Abstract
We designed and successfully implemented the use of in situ-synthesized 45-mer oligonucleotide DNA microarrays (XeoChips) for genome-wide expression profiling of Burkholderia xenovorans LB400, which is among the best aerobic polychlorinated biphenyl degraders known so far. We conducted differential gene expression profiling during exponential growth on succinate, benzoate, and biphenyl as sole carbon sources and investigated the transcriptome of early-stationary-phase cells grown on biphenyl. Based on these experiments, we outlined metabolic pathways and summarized other cellular functions in the organism relevant for biphenyl and benzoate degradation. All genes previously identified as being directly involved in biphenyl degradation were up-regulated when cells were grown on biphenyl compared to expression in succinate-grown cells. For benzoate degradation, however, genes for an aerobic coenzyme A activation pathway were up-regulated in biphenyl-grown cells, while the pathway for benzoate degradation via hydroxylation was up-regulated in benzoate-grown cells. The early-stationary-phase biphenyl-grown cells showed similar expression of biphenyl pathway genes, but a surprising up-regulation of C1 metabolic pathway genes was observed. The microarray results were validated by quantitative reverse transcription PCR with a subset of genes of interest. The XeoChips showed a chip-to-chip variation of 13.9%, compared to the 21.6% variation for spotted oligonucleotide microarrays, which is less variation than that typically reported for PCR product microarrays.
Many bacteria, most of which belong to the beta subclass of the Proteobacteria and the Actinobacteria (20, 42), can grow aerobically with biphenyl as a sole carbon source and can cometabolically degrade polychlorinated biphenyls (PCBs) (1, 4), which are among the most problematic environmental pollutants (29). Burkholderia xenovorans strain LB400 (17) is one of the most-studied and effective aerobic PCB degraders known and is able to cometabolically degrade up to hexachlorinated biphenyls (3, 8, 12, 18, 22, 31-33). Although the operon organization for the upper biphenyl degradation pathway (the upper Bph pathway) has been extensively documented, there are contradictory reports on its regulation, suggesting that there is constitutive (5, 25) or biphenyl-induced expression (2, 12, 27). The enzymatic reactions of the upper Bph pathway result in formation of benzoate and pentadiene or their chlorinated derivatives. Pentadiene and possibly some chlorinated derivatives can be metabolized by proteins encoded by the lower Bph pathway, which are often in the same operon (21, 24), while the traditional catechol-β-ketoadipate pathway (19) is believed to be responsible for the degradation of benzoate. Recently, however, an alternative aerobic benzoate pathway via coenzyme A (CoA) activation has been characterized in Azoarcus evansii by Gescher et al. (15), and these authors noted that there are two homologous copies of this pathway in the LB400 genome (15).
In collaboration with our laboratory, a draft sequence of LB400's genome was generated by The Joint Genome Institute. The latest annotated assembly (JGI/ORNL annotation December 2003) suggests that the total genome size is ∼9.7 Mbp (50 major contigs) and that there are 9,851 open reading frames (ORFs). Despite years of research on this PCB-degrading organism, very little is known about its general physiology, metabolism, biochemistry, and management of the metabolic warehouse of the large genome. To enhance our knowledge of this organism and, more specifically, how its genomic context allows it to be successful as a PCB degrader, a microarray containing one 45-mer oligonucleotide probe per ORF was constructed to explore genes that are potentially important in the biodegradation of PCBs. In collaboration with Xeotron Corporation (Houston, Tex.), we evaluated and used their new light-directed, in situ-synthesized microfluidic microarray platform (XeoChip) (14) for this purpose. We investigated the whole-genome expression patterns when LB400 was grown on biphenyl or benzoate compared to the patterns when the organism was grown on succinate. Furthermore, we investigated the expression patterns in the early stationary phase of biphenyl-grown cells. We validated our array results by using the data for a subset of genes measured by quantitative reverse transcription PCR (Q-RT-PCR).
MATERIALS AND METHODS
Bacterial strain and genome sequence.
B. xenovorans strain LB400 was originally isolated from a PCB-contaminated New York state landfill site (3), and an 8× coverage sequence draft was produced by Department of Energy's Joint Genome Institute and automatically annotated by Oak Ridge National Laboratory's Computational Genomics Group. The annotation used in this work is available at http://cme.msu.edu/lb400/LB400/Annotation.txt (JGI/ORNL annotation May 2001). The latest sequence and draft analysis is available at http://genome.jgi-psf.org/draft_microbes/burfu/burfu.home.html.
Media and growth conditions.
LB400 was grown in liquid K1 mineral medium (46) supplemented with succinate (10 mM), benzoate (5 mM), or particulate biphenyl (5 mM; S = 6.99 × 10−3 g/liter) as the sole carbon source. Cells were grown at 29 ± 1°C in 200-ml batch cultures in 500-ml Erlenmeyer flasks on a rotary shaker at 250 rpm. At the start of each experiment, a glycerol stock culture was grown on R2A (Difco) agar plates, and biomass from pooled colonies was grown on a K1 agar plate containing biphenyl. K1 liquid medium containing succinate (reference conditions, transfer 1) was inoculated with pooled colonies from this plate and grown until the stationary phase. Two subsequent transfers were made in succinate medium prior to harvest (reference conditions, transfer 3). Cells from transfer 2 in K1 medium containing succinate were used to inoculate liquid K1 medium containing biphenyl and liquid K1 medium containing benzoate (treatment conditions, transfer 1), and cells were harvested after two more transfers in K1 medium containing biphenyl and liquid K1 medium containing benzoate (treatment conditions, transfer 3), respectively. Transfer 3 for each carbon source (reference and treatment) was done in duplicate to obtain biological replicates. The two biological replicates of succinate-, benzoate-, and biphenyl-grown cells were harvested at the maximum growth rates (0.40, 0.27, and 0.23 cell division/h, respectively) at optical densities of 0.45, 0.35, and 0.35, respectively. Additionally, two biological replicates of biphenyl-grown cells were harvested within 1-log-phase generation time after the start of the stationary phase, at an optical density of ∼0.75. We termed this sampling period the early stationary phase because the growth rate decreased to a much lower value after this point although the cells eventually reached an optical density of ∼1.2.
RNA extraction and labeling.
RNAlater (Ambion) was added at a 1:1 ratio to a culture to protect RNA against degradation. Bacterial cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C. The RNA was extracted with an RNeasy RNA extraction kit (QIAGEN), and the remaining DNA was removed by 30 min of incubation at room temperature with 1.5 U of DNase I (10 U/μl; Roche) per μg of nucleic acid. The integrity of the RNA and the absence of DNA were verified by 1.2% agarose gel electrophoresis.
For direct incorporation (succinate versus early-stationary-phase biphenyl), 25 μg of total RNA was labeled by using the Xeotron cDNA labeling protocol (www.xeotron.com), except that 30 μg of random hexamers (Invitrogen) was used instead of poly(dT) primers. Reaction cleanup was performed by using the Qiaquick PCR purification system protocol (QIAGEN). Amino-allyl labeling (all other hybridizations) was performed by using a protocol adapted from a protocol of The Institute for Genomic Research (http://www.tigr.org/tdb/microarray/protocolsTIGR.shtml). Briefly, 2 μg of total RNA was labeled overnight at 45°C by using 6 μg of random primers (Invitrogen), amino-allyl-labeled dUTP (Sigma), and Superscript III reverse transcriptase (Invitrogen), and subsequently reactive Cy5 or Cy3 fluorophores (Amersham) were coupled to the amino-allyl groups. Purification after enzymatic incorporation and chemical coupling was performed by using QiaQuick PCR purification columns (QIAGEN) as described in The Institute for Genomic Research protocol. The quantity of labeled cDNA and the fluororophore incorporation efficiency were determined by using UV-visible spectrophotometry.
Microarray design and synthesis.
The LB400 genomic microarray consists of one unique 45-mer (specificity checked by BLAST versus the LB400 genome) for each of the 9,511 annotated ORFs (JGI/ORNL annotation December 2000b), designed by using OligoArray (30), with a melting temperature of 86 to 92°C and a G+C content of 50 to 60% and subsequently synthesized in situ by Xeotron by using their 4,000-chamber (spot) format. The genome is represented by a genomic set consisting of three chips, each containing ∼4,000 synthesized 45-mers. A total of 1,601 probes on chip 1 and chip 3 of the genomic set are identical and used as a chip-to-chip replication control in each experiment. For comparison tests of the XeoChips, we used a spotted 70-mer oligonucleotide microarray that contained triplicate spots of 507 probes (the probe for a gene did not necessarily overlap with the XeoChip probe for that gene) targeting a random set of the genes of LB400 (J. Park, T. V. Tsoi, S. A. Hashsham, J. R. Cole, S. Callister, and J. M. Tiedje, Abstr. NIEHS Symp. Bioremediat. Biodegradat., abstr. 23, 2002). Production was based on optimized protocols described previously (9).
Hybridization, scanning, and data analysis.
All buffers were passed through a 0.22-μm-pore-size filter to keep particulate matter out of the microfluidic channels. XeoChips were placed in Xeotron hybridization adaptors and prehybridized with a solution containing 25% formamide (Ambion), 6× SSPE (pH 6.6; Ambion), and 1 mg of acetylated bovine serum albumin per ml (Ambion) for 5 min at a flow rate of 100 μl/min (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA). A 100-μl hybridization solution consisting of 25% formamide, 6× SSPE (pH 6.6), 1 mg of bovine serum albumin per ml, and labeled target containing 100 pmol of each dye was denatured at 95°C for 3 min, snap cooled on ice, filtered by using Spin-X 0.22-μm-pore-size spin column filters (Corning), and hybridized by circulating the hybridization mixture for 18 h at 32°C at a flow rate of 100 μl/min. Both solutions were stripped of dissolved gases by flushing them with nitrogen for 2 min prior to the denaturation step. After hybridization, each chip was washed at a flow rate of 100 μl/min with 6× SSPE (pH 6.6) for 5 min at 32°C and subsequently with 1× SSPE (pH 6.6) for 5 min at 32°C. The spotted 70-mer microarray was hybridized as described previously (9).
Fluorescence intensity data were acquired by using an Axon 4000A laser scanner (Axon Laboratories) and, for XeoChips, were subsequently imported in ArrayPro 4.0 (Media Cybernetics, L.P.) to extract the 635-nm (Cy5) and 532-nm (Cy3) signals. Based on an analysis by Fang et al. (13), signals without background subtraction were used for both platforms. Spotted microarray data were extracted by using Genepix Pro 3.0 (Axon Laboratories). Median signals for each channel (Cy5 and Cy3) were imported into GeneSpring 5.0 (Silicon Genetics) and normalized by using Lowess intensity-dependent normalization. P values were calculated by using GeneSpring's cross gene error model and the Student t test algorithm based on the variation between log2 ratio values of biological replicates. Plots of M [log2 (Cy5/Cy3)] versus A [log2 (√(Cy5 × Cy3)] were drawn as described by Dudoit el al (11) by using all data from one experiment grouped per chip. Chip-to-chip reproducibility was determined by calculating the coefficient of variation (CV) (CV = [standard deviation/mean] × 100) for the ratios for identical probes on separate microarrays (i.e., separate slides for spotted microarrays [1,536 probes] and chip 1 and chip 3 of a genomic set for XeoChips [1,601 probes]). The CV for biological reproducibility for XeoChip experiments was calculated separately for ratios of hybridizations with dye-swapped labeled biological replicate samples for all probes (9,511 probes), all nonreplicated probes (7,910 probes), and all replicated probes (1,601 probes).
Quantitative real-time PCR.
Triplicate Q-RT-PCR runs were performed for 25 genes by using the same RNA samples that were used for the microarray hybridizations. One microgram of total RNA was converted into cDNA, and 1/500 to 1/1,000 of the sample was utilized for a 40-cycle, two-step PCR with an ABI 7900HT (Applied Biosystems, Foster City, Calif.) by using 1× SYBR Green master mixture (Applied Biosystems) and each primer at a concentration of 125 nM (sequences available on request). Amplicon size (80 to 100 bp) and reaction specificity were confirmed by agarose gel electrophoresis and product dissociation curves. The number of target copies in each sample was interpolated from the detection threshold value by using a purified PCR product standard curve for bphA, which was constructed for each Q-RT-PCR run. l6S rRNA expression was measured as an internal control, and the measured internal control signal was used to normalize variations due to different reverse transcription efficiencies.
RESULTS
XeoChip data quality.
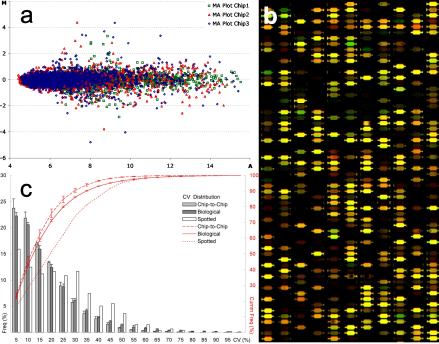
Normalized data quality was analyzed by using M-versus-A plots for each of the three chips of the genomic set, which indicated absence of chip-dependent and signal intensity-dependent biases (Fig. 1a). Furthermore, by comparing the signal ratios of each experiment for 1,601 identical probes on chips 1 and 3, we measured the XeoChip (Fig. 1b) chip-to-chip reproducibility (Fig. 1c). The average CV was 13.9% ± 0.7% with a 90% percentile of 29.0% ± 1.4%, compared to an average CV for two hybridizations to the 1,536 probes of the spotted microarrays of 21.6% and a 90% percentile of 43.7%. The average CVs for dye-swapped biological replicates on XeoChips were 15.6% ± 0.8%, 16.0% ± 0.7%, and 13.2% ± 1.3% for all probes, the nonreplicated probes, and the replicated probes, respectively (Fig. 1c). The ratios of genes that were differentially expressed more than twofold correlated with r values of 0.98 (chip to chip) and 0.95 (biological).
FIG. 1.
(a) Plots of M [log2 (Cy5/Cy3)] versus A [log2 ((Cy5 × Cy3)] per chip for a typical XeoChip hybridization, after Lowess normalization. (b) Image of a XeoChip (4,000-chamber [spot] platform) after hybridization. (c) Chip-to-chip reproducibility and biological reproducibility of XeoChip platform determined by using data from all hybridizations: frequency (Freq) and cumulative frequency (Cumm Freq) of the CVs of ratios between chips hybridized with same hybridization mixture (chip to chip), of ratios between nonreplicated probes of biological dye-swapped replicates (biological), and of ratios on spotted microarrays hybridized with the same hybridization mixture (spotted). The error bars indicate standard errors between experiments.
Evaluation of the expression ratios by Q-RT-PCR.
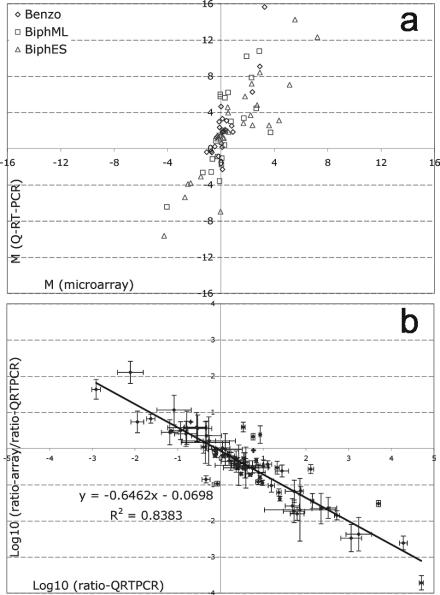
When microarray data were compared with Q-RT-PCR results for the three experiments, 61 of 75 measurements (81%) were in qualitative correspondence (Fig. 2a) (i.e., they were consistent in suggesting up- or down-regulation). When a twofold threshold was used for both assays, only one false positive (compared to Q-RT-PCR) resulted from the microarray analysis. Linear regression analysis indicated that the bias between microarray and Q-RT-PCR ratios correlated well with the ratio of gene expression as determined by Q-RT-PCR (Fig. 2b).
FIG. 2.
Evaluation of microarray results by Q-RT-PCR. (a) Log2 ratio of microarray versus log2 ratio of Q-RT-PCR for each experiment, averaged over biological replications. (b) Bias between microarray and Q-RT-PCR data. The ratios determined by Q-RT-PCR [log10 (ratio-QRTPCR)] are plotted against the ratio of ratios obtained by using microarrays and Q-RT-PCR [log10 (ratio-array)/(ratio-QRTPCR)]. The error bars indicate one standard deviation for biological replicates. Benzo, benzoate-grown cells; BiphML, mid-log-phase biphenyl-grown cells; BiphES, early-stationary-phase biphenyl-grown cells.
Global summary of differential expression.
Relative to succinate-grown cells, the least differential expression (threshold, more than twofold up- or down-regulation on average for biological replicates) was observed for benzoate-grown cells (9 genes were down-regulated and 46 genes were up-regulated), while the values were slightly higher for mid-log-phase biphenyl cells (70 genes were down-regulated and 53 genes were up-regulated) and rose significantly when we looked at the early-stationary-phase biphenyl cells (480 genes were down-regulated and 276 genes were up-regulated). Based on a >2× background (signal in empty XeoChip chambers) threshold, a signal was detected for ∼45% of the genes under either condition. Functional classification of differentially expressed genes based on clusters of orthologous groups of proteins (COGs) (Table 1) indicated resource reallocations depending on the growth substrate and phase.
TABLE 1.
Summary of differential expression based on COG protein classification
| COG(c) | Description | Down-regulated genesa
|
Up-regulated genesa
|
Distribution in LB400 genome (%)b | ||||
|---|---|---|---|---|---|---|---|---|
| Benzo | BiphML | BiphES | Benzo | BiphML | BiphES | |||
| C | Energy production and conversion | 0.0 (0) | 15.7 (11) | 11.7 (56) | 8.7 (4) | 9.4 (5) | 10.5 (29) | 5.9 (591) |
| E | Amino acid transport and metabolism | 0.0 (0) | 7.1 (5) | 10.4 (50) | 4.3 (2) | 9.4 (5) | 5.8 (16) | 6.5 (653) |
| F | Nucleotide transport and metabolism | 0.0 (0) | 1.4 (1) | 3.1 (15) | 0.0 (0) | 1.9 (1) | 0.7 (2) | 0.9 (90) |
| G | Carbohydrate transport and metabolism | 0.0 (0) | 1.4 (1) | 2.5 (12) | 2.2 (1) | 3.8 (2) | 4.7 (13) | 4.9 (497) |
| H | Coenzyme metabolism | 11.1 (1) | 1.4 (1) | 3.5 (17) | 0.0 (0) | 1.9 (1) | 1.8 (5) | 2.1 (212) |
| I | Lipid metabolism | 0.0 (0) | 2.9 (2) | 2.7 (13) | 10.9 (5) | 5.7 (3) | 2.2 (6) | 3.4 (339) |
| J | Translation, ribosomal structure, and biogenesis | 33.3 (3) | 28.6 (20) | 17.1 (82) | 2.2 (1) | 0.0 (0) | 0.7 (2) | 2.1 (213) |
| K | Transcription | 11.1 (1) | 2.9 (2) | 3.3 (16) | 2.2 (1) | 11.3 (6) | 5.4 (15) | 6.7 (672) |
| L | DNA replication, recombination, and repair | 0.0 (0) | 1.4 (1) | 1.3 (6) | 0.0 (0) | 3.8 (2) | 3.3 (9) | 3.0 (300) |
| D | Cell division and chromosome partitioning | 0.0 (0) | 1.4 (1) | 0.4 (2) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.5 (52) |
| M | Cell envelope biogenesis, outer membrane | 11.1 (1) | 5.7 (4) | 4.6 (22) | 4.3 (2) | 5.7 (3) | 2.2 (6) | 4.3 (430) |
| N | Cell motility and secretion | 0.0 (0) | 0.0 (0) | 0.6 (3) | 0.0 (0) | 3.8 (2) | 0.7 (2) | 0.3 (26) |
| O | Posttranslational modification, protein turnover, chaperones | 0.0 (0) | 2.9 (2) | 6.3 (30) | 0.0 (0) | 3.8 (2) | 2.2 (6) | 2.0 (205) |
| P | Inorganic ion transport and metabolism | 0.0 (0) | 0.0 (0) | 4.0 (19) | 0.0 (0) | 3.8 (2) | 4.0 (11) | 3.9 (394) |
| Q | Secondary metabolite biosynthesis, transport, and catabolism | 0.0 (0) | 2.9 (2) | 1.7 (8) | 19.6 (9) | 3.8 (2) | 2.2 (6) | 2.2 (226) |
| T | Signal transduction mechanisms | 11.1 (1) | 0.0 (0) | 1.9 (9) | 0.0 (0) | 0.0 (0) | 4.0 (11) | 3.1 (314) |
| R | General function prediction only | 0.0 (0) | 8.6 (6) | 8.5 (41) | 19.6 (9) | 15.1 (8) | 14.9 (41) | 12.0 (1,208) |
| S | Function unknown | 0.0 (0) | 0.0 (0) | 5.2 (25) | 13.0 (6) | 3.8 (2) | 6.9 (19) | 5.7 (574) |
| No | Hypothetical | 22.2 (2) | 15.7 (11) | 11.3 (54) | 13.0 (6) | 13.2 (7) | 27.9 (77) | 30.5 (3,073) |
| J,K,L | Information storage and processing | 44.4 (4) | 32.9 (23) | 21.7 (104) | 4.3 (2) | 15.1 (8) | 9.4 (26) | 11.8 (1,185) |
| D,O,M,N,P,T | Cell processes | 22.2 (2) | 10.0 (7) | 17.7 (85) | 4.3 (2) | 17.0 (9) | 13.0 (36) | 14.1 (1,421) |
| C,G,E,F,G,H,I,Q | Metabolism | 11.1 (1) | 32.9 (23) | 35.6 (171) | 45.7 (21) | 35.8 (19) | 27.9 (77) | 25.9 (2,608) |
| R,S | Poorly characterized | 0.0 (0) | 8.6 (6) | 13.8 (66) | 32.6 (15) | 18.9 (10) | 21.7 (60) | 17.7 (1,782) |
| No | No match | 22.2 (2) | 15.7 (11) | 11.3 (54) | 13.0 (6) | 13.2 (7) | 27.9 (77) | 30.5 (3,073) |
The values are the percentages of up- or down-regulated genes (based on the total number of up- or down-regulated genes) for the following different conditions: benzoate-grown cells (Benzo), mid-log-phase biphenyl-grown cells (BiphML), and early-stationary-phase biphenyl-grown cells (BiphES). The values in parentheses are the absolute numbers of differentially expressed genes.
Percentage of the 10,069 genes in the genome (actual number of genes).
Biphenyl and benzoate degradation.
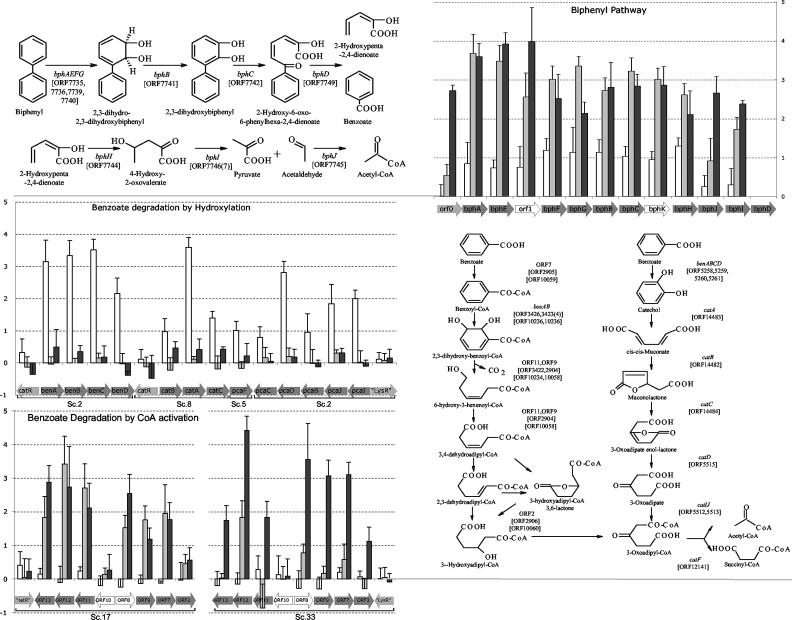
Using the differential expression data, we located the metabolic pathways relevant to biphenyl and benzoate metabolism (Fig. 3). While most biphenyl pathway genes were also expressed when LB400 was growing on succinate, the pathway expression signals increased significantly (P < 0.05) in early-stationary-phase, log-phase biphenyl-grown, and benzoate-grown cells (8.73- ± 0.64-, 7.29- ± 0.56-, and 1.84- ± 0.20-fold, respectively).
FIG. 3.
Biphenyl and benzoate metabolic pathways. Genes, whose annotations were manually verified by using BLAST, are presented as they are organized on the chromosome (position, orientation, and scaffold) for catabolic genes (dark grey arrows), putative regulatory genes (light grey arrows), and ORFs with unknown functions (open arrows). The ORF numbers in LB400 corresponding to the catabolic genes are indicated in the pathways. The graphs show the M values [log2 (test condition/succinate)] for benzoate-grown cells (open bars), log-phase biphenyl-grown cells (grey bars), and early-stationary-phase biphenyl-grown cells (black bars). The error bars indicate the standard errors (standard deviation/n) for the biological replications. Genes without data were not included in the sequence draft and are therefore not present on the microarray, while two neighboring ORFs were annotated as bphI due to replication during genome assembly. log, log phase; stat, stationary phase.
Different benzoate degradation pathways were observed to be up-regulated depending on the carbon source used. In benzoate-grown cells, significant (P < 0.05) up-regulation of all structural genes of the benzoate degradation by the hydroxylation pathway (through catechol 1,2-dioxygenase) (Fig. 3) was obvious, while biphenyl-grown cells showed no differential expression of any of these genes, although the more sensitive Q-RT-PCR analysis indicated that this pathway was also slightly activated under biphenyl growth conditions. In contrast, we observed significant up-regulation (P < 0.05) of one copy of the two aerobic pathways for benzoate degradation by CoA activation in biphenyl-grown cells and not in benzoate-grown cells (Fig. 3). It is noteworthy that no evidence of a catechol 2,3-dioxygenase was found in the 50 contigs of the LB400 genome.
Growth phase-dependent metabolism.
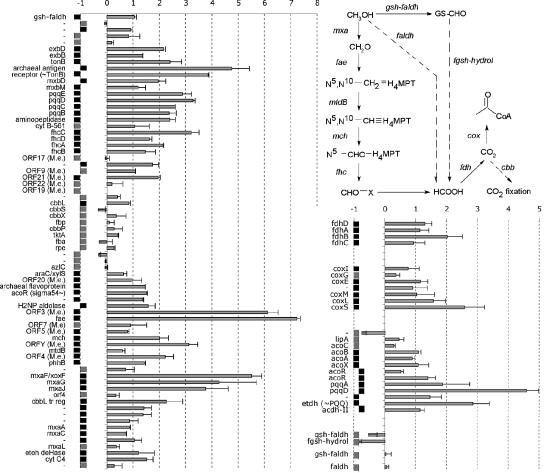
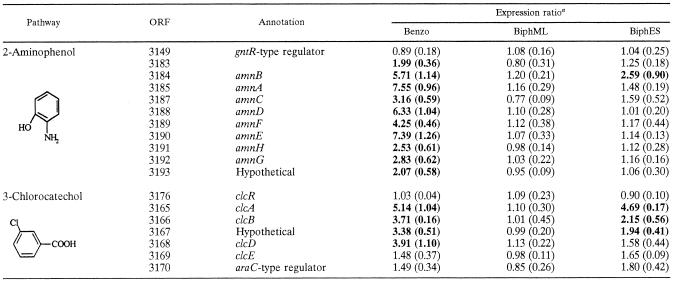
The most striking observation for the expression patterns in the growth phase is the up-regulation of genes coding for a homolog of methanol dehydrogenase (xoxF), biosynthesis of its (and other quinoproteins') cofactor pyrroloquinoline quinone (PQQ), and a tetrahydromethanopterin-dependent oxidative pathway for conversion of its product, formaldehyde, to CO2 and further conversion to acetyl-CoA by carbon monoxide dehydrogenase (Fig. 4). Additionally, up-regulation of the chlorocatechol pathway (Table 2) was observed, which was not detected during exponential growth. When cells were grown on benzoate, however, the chlorocatechol pathway (as well as the 2-aminophenol pathway) was significantly (P < 0.05) up-regulated relative to the expression during growth with succinate (Table 2).
FIG. 4.
C1 metabolic pathways induced in early-stationary-phase biphenyl-grown cells and their organization on the chromosome, represented by squares left and right of the center based on their orientation. Squares representing genes that are significantly up-regulated in early-stationary-phase biphenyl-grown cells are black. The plot shows M values and standard errors for biological replicates. The solid lines in the pathway indicate up-regulated steps. The gene names (annotations were manually verified by BLAST) are those found in GenBank, except for the following names: gsh-faldh (glutathione-dependent formaldehyde dehydrogenase), fgsh-hydrol (formylglutathione hydrolase), M.e. (Methylobacterium extorquens), tr reg (transcriptional regulator), deHase (dehydrogenase), etoh deHase and etdh (ethanol dehydrogenase), and acdh II (type II acetaldehyde dehydrogenase). H4MPT, tetrahydromethanopterin.
TABLE 2.
Expression ratios for 2-aminophenol and 3-chlorocatechol pathways (annotation manually verified by BLAST) as measured for benzoate-grown, mid-log-phase biphenyl-grown, and early-stationary-phase biphenyl-grown cells relative to succinate-grown cells
The values in parentheses are standard errors for biological replicates. Boldface type indicates values for genes that are significantly differentially expressed (P < 0.05) for each condition. Benzo, benzoate-grown cells, BiphML, mid-log-phase biphenyl-grown cells; BiphES, early-stationary-phase biphenyl-grown cells.
Transport and regulation.
Based on the current annotation, 1,399 (13.9%) of the ORFs are involved in a transport function, while 956 ORFs (9.5%) are involved in a regulatory function. The most differential expression in these classes of genes was observed after the transition to the stationary phase (for regulation, 24 genes were up-regulated and 28 genes were down-regulated; for transport, 42 genes were up-regulated and 55 genes were down-regulated). In Table 3 we list the genes possibly involved in benzoate or biphenyl metabolism, based on their expression patterns. Differences between the COG distributions of up- and down-regulated genes indicated the importance of transcriptional regulation (subgroup of COG K), carbohydrate transport (COG G), and to some extent amino acid transport (especially branched) (COG E) for biphenyl-grown cells (Table 1). Interestingly, in the early-stationary-phase biphenyl-grown sample, σ54, a σ54 modulation protein, and 2 of 13 σ54-dependent transcriptional regulators were up-regulated (Table 4).
TABLE 3.
Summary of differential expression, expressed as ratios relative to succinate expression, for transport and regulation specific to biphenyl or benzoate (annotation manually verified by using BLAST)
| ORF | Annotation | Function | Regulation of expression | Expression ratioa
|
||
|---|---|---|---|---|---|---|
| Benzo | BiphML | BiphES | ||||
| 1980 | Extracellular ligand-binding receptor (livK) | Transport | Down | 0.56 (0.25) | 0.38 (0.23) | 0.20 (0.04) |
| 2329 | Porin, gram-negative type (ompC) | Transport | Down | 0.62 (0.22) | 0.06 (0.13) | 0.05 (0.13) |
| 10345 | Probable amino acid-binding periplasmic (pbp) ABC transporter protein (livK) | Transport | Down | 1.11 (0.45) | 0.29 (0.25) | 0.37 (0.12) |
| 13513 | MFS transporter (partial) | Transport | Down | 0.78 (0.22) | 0.15 (0.12) | 0.09 (0.04) |
| 13514 | MFS transporter (partial) | Transport | Down | 0.78 (0.22) | 0.15 (0.12) | 0.09 (0.04) |
| 12907 | Bacterial regulatory protein (lysR) | Regulator | Down | 0.81 (0.32) | 0.47 (0.39) | 0.41 (0.24) |
| 7315 | Carbohydrate-selective porin (oprB) | Transport | Down | 1.10 (0.38) | 0.34 (0.22) | 0.87 (0.36) |
| 9981 | Membrane protein involved in aromatic hydrocarbon degradation (todX) | Transport | Down | 0.84 (0.20) | 0.43 (0.34) | 0.86 (0.29) |
| 12874 | Bacterial regulatory protein (gntR/fadR) | Regulator | Down | 0.80 (0.43) | 0.35 (0.24) | 0.83 (0.47) |
| 7135 | Porin, gram-negative type (ompC) | Transport | Down | 0.50 (0.21) | 1.01 (0.07) | 1.38 (0.62) |
| 13094 | ABC transporter, iron transport (fepC) | Transport | Down | 0.49 (0.09) | 0.71 (0.32) | 0.52 (0.11) |
| 1226 | Putative porin signal peptide protein (ompC) | Transport | Up | 0.84 (0.45) | 2.85 (0.81) | 5.83 (1.42) |
| 1937 | Putative substrate-binding periplasmic (pbp) ABC transporter protein (livK) | Transport | Up | 1.10 (0.13) | 3.59 (0.57) | 4.92 (1.22) |
| 7737 | Conserved hypothetical protein (orf1, downstream of bphE) | Transport | Up | 1.69 (0.57) | 5.89 (1.26) | 15.82 (3.03) |
| 10382 | Extracellular ligand-binding receptor (livK) | Transport | Up | 1.00 (0.10) | 3.11 (0.46) | 3.29 (1.25) |
| 13684 | Possible porin signal peptide protein (ompC, partial) | Transport | Up | 1.14 (0.38) | 2.53 (0.83) | 4.66 (1.23) |
| 10103 | Bacterial regulatory protein (lysR) | Regulator | Up | 0.97 (0.06) | 2.38 (0.42) | 12.02 (1.91) |
| 668 | Lipoprotein, involved in inorganic ion transport (nlpA) | Transport | Up | 1.04 (0.55) | 2.76 (0.66) | 1.93 (0.63) |
| 14292 | ABC transporter (uup) | Transport | Up | 0.83 (0.34) | 2.59 (0.30) | 1.25 (0.85) |
| 5285 | RNA polymerase σ24 (rpoE) | Regulator | Up | 1.03 (0.06) | 2.17 (0.52) | 1.28 (0.63) |
| 6767 | Bacterial regulatory protein (lysR) | Regulator | Up | 1.46 (0.14) | 2.12 (0.24) | 1.25 (0.06) |
| 12574 | Bacterial regulatory proteins, IclR family (pcaR) | Regulator | Up | 0.94 (0.27) | 2.03 (0.21) | 1.13 (0.33) |
| 4744 | Putative porin signal peptide protein (ompC, partial) | Transport | Up | 2.94 (0.61) | 0.53 (0.33) | 1.10 (0.48) |
| 4746 | Putative porin signal peptide protein (ompC, partial) | Transport | Up | 2.00 (0.53) | 0.70 (0.50) | 1.19 (0.63) |
| 12426 | Bacterial regulatory protein (arsR) | Regulator | Up | 2.15 (0.20) | 1.11 (0.06) | 1.07 (0.18) |
Values in parentheses are standard errors for biological replicates. Boldface type indicates values for genes that are significantly differentially expressed (P < 0.05). Benzo, benzoate-grown cells; BiphML, mid-log-phase biphenyl-grown cells; BiphES, early-stationary-phase biphenyl-grown cells.
TABLE 4.
σ54 transcription factor and its dependent transcriptional regulators, based on a GAFTGA amino acid motif searcha
| ORF | Annotationc or probable system controlled by σ54-dependent regulator | Expression ratiob
|
||
|---|---|---|---|---|
| Benzo | BiphML | BiphES | ||
| 15698 | Probable RNA polymerase σN (σ54) | 0.78 (0.00) | 1.26 (0.42) | 3.22 (0.40) |
| 15699 | Putative σ54 modulation protein | 0.83 (0.00) | 0.96 (0.29) | 2.01 (0.90) |
| 562 | nif operon | 0.88 (0.00) | 1.01 (0.09) | 1.27 (0.37) |
| 5028 | Unknown | 0.92 (0.11) | 1.09 (0.10) | 1.79 (0.72) |
| 5125 | Aldehyde dehydrogenase | 0.92 (0.20) | 1.11 (0.36) | 1.31 (0.38) |
| 6037 | Unknown | |||
| 8912 | Short-chain dehydrogenase | 0.80 (0.24) | 0.86 (0.29) | 1.43 (0.24) |
| 8913 | Methanol dehydrogenase, heavy chain (mxaF) | 0.80 (0.28) | 0.85 (0.38) | 2.62 (0.33) |
| 9390 | Unknown | 1.06 (0.00) | 1.09 (0.20) | 0.93 (0.07) |
| 12676 | Unknown | 1.03 (0.25) | 1.81 (0.52) | 1.10 (0.22) |
| 12776 | Methanol dehydrogenase, heavy chain (mxaF) | 0.91 (0.22) | 1.19 (0.12) | 2.85 (0.20) |
| 14211 | C4-dicarboxylate transport transmembrane protein | 0.96 (0.25) | 1.00 (0.12) | 0.89 (0.30) |
| 3525 | Glutamine synthetase (nitrogen assimilation) | 1.57 (0.35) | 1.10 (0.25) | 1.11 (0.29) |
| 4602 | Unknown | 0.87 (0.25) | 0.81 (0.07) | 1.00 (0.50) |
| 4655 | Unknown | 1.11 (0.08) | 0.83 (0.16) | 1.06 (0.08) |
See reference 35.
Values in parentheses are standard errors for biological replicates. Boldface type indicates values for genes that are significantly differentially expressed (P < 0.05). Benzo, benzoate-grown cells; BiphML, mid-log-phase biphenyl-grown cells; BiphES, early-stationary-phase biphenyl-grown cells.
Only ORFs 15698 and 15699 have annotations.
DISCUSSION
Our microarray analyses allowed us to place biphenyl degradation in a genomic context so that degradation of biphenyl and its metabolite benzoate, as well as associated cell physiology adjustments, can be more comprehensively understood. Also, this more complete understanding is a useful stepping stone for further studies on genome-wide effects of PCBs and PCB (co)metabolism in LB400. The most obvious growth substrate- and growth phase-dependent resource reallocations were found for ribosomal biogenesis, the down-regulation of which (number of genes and extent of down-regulation) was positively correlated with growth rate reduction, energy production and conversion, transport, and transcriptional regulation. This and especially the large proportion of differentially expressed genes with poorly characterized functions (∼30%) suggest that there is more to efficiently degrading biphenyl than expressing the biphenyl pathway genes.
By linking our data to previous reports, we can propose a refinement of the regulatory network of the biphenyl pathway in LB400. The observed equal expression of ORF0 on benzoate and on succinate and its up-regulation on biphenyl confirmed Erickson and Mondello's (12) report, based on S1 nuclease mapping, of a biphenyl-inducible promoter upstream of ORF0. These authors also suggested that there are two constitutive σ70-dependent promoters upstream of bphA1. However, mutational studies and Northern blot analyses performed by Beltrametti et al. (2), who proposed that ORF0 is a positive regulator of bphA1A2, and our observation of increased expression ratios of the bph genes with increased ORF0 expression indicate that there is a positive regulatory function of ORF0 induced by biphenyl. The similarity in expression patterns among ORF0, bphJ, and bphI and possibly bphD suggests that ORF0 is an inducer of bphJ, bphI, and bphD. This is similar in part to what has been suggested for Pseudomonas pseudoalcaligenes KF707, whose bph genes exhibit 98 to 100% sequence similarity to LB400 bph genes (BLAST), except for ORF0 (86%), and are organized just like the LB400 bph genes (41). In KF707, bphR1 (formerly ORF0) was identified as a transcriptional regulator for itself and the lower biphenyl pathway genes, as well as bphD, while bphR2, which is not located near the bph cluster, is a necessary LysR-type regulator controlling all bph genes (40). Although the protein sequences are not significantly similar (15%), a LysR-type transcriptional regulator of LB400 (ORF 10103), located elsewhere in the genome, was up-regulated under both biphenyl growth conditions, possibly acting in the role of bphR2.
Based on sequence information, LB400 has two candidate benzoate degradation pathways, one of which, an aerobic degradation pathway with CoA activation (15), is present in two copies, which exhibit 70 to 86% nucleotide sequence similarity (ClustalW). The different pathway choices for benzoate degradation, depending on the carbon source added, and specifically the induction of at least one gene cluster encoding the CoA pathway during growth on biphenyl indicate the presence of a carbon source-specific regulatory network. Apart from pcaR, a positive regulator of the β-ketoadipate pathway, the microarray failed to detect the Q-RT-PCR-measured up-regulation of the catechol-β-ketoadipate pathway in biphenyl-grown cells compared to succinate-grown cells due to the pathway's background-level expression. Based on the Q-RT-PCR data, which indicated that there was ∼1,000-fold-lower expression of this pathway in biphenyl-grown cells than in benzoate-grown cells, and the knowledge that one of the pathway's intermediates, cis,cis-muconate, positively feeds back, inducing catA expression (19), we hypothesize that catechol-β-ketoadipate pathway expression is positively correlated with the benzoate concentration in the medium, which can be assumed to be significantly lower when cells are growing on biphenyl than when cells are growing on the same concentration of benzoate. Instead of being induced by benzoate, CoA pathway regulation could be dependent on biphenyl, one of the metabolic intermediates, or a lower oxygen concentration due to upper Bph pathway dioxygenase (BphA/BphC) oxygen consumption. Different promoter regions and transcriptional regulators, which are neighbors of the two CoA pathway copies, offer a potential explanation for the differential expression of the two copies. The substrate specificities of the enzymes, especially for chlorinated benzoates, remain to be determined. Also, the LysR-type regulator (ORF 10103) up-regulated under both biphenyl growth conditions and an ArsR-type regulator (ORF 12464) that is specifically up-regulated in benzoate-grown cells are targets for further research on regulation of these pathways.
As expected due to the growth phase transition, the largest amount of differentially expressed genes was observed in the early-stationary-phase sample (36, 37, 39). Remarkably, a large number of the up-regulated genes were involved in C1 metabolism, specifically the oxidation of methanol (or possibly a methoxylated carbon source) to CO2 (38) and possibly further acetogenic assimilation to acetyl-CoA (10). Burkholderia species have been shown to assimilate C1 compounds (26, 28), and considering the ubiquitous presence of C1 compounds from microbial degradation of plant aromatic compounds present in the rhizosphere, one of the typical niches of Burkholderia species (7), the presence of these pathways in LB400 could provide an ecological advantage. As for regulation, the coexpression of σ54, which is involved in regulation of a wide range of molecular processes (6), as well as two σ54-dependent transcriptional regulators, which are located next to PQQ biosynthesis genes and tetrahydromethanopterin-dependent formaldehyde oxidation pathway genes, suggests that this metabolic shift to C1 metabolism is σ54 dependent. The two transcriptional regulators were previously linked to mxaF (35), the homolog of which (xoxF) has several potential σ54-dependent promoters in LB400 (data not shown). A last element in the regulation of this metabolic switch is the up-regulated two-component mxbM-mxbD regulatory system, which has been reported to regulate both PQQ biosynthesis and methanol dehydrogenase (16).
As described above, the most significant change in metabolism-related expression was the activation of a methylotrophic metabolic pathway. No involvement of the methylotrophic enzymes with biphenyl or PCB degradation has been reported thus far. The activation of C1 metabolic pathways, as well as the multitude of genes with transport annotation, could be an indication of LB400 switching to a scavenging mode and therefore turning to less energy-efficient, although more available, carbon sources. The wide diversity of up-regulated functions in the cells, the distribution of which is highly similar to the genome's contents, could be interpreted as another indication of this switch to scavenging. It is interesting that the C1 pathway coexpression with the core PCB degradation pathway fits well with the hypothesis that PCB degradation pathways evolved from (often methoxylated) plant aromatic degradation pathways (34).
In concert with this work we documented the performance of a new type of microarray especially suited to microbiological applications. Based on M-A plots, which have been shown to be useful for detecting artifacts and the signal dependency of ratios and for general normalization quality evaluation (11, 43), GeneSpring's Lowess normalization of data proved to be sufficient when the new microarrays were used. The average chip-to-chip CV compared favorably to the values reported for cDNA microarrays (10 to 25%) (44), as well as 70-mer oligonucleotide microarrays which we tested ourselves. A similar correlation was observed between significantly differentially expressed genes and ink jet technology arrays (r = 0.98) (23). High reproducibility and chip hybridization homogeneity due to forced flow of the hybridization mixture resulted in more reliable data, with less replication. The most prominent advantage of these chips lies in their design flexibility, which allows easy, fast, and low-cost alterations to the chip design simply by providing a new spreadsheet of probe sequences and locations to the in situ chip synthesizer (14). This should be useful when genome closure is attained and annotation updates are made, and it also offers opportunities for microarrays that target more complex biological problems, where unpredictability of bacterial diversity requires iterative probe design capability.
The qualitative correspondence of most of the evaluated microarray results with Q-RT-PCR results indicated the reliability of XeoChips for identifying differentially expressed genes. Sixty percent of the noncorrespondence could be explained by low signal detection at the probes, due to low transcript levels and the relative insensitivity of the microarray assay. Importantly, only one false positive (type I error) in 75 comparisons was observed, while the remainder of noncorresponding results were due to false negatives compared to the more sensitive Q-RT-PCR. These results make sense in the context of the Q-RT-PCR-versus-microarray bias (i.e., underestimation of ratios by the microarray or overestimation by the Q-RT-PCR). This observation is consistent with a previous report (45) which showed that there is a bias for cDNA microarray and Q-RT-PCR data that follows a consistent trend.
This study confirmed that the new and flexible in situ-synthesized XeoChip is a reliable DNA microarray platform for bacterial genomics research. Discoveries made by using this approach, such as LB400's benzoate metabolism and early-stationary-phase C1 metabolism, when linked to physiological data, could result in new hypotheses regarding the underlying molecular mechanisms for improved biodegradation of recalcitrant pollutants.
Acknowledgments
This work was supported by Superfund Basic Research Program grant P42 ES 04911-12 from the U.S. National Institute of Environmental Health Sciences and by the Microbial Genome Program of the U.S. Department of Energy. V. J. Denef is an aspirant of the Fund for Scientific Research, Flanders, Belgium (FWO-Vlaanderen).
We acknowledge Lindsay Eltis for suggestions on regulation of benzoate metabolism, Christopher Marx for discussions on C1 metabolism, and Benli Chai for bioinformatic support.
REFERENCES
- 1.Bedard, D. L., R. Unterman, L. H. Bopp, M. J. Brennan, M. L. Haberl, and C. Johnson. 1986. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl. Environ. Microbiol. 4:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltrametti, F., D. Reniero, S. Backhaus, and B. Hofer. 2001. Analysis of transcription of the bph locus of Burkholderia sp. strain LB400 and evidence that the ORF0 gene product acts as a regulator of the bphA1 promotor. Microbiology 147:2169-2182. [DOI] [PubMed] [Google Scholar]
- 3.Bopp, L. H. 1986. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J. Ind. Microbiol. 1:23-29. [Google Scholar]
- 4.Boyle, A. W., C. J. Silvin, J. P. Hassett, J. P. Nakas, and S. W. Tanenbaum. 1992. Bacterial PCB degradation. Biodegradation 3:285-298. [Google Scholar]
- 5.Brazil, G. M., L. Kenefick, M. Callanan, A. Haro, V. De Lorenzo, D. N. Dowling, and F. O'Gara. 1995. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl. Environ. Microbiol. 61:1946-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck, M., M. T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 (σN) transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:-729. [DOI] [PubMed] [Google Scholar]
- 8.Dai, S., F. H. Vaillancourt, H. Maaroufi, N. M. Drouin, D. B. Neau, V. Snieckus, J. T. Bolin, and L. D. Eltis. 2002. Identification and analysis of a bottleneck in PCB biodegradation. Nat. Struct. Biol. 9:934-939. [DOI] [PubMed] [Google Scholar]
- 9.Denef, V. J., J. Park, J. L. M. Rodrigues, T. V. Tsoi, S. A. Hashsham, and J. M. Tiedje. 2003. Validation of a more sensitive method for using spotted oligonucleotide DNA microarrays for functional genomics studies on bacterial communities. Environ. Microbiol. 5:933-943. [DOI] [PubMed] [Google Scholar]
- 10.Diekert, G., and G. Wohlfarth. 1994. Metabolism of homocetogens. Antonie Leeuwenhoek 66:209-221. [DOI] [PubMed] [Google Scholar]
- 11.Dudoit, S., Y. H. Yang, M. J. Callow, and T. P. Speed. 2002. Statistical methods for identifying genes with differential expression in replicated cDNA microarray experiments. Stat. Sin. 12:111-139. [Google Scholar]
- 12.Erickson, B. D., and F. J. Mondello. 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol. 174:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang, Y., A. Brass, D. C. Hoyle, A. Hayes, A. Bashein, S. G. Oliver, D. Waddington, and M. Rattray. 2003. A model-based analysis of microarray experimental error and normalisation. Nucleic Acids Res. 31:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, X., E. LeProust, H. Zhang, O. Srivannavit, E. Gulari, P. Yu, C. Mishiguchi, Q. Xiang, and X. Zhou. 2001. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 29:4744-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gescher, J., A. Zaar, M. Mohamed, H. Schägger, and G. Fuchs. 2002. Genes coding for a new pathway of aerobic benzoate metabolism in Azoarcus evansii. J. Bacteriol. 184:6301-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodwin, P. M., and C. Anthony. 1998. The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv. Microb. Physiol. 40:1-80. [DOI] [PubMed] [Google Scholar]
- 17.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. Classification of the PCB- and biphenyl degrading strain LB400 and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 18.Haddock, J. D., L. M. Nadim, and D. T. Gibson. 1993. Oxidation of biphenyl by a multicomponent enzyme system from Pseudomonas sp. strain LB400. J. Bacteriol. 175:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez, B. S., J. J. Arensdorf, and D. D. Focht. 1995. Catabolic characteristics of biphenyl-utilizing isolates which cometabolize PCBs. Biodegradation 6:75-82. [Google Scholar]
- 21.Hofer, B., S. Backhaus, and K. N. Timmis. 1994. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene 144:9-16. [DOI] [PubMed] [Google Scholar]
- 22.Hofer, B., L. D. Eltis, D. N. Dowling, and K. N. Timmis. 1993. Genetic analysis of a Pseudomonas locus encoding a pathway for biphenyl/polychlorinated biphenyl degradation. Gene 130:47-55. [DOI] [PubMed] [Google Scholar]
- 23.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 24.Kikuchi, Y., Y. Yasukoshi, Y. Nagata, M. Fukuda, and M. Takagi. 1994. Nucleotide sequence and functional analysis of the meta-cleavage pathway involved in biphenyl and polychlorinated biphenyl degradation in Pseudomonas sp. strain KKS102. J. Bacteriol. 176:4269-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Master, E. R., and W. W. Mohn. 2001. Induction of bphA, encoding biphenyl dioxygenase, in two polychlorinated biphenyl-degrading bacteria, psychrotolerant Pseudomonas strain Cam-1 and mesophilic Burkholderia strain LB400. Appl. Environ. Microbiol. 67:2669-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsui, R., Y. Kusano, H. Yurimoto, Y. Sakai, N. Kato, and M. Tanaka. 2003. Formaldehyde fixation contributes to detoxification for growth of a nonmethylotroph, Burkholderia cepacia TM1, on vanillic acid. Appl. Environ. Microbiol. 69:6128-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondello, F. J. 1989. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J. Bacteriol. 171:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radajewski, S., G. Webster, D. S. Reay, S. A. Morris, P. Ineson, D. B. Nedwell, J. I. Prosser, and J. C. Murrell. 2002. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331-2342. [DOI] [PubMed] [Google Scholar]
- 29.Robertson, L. W., and L. G. Hansen. 2001. PCBs: recent advances in environmental toxicology and health effects. The University Press of Kentucky, Lexington.
- 30.Rouillard, J. M., C. J. Herbert, and M. Zuker. 2002. OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics 18:486-487. [DOI] [PubMed] [Google Scholar]
- 31.Seah, S. Y. K., G. Labbé, S. Nerdinger, M. R. Johnson, V. Snieckus, and L. D. Eltis. 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 275:15701-15708. [DOI] [PubMed] [Google Scholar]
- 32.Seeger, M., K. N. Timmis, and B. Hofer. 1995. Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorobiphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl. Environ. Microbiol. 61:2654-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeger, M., M. Zielinski, K. N. Timmis, and B. Hofer. 1999. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 65:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer, A. C., D. E. Crowley, and I. P. Thompson. 2003. Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol. 21:123-130. [DOI] [PubMed] [Google Scholar]
- 35.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vorholt, J. A. 2002. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch. Microbiol. 178:239-249. [DOI] [PubMed] [Google Scholar]
- 39.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe, T., H. Fujihara, and K. Furukawa. 2003. Characterization of the second LysR-type regulator in the biphenyl-catabolic gene cluster of Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 185:3575-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe, T., I. Ryuichi, N. Kimura, and K. Furukawa. 2000. Versatile transcription of biphenyl catabolic bph operon in Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 275:31016-31023. [DOI] [PubMed] [Google Scholar]
- 42.Williams, W. A., J. H. Lobos, and W. E. Cheetham. 1997. A phylogenetic analysis of aerobic polychlorinated biphenyl-degrading bacteria. Int. J. Syst. Bacteriol. 47:207-210. [DOI] [PubMed] [Google Scholar]
- 43.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yue, H., P. S. Eastman, B. B. Wang, J. Minor, M. H. Doctolero, R. L. Nuttall, R. Stack, J. W. Becker, J. R. Montgomery, M. Vainer, and R. Johnston. 2001. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 29:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaitsev, G. M., and Y. N. Karasevich. 1985. Primary steps in metabolism of 4-chlorobenzoate in Arthrobacter globiformis. Mikrobiologiya 50:423-428. [Google Scholar]