Anaeromyxobacter-related 16S rRNA genes remained constant and the number of Paenibacillus-Brevibacillus-related 16S rRNA genes increased.
TABLE 4.
MPN-PCR assay results
| Borehole set | Organism(s) probed | No. of copies of 16S RNA gene/g of sediment (95% confidence limits)a
|
|
|---|---|---|---|
| Before biostimulation | After biostimulation | ||
| A | Geobacter sp. | 5.38 × 103 (4.23 × 103-2.63 × 104) | 2.63 × 105 (2.19 × 105-8.50 × 105) |
| B | Geobacter sp. | 5.38 × 103 (4.23 × 103-2.63 × 104) | 1.50 × 103 (1.06 × 103-7.38 × 103) |
| C | Geobacter sp. | 2.25 × 103 (1.80 × 103-8.85 × 103) | 1.50 × 103 (1.06 × 103-7.38 × 103) |
| D | Geobacter sp. | 6.50 × 102 (4.25 × 102-1.30 × 103) | 2.88 × 104 (2.38 × 104-1.50 × 105) |
| A | Anaeromyxobacter sp. | 2.63 × 104 (2.10 × 104-1.03 × 105) | 1.16 × 103 (9.75 × 102-5.91 × 103) |
| B | Anaeromyxobacter sp. | 2.63 × 104 (2.10 × 104-1.03 × 105) | 1.13 × 102 (1.00 × 102-5.63 × 102) |
| C | Anaeromyxobacter sp. | 3.45 × 103 (2.25 × 103-1.46 × 104) | 5.38 × 103 (4.50 × 103-1.79 × 104) |
| D | Anaeromyxobacter sp. | 4.25 × 104 (3.18 × 104-1.65 × 105) | 1.16 × 103 (9.75 × 102-5.91 × 103) |
| A | Paenibacillus-Brevibacillus spp. | 1.16 × 103 (9.75 × 102-5.91 × 103) | 1.43 × 102 (9.3 × 101-6.07 × 102) |
| B | Paenibacillus-Brevibacillus spp. | 1.16 × 103 (9.75 × 102-5.91 × 103) | 1.00 × 100 |
| C | Paenibacillus-Brevibacillus spp. | 1.00 × 100 | 6.98 × 103 (5.85 × 103-3.55 × 104) |
| D | Paenibacillus-Brevibacillus spp. | 2.35 × 104 (1.65 × 104-7.85 × 104) | 1.88 × 102 (1.5 × 102-7.38 × 102) |
Quantitative PCR assay results obtained before and after biostimulation of contaminated sediment with glucose (comparing FB32 to nearby FB45 [borehole set A], FB32 to nearby FB46 [borehole set B], and FB33 to nearby FB47 [borehole set C]) and ethanol (comparing FB34 to nearby FB49 [borehole set D]) are shown.
DISCUSSION
Few previous studies have applied cultivation-independent approaches for in-depth characterization of metal-reducing communities in subsurface sediment (18, 46, 47, 51). In order to develop effective bioremediation strategies for contaminant metals, the in situ microbial communities likely to catalyze metal reduction need to be understood. The change in the general bacterial community composition brought on by biostimulation must also be understood to place the metal-reducing organisms into the context of competing heterotrophs. We conducted a field experiment on the in situ bioremediation of subsurface environments at the U.S. Department of Energy Natural and Accelerated Bioremediation Research FRC, Oak Ridge, Tenn., where subsurface sediments are cocontaminated with high levels of U(VI) and nitrate. The dominant contaminants in FRC sediment include radionuclides [U(VI), technetium], toxic metals (nickel, aluminum, barium, chromium, mercury), chelating agents (EDTA), chlorinated hydrocarbons (TCE, perchloroethylene), polychlorinated biphenyls, and fuel hydrocarbons (toluene, benzene) (http://www.lbl.gov/NABIR/). This combination of a low pH and high concentrations of nitrate, radionuclides, and organic contaminants in an aerobic subsurface environment is representative of many sites within the U.S. nuclear weapon complex managed by the Department of Energy (39, 45). Therefore, our results are not only important for bioremediation research at the FRC but can also be applied to other sites.
Cloning and sequencing techniques provided measurements of the bacterial 16S rRNA gene compositions of FRC sediments before and after biostimulation experiments. Before biostimulation, the contaminated FRC subsurface was likely carbon limited (20) and contained an extremely high concentration of nitrate (Table 3) (http://www.lbl.gov/NABIR/). Over a 3.5-month period during the in situ biostimulation experiment, the microbial populations responded to an increased pH (Table 3) and increased carbon substrate availability. The environment within the FRC sediment was no longer inhibited by acidic pH or carbon limitations, and sedimentary organisms were able to compete for electron donors and acceptors. Before sediment biostimulation, microorganisms within the α-Proteobacteria subdivision made up 57% of clone libraries, with almost half of those sequences closely related to Methylobacterium species (Fig. 2 and 4). As previously mentioned, M. mesophilicum was detected within a group of bacteria with the ability to degrade EDTA-metal complexes (58). EDTA forms stable, water-soluble complexes with metals, including U(VI), hindering their adsorption to sediment surfaces. Thus, another approach for limiting the migration of complexed U(VI) at the FRC may be to biodegrade co-occurring organic ligands (15). Owing to cocontamination of sediments with synthetic chelators, EDTA may have been one of the few carbon sources available before biostimulation experiments, thus encouraging the growth of microorganisms capable of EDTA degradation. Chlorinated compounds were also available, perhaps supporting the presence of dechlorinating organisms, such as M. dichloromethanicum. Thus, the dominance of Methylobacterium-type sequences may have been due to their ability to survive by using the limited growth substrates available in the contaminated sediment. In fact, before biostimulation, 43% of the clone libraries were made up of sequences related to dechlorinating organisms and 22% of the sequences were related to nitrate-reducing organisms (Table 5). This could be expected, as both nitrate and halogenated compounds are common contaminants present at elevated concentrations in the subsurface of the FRC (http://www.lbl.gov/NABIR/). Not surprisingly, sequences related to organisms that have adapted to survival in low-nutrient environments were also present in FRC sediment before biostimulation, comprising about 22% of the clone libraries. Almost all of the sequences within the clone library before biostimulation were related to organisms with the ability to reduce nitrate, dechlorinate, or survive in low-nutrient environments. It should be noted that the above-mentioned sequences may be located within less acidic microsites of the contaminated sediment, as our measurement reflects the bulk pH of the contaminated sediment.
TABLE 5.
FRC contaminants and potential bioremediating organisms
| Contaminant | Physiological potential | Potential bioremediating organism (reference) | Clone library
|
|
|---|---|---|---|---|
| % Before | % After | |||
| U(VI) | Reduction and immobilization | Geobacter sp. (30) | 4.5 | 37.0 |
| by FeRB | Anaeromyxobacter dehalogenans (16) | |||
| Desulfitobacterium metallireducens (12) | ||||
| Reduction and immobilization | Clostridium beijerinckii (55) | 5.7 | 10.5 | |
| by fermentative FeRB | Serratia proteamaculans (30) | |||
| Nitrate | Reduction | Pseudomonas stutzeri (39) | 22.0 | 27.1 |
| Alcaligenes defragrans (17) | ||||
| Ralstonia pickettii (41) | ||||
| Anaeromyxobacter dehalogenans (48) | ||||
| Denitrifying Fe-oxidizing clone (53) | ||||
| Paenibacillus sp. (50) | ||||
| Chlorinated hydrocarbons | Dechlorination | Methylobacterium dichloromethanicum (21) | 42.5 | 34.4 |
| Anaeromyxobacter dehalogenans (48) | ||||
| Clone from TCE-contaminated site (A. B. Carroll and S. H. Zinder, unpublished data) | ||||
| Dechloromonas sp. (30) | ||||
| Polychlorinated biphenyls | Dechlorination | Acidosphaera rubrifaciens (40) | 14.9 | 2.2 |
| Caulobacter leidyi (40) | ||||
| Fuel hydrocarbons | Degradation | Burkholderia sp. N2P5 (38) | 5.7 | 14.9 |
| Sphingomonas paucimobilis (38) | ||||
We hypothesized that metal-reducing members of the δ-Proteobacteria subdivision would make up a substantial proportion of sedimentary microbial communities after in situ biostimulation. Our hypothesis was upheld, as clone libraries were dominated by members of the δ-Proteobacteria subdivision, which made up about 40% of the 16S rRNA gene sequences after in situ biostimulation (Fig. 4). Cloned sequences within the δ-Proteobacteria subdivision were all closely related to members of the Geobacteraceae family or the Anaeromyxobacter genus. Interestingly, the δ-Proteobacteria subdivision sequences that were found to increase considerably after biostimulation are closely related species that have the ability to use several types of electron acceptors. Certain members of the Geobacteraceae family have been known to reduce both nitrate and Fe(III) (29). Anaeromyxobacter species can also utilize several electron acceptors, including nitrate, Fe(III), and chlorinated hydrocarbons (16, 48). It appears that organisms with the ability to utilize multiple FRC contaminants as electron donors or acceptors are able to outcompete other organisms in the acidic subsurface. Other than members of the δ-Proteobacteria subdivision, sequences related to species such as Serratia proteamaculans and Clostridium beijerinckii, fermentative Fe(III)-reducing organisms (30, 55), increased from 5.7 to 10.5% of the clone libraries after biostimulation (Table 5). Sequences related to both dissimilatory and fermentative FeRB showed the largest increase (10.2 to 47.5%) of sequences identified through cloning and sequencing after biostimulation. The second largest increase (5.7 to 14.9%) was that of sequences related to Sphingomonas paucimobilis and Burkholderia sp. strain N2P5, two species capable of degrading polycyclic aromatic hydrocarbons in fuel hydrocarbons (38), another significant contaminant at the FRC. Thus, a qualitative change in microbial community structure was observed after biostimulation, as the dominance of sequences related to members of the α-Proteobacteria subdivision in contaminated sediment shifted to sequences related to members of the δ-Proteobacteria subdivision in biostimulated sediment.
In contrast, no sequences detected in the clone library from the uncontaminated background sediment were related to the metal-reducing members of the δ-Proteobacteria subdivision (Fig. 3). In fact, the most common 16S rRNA gene sequences retrieved from the background sediment were either within the β-Proteobacteria subdivision or not closely related to any previously identified organisms (Fig. 2 and 3). Most of the sequences retrieved from the background sediment were related to chemoorganotrophic and chemolithotrophic organisms.
We sought to quantify structure-function relationships of metal-reducing microbial groups in the FRC sediment. Although it would be difficult to state absolute quantification of Geobacteraceae species in the FRC sediment, it is clear that a large increase in 16S rRNA gene sequences closely related to this family did occur in one-half of the sites studied (Table 4) over a 3.5-month period in parallel with extensive nitrate and metal reduction (Table 3; Fig. 1). 16S rRNA gene sequences closely related to members of the Geobacteraceae family have been detected in several types of contaminated environments, including the subsurface (3, 18, 46, 47). For example, Geobacter sp. was identified in the Fe(III)-reducing zones of a petroleum-contaminated site and a landfill leachate-polluted aquifer (46, 47). Other studies involving U(VI)-contaminated aquifer sediments at neutrophilic pH revealed an enrichment of Geobacteraceae in carbon-amended sediment incubations (18) and during groundwater treatment (3), suggesting that members of the family Geobacteraceae were responsible for substantial Fe(III) and U(VI) reduction in the subsurface. In the presence of sufficient carbon substrate, the Geobacteraceae group appears to be important in a variety of contaminated subsurface environments, perhaps because of its ability to utilize the abundant electron acceptors present. A close correspondence between microbial activity and an increase in 16S rRNA gene sequences related to members of the Geobacteraceae family suggests that Geobacter-type organisms are important metal reducers in the acidic subsurface of the FRC. Although Geobacteraceae sequences increased in half of the cores tested, other microbial groups must have been involved in bioremediation at the two remaining boreholes. Thus, it will be important to expand our studies to other organisms responding to biostimulation in acidic subsurface sediments.
We hypothesized that electron acceptor utilization was limited by low pH and a paucity of carbon substrates in the contaminated FRC subsurface. Our hypothesis was confirmed, as the abundance and diversity of organisms changed after biostimulation, when the subsurface was neutralized and carbon was added. Both qualitative and quantitative shifts in microbial communities were detected in our investigation. Results from cloning and sequencing of community DNA paralleled those of quantitative PCR to show that stimulation of members of the δ-Proteobacteria subdivision represented the largest change in the microbial community structure. However, in contrast to the MPN-PCR results (Table 4), clone libraries indicated that A. dehalogenans was a prominent member of the biostimulated microbial community (Fig. 3). This could be due to cloning bias as a result of the nucleotide composition of Anaeromyxobacter-type sequences. Yet, A. dehalogenans sequences were also the dominant sequences detected in Fe(III)-reducing enrichments in a previous study of contaminated FRC sediment (42). Both geochemical and microbial community analyses demonstrated that the FRC subsurface sediments contain a large amount of heterogeneity, even within cores. Thus, we cannot rule out the importance of this newly described metal reducer during biostimulation.
Future studies should focus on the active members of microbial populations that are stimulated through in situ carbon source addition. By using RNA (rather than DNA) as a nucleotide template, the active indigenous microbial groups involved in bioremediation processes could be better resolved.
Acknowledgments
This research was funded by the Natural and Accelerated Bioremediation Research (NABIR) Program, Biological and Environmental Research (BER), U.S. Department of Energy (grant DE-FG02-OOER62986).
We thank Harold J. Adams for help in sample processing and Steve Miller at the Florida State University Sequencing Facility. We also thank Dave Watson, Lee Krumholz, and Barry Kinsall for help with sediment sampling and shipment.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.American Public Health Association. 1969. Standard methods for the examination of water and wastewater, including bottom sediments and sludge, p. 604-609. American Public Health Association, Washington, D.C.
- 3.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braman, R. S., and S. A. Hendrix. 1989. Nanogram nitrate and nitrate determination in environmental and biological materials by vanadium(III) reduction with chemiluminescence detection. Anal. Chem. 61:2715-2718. [DOI] [PubMed] [Google Scholar]
- 5.Brina, R., and A. G. Miller. 1992. Direct detection of trace levels of uranium by laser-induced kinetic phosphorimetry. Anal. Chem. 64:1413-1418. [Google Scholar]
- 6.Chang, Y., A. D. Peacock, P. E. Long, J. R. Stephen, J. P. McKinley, S. J. Macnaughton, A. K. M. Hussain, A. M. Saxton, and D. C. White. 2001. Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl. Environ. Microbiol. 67:3149-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels, L., R. S. Hanson, and J. A. Phillips. 1994. Chemical analysis, p. 512-554. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.) Methods for general and molecular bacteriology, American Society for Microbiology, Washington, D.C.
- 9.DiChristina, T. J. 1992. Effects of nitrate and nitrite on dissimilatory iron reduction by Shewanella putrefaciens 200. J. Bacteriol. 174:1891-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dollhopf, S. L., S. A. Hashsham, F. B. Dazzo, R. F. Hickey, C. S. Criddle, and J. M. Tiedje. 2001. The impact of fermentative organisms on carbon flow in methanogenic systems under constant low substrate conditions. Appl. Microbiol. Biotech. 56:531-538. [DOI] [PubMed] [Google Scholar]
- 11.Fernández, A., S. Huang, S. Seston, J. Xing, R. Hickey, C. Criddle, and J. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finneran, K. T., H. M. Forbush, C. Gaw VanPraagh, and D. R. Lovley. 2002. Desulfitobacterium metallireducens sp. nov., an anaerobic bacterium that couples growth to the reduction of metals and humic acids, as well as chlorinated compounds. Int. J. Syst. Evol. Microbiol. 52:1929-1935. [DOI] [PubMed] [Google Scholar]
- 13.Finneran, K. T., M. E. Housewright, and D. R. Lovley. 2002. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediment. Environ. Microbiol. 4:510-516. [DOI] [PubMed] [Google Scholar]
- 14.Fredslund, L., F. Ekelund, C. S. Jacobsen, and K. Johnsen. 2001. Development and application of a most-probable-number-PCR assay to quantify flagellate populations in soil samples. Appl. Environ. Microbiol. 67:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganesh, R., K. G. Robinson, G. D. Reed, and G. S. Sayler. 1997. Reduction of hexavalent uranium from organic complexes by sulfate- and iron-reducing bacteria. Appl. Environ. Microbiol. 63:4385-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, Q., and R. A. Sanford. 2003. Characterization of Fe(III) reduction by chlororespiring Anaeromyxobacter dehalogenans. Appl. Environ. Microbiol. 69:2712-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyen, U., and J. Harder. 1998. Cometabolic isoterpinolene formation from isolimonene by denitrifying Alcaligenes defragrans. FEMS Microbiol. Lett. 169:67-71. [Google Scholar]
- 18.Holmes, D. E., K. T. Finneran, R. A. O'Neil, and D. R. Lovley. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl. Environ. Microbiol. 68:2300-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurt, R. A., X. Qiu, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Istok, J. D., J. M. Senko, L. R. Krumholz, D. Watson, M. A. Bogle, A. Peacock, Y.-J. Chang, and D. C. White. 2004. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ. Sci. Technol. 38:468-475. [DOI] [PubMed] [Google Scholar]
- 21.Kayser, M. F., Z. Ucurum, and S. Vuilleumier. 2002. Dichloromethane metabolism and C1 utilization genes in Methylobacterium strains. Microbiology 148:1915-1922. [DOI] [PubMed] [Google Scholar]
- 22.Kersters, K. P., P. De Vos, M. Gillis, J. Swings, P. VanDamme, and E. Stackebrandt. 28March2003. Introduction to the proteobacteria. In K. P. Kersters et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community. 3rd ed, release 3.12. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 23.Kostka, J. E., and G. W. Luther III. 1994. Partitioning and speciation of solid phase iron in saltmarsh sediments. Geochim. Cosmochim. Acta 58:1701-1710. [Google Scholar]
- 24.Kostka, J. E., D. D. Dalton, H. Skelton, S. L. Dollhopf, and J. W. Stucki. 2002. Growth of iron(III)-reducing bacteria on clay minerals as the sole electron acceptor and comparison of growth yields on a variety of oxidized iron forms. Appl. Environ. Microbiol. 68:6256-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laukova, A., P. Jurisce, Z. Vasilkova, and I. Papajova. 2000. Treatment of sanitary-important bacteria by bacteriocin substance V24 in cattle dung water. Lett. Appl. Microbiol. 30:402-405. [DOI] [PubMed] [Google Scholar]
- 26.Liger, E., L. Charlet, and P. Van Chappellen. 1999. Surface catalysis of uranium(VI) reduction by iron(II). Geochim. Cosmochim. Acta 63:2939-2955. [Google Scholar]
- 27.Liu, Y., D. L. Balkwill, H. C. Aldrich, G. R. Drake, and D. R. Boone. 1999. Characterization of the anaerobic propionate-degrading syntrophs Smithella propionica gen. nov., sp. nov. and Syntrophobacter wolinii. Int. J. Syst. Bacteriol. 49:545-556. [DOI] [PubMed] [Google Scholar]
- 28.Lovley, D. R. 1991. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol. Rev. 55:259-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovley, D. R. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Ind. Microbiol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 30.Lovley, D. R. 2000. Fe(III) and Mn(IV) reduction, p. 3-30. In D. R. Lovley (ed.) Environmental microbe-metal interactions. ASM Press, Washington, D.C.
- 31.Lovley, D. R. 1December2000, posting date. Dissimilatory Fe(III)- and Mn(IV)-reducing prokaryotes. In D. R. Lovley et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.4. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 32.Lovley, D. R. 2002. Dissimilatory metal reduction: from early life to bioremediation. ASM News 68:231-237. [Google Scholar]
- 33.Lovley, D. R., E. E. Roden, E. J. P. Phillips, and J. C. Woodward. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar. Geol. 113:41-53. [Google Scholar]
- 34.Lovley, D. R., and R. T. Anderson. 2000. Influence of dissimilatory metal reduction on fate of organic and metal contaminants in the subsurface. Hydrogeol. J. 8:77-88. [Google Scholar]
- 35.Madigan, M., J. M. Martinko, and J. Parker (ed.). 2000. Brock biology of microorganisms, 9th ed., p. 608. Prentice-Hall, Inc., Upper Saddle River, N.J.
- 36.McLean, E. O. 1982. Soil pH and lime requirement, p. 199-209. In A. L. Page, R. H. Miller, and D. R. Keeney (ed.), Methods of soil analysis, part 2, chemical and microbiological properties—agronomy monograph no. 9, 2nd ed. ASA-SSSA, Madison, Wis.
- 37.Mohagheghi, A., D. M. Updegraff, M. B. Goldhaber. 1985. The role of sulfate reducing bacteria in the deposition of sedimentary uranium ores. Geomicrobiol. J. 4:153-173. [Google Scholar]
- 38.Mueller, J. G., R. Devereux, D. L. Santavy, S. E. Lantz, S. G. Willis, and P. H. Pritchard. 1997. Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Antonie van Leeuwenhoek 71:329-343. [DOI] [PubMed] [Google Scholar]
- 39.Natural and Accelerated Bioremediation Research Program. 2003. Bioremediation of metals and radionuclides…what it is and how it works, p. 1-78. In T. C. Hazen, S. M. Benson, F. B. Metting, B. Faison, A. C. Palmisano, and J. McCullough (ed.), NABIR primer, 2nd ed. Lawrence Berkeley National Laboratory, Berkeley, Calif.
- 40.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, J., J. J. Kukor, and L. M. Abriola. 2002. Characterization of the adaptive response to trichloroethylene-mediated stresses in Ralstonia pickettii PKO1. Appl. Environ. Microbiol. 68:5231-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrie, L., N. N. North, S. L. Dollhopf, D. L. Balkwill, and J. E. Kostka. 2003. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium U(VI). Appl. Environ. Microbiol. 69:7467-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabus, R., T. Hansen, and F. Widdel. 8September2000, posting date. Dissimilatory sulfate- and sulfur-reducing prokaryotes. In R. Rabus et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.3. [Online.] Springer-Verlag, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 44.Rappe, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 45.Riley, R. G., and J. Zachara. 1992. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface research. DOE/ER-0547T. U.S. Department of Energy, Washington, D.C.
- 46.Röling, W. F. m., B. M. van Breukelen, M. Braster, B. Lin, and H. W. van Verseveld. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanford, R. A., J. R. Cole, and J. M. Tiedje. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Senko, J. M, J. D. Istok, J. M. Suflita, and L. R. Krumholz. 2002. In-situ evidence for uranium immobilization and remobilization. Environ. Sci. Technol. 36:1491-1496. [DOI] [PubMed] [Google Scholar]
- 50.Shida, O., H. Takagi, K. Kadowaki, L. K. Nakamura, and K. Komagata. 1997. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int. J. Syst. Bacteriol. 47:289-298. [DOI] [PubMed] [Google Scholar]
- 51.Snoeyenbos-West, O. L., K. P. Nevin, R. T. Anderson, and D. R. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 52.Stookey, L. L. 1970. Ferrozine: a new spectrophotometric reagent for iron. Anal. Chem. 42:779-781. [Google Scholar]
- 53.Straub, K. L., M. Benz, B. Schink, and F. Widdel. 1996. Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl. Environ. Microbiol. 62:1458-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strunk, O., and W. Ludwig. 1997. ARB: software for phylogenetic analysis. Technical University of Munich, Munich, Germany.
- 55.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl. Environ. Microbiol. 69:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1988. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tebo, B. M., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193-198. [Google Scholar]
- 58.Thomas, R. A. P., K. Lawlor, M. Bailey, and L. E. Macaskie. 1998. Biodegradation of metal-EDTA complexes by an enriched microbial population. Appl. Environ. Microbiol. 64:1319-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]