Abstract
Vectors mediating strong, durable, and tissue-specific transgene expression are mandatory for safe and effective gene therapy. In settings requiring systemic vector administration, the availability of suited vectors is extremely limited. Here, we present a strategy to select vectors with true specificity for a target tissue from random peptide libraries displayed on adeno-associated virus (AAV) by screening the library under circulation conditions in a murine model. Guiding the in vivo screening by next-generation sequencing, we were able to monitor the selection kinetics and to determine the right time point to discontinue the screening process. The establishment of different rating scores enabled us to identify the most specifically enriched AAV capsid candidates. As proof of concept, a capsid variant was selected that specifically and very efficiently delivers genes to the endothelium of the pulmonary vasculature after intravenous administration. This technical approach of selecting target-specific vectors in vivo is applicable to any given tissue of interest and therefore has broad implications in translational research and medicine.
Introduction
The lack of vectors mediating strong and durable gene expression specifically in a single tissue of interest after simple intravenous injection is a major obstacle on the way to success in gene therapy.
The vascular endothelium expresses tissue-specific receptors, which enables the specific homing of circulating ligands under normal and pathologic conditions. This vascular heterogeneity has been characterized extensively and ligands that home to defined tissues after intravenous injection have been isolated by screening phage display peptide libraries in vivo.1,2,3,4,5,6 Exploiting such tissue-specific ligand-receptor pairs for the targeting of drugs or genes3,4,5,7,8 holds major promise. Nevertheless, it has been difficult to use such ligands for tissue-specific gene transfer by viral vectors suitable to be used in clinical settings.
Adeno-associated virus (AAV)-based vectors are considered safe and effective, they have been used in many clinical trials9,10,11 and one of them has been even approved as first commercial gene therapy drug in the European Union.12The various AAV serotypes vastly differ in their tropism13,14,15,16,17 but none is specific towards only one single tissue after systemic administration,17 the most favorable way of vector administration for the majority of gene therapy applications. Mixing capsid subunits of different AAV serotypes can redirect and improve transduction of target tissues to some extent.18,19,20,21,22,23,24,25,26,27,28 Due to the limited diversity of pre-existing capsid proteins, however, real vector specificity has rarely been achieved by this approach. Many of the targeting peptides identified by phage display or other ligands have been inserted into the structural context of the AAV capsid.5,6,29,30,31,32,33,34 While this can improve transduction properties in certain settings, such vectors lack true tissue specificity because the properties of incorporated peptides can change with their surrounding structural context. Random AAV display peptide libraries may circumvent this problem as they allow the selection of targeting ligands directly in the structural constraint of the AAV surface.35,36,37 Several vector capsids that mediate improved transduction of their target cells have been selected from such libraries in vitro35,36,37,38,39,40,41,42 and in vivo.39,43,44,45,46 However, none of these peptide library approaches have yet established true vector specificity after systemic administration, except for tropism towards the heart44 the transduction of which may be an indirect effect of manipulations of the AAV2 capsid region mediating primary receptor attachment.47 Overall, the number of approaches that have employed AAV library selection39,44,46 or directed evolution25,26,27,28 upon systemic administration in vivo is limited, which might explain that tissue-selectivity in vivo has rarely been achieved yet.
In this study, we demonstrate that real vector specificity and high transduction efficiency in vivo can indeed be obtained by screening AAV display peptide libraries in a murine model. We guided our selection by next-generation sequencing (NGS) analysis of enriched clones that were recovered during several selection rounds in an in vivo screening protocol in mice. Choosing the therapeutically highly relevant pulmonary vasculature as target tissue, we were able to select an AAV capsid showing unprecedentedly efficient and tissue-specific transduction patterns, based on organ-specific vector homing to the endothelium after intravenous administration. This strategy does not only have practical implications for the field of pulmonary medicine as it may also be applicable to any other tissue of interest for in vivo gene delivery.
Results
Screening the random AAV-display peptide library for lung-targeted capsids in vivo yields a distinct peptide sequence
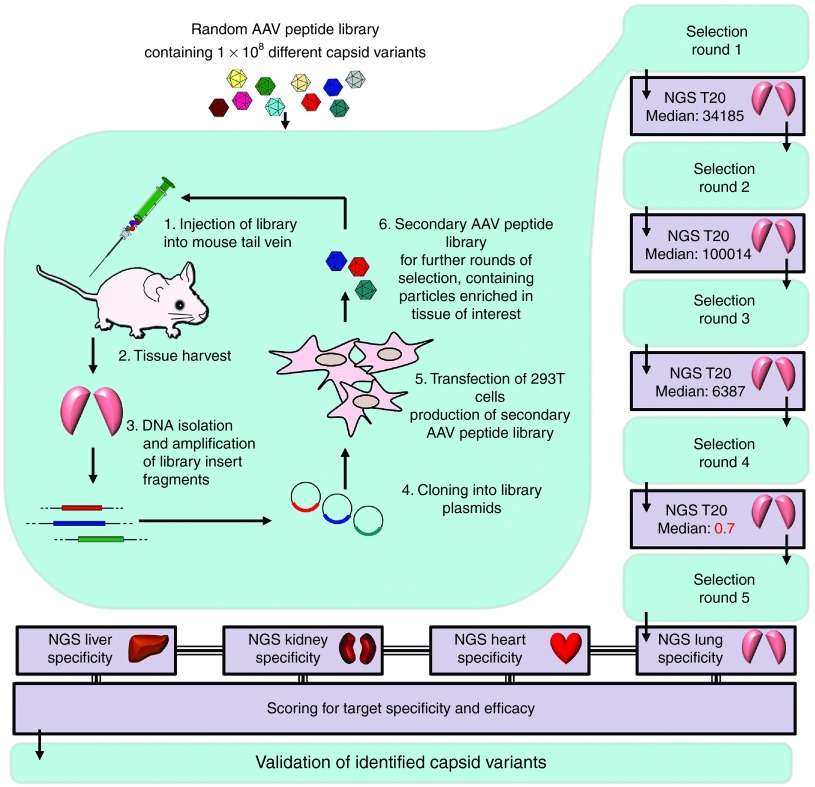
To select specific tissue-targeted AAV2 capsids, an AAV2-displayed random heptamer peptide library was screened in vivo in mice, choosing the lung as the target of interest. Library particles were injected intravenously and given 2 days for homing and target cell infection under normal circulation conditions. The lung was removed and total DNA including the lung-homing library particles' genomes was isolated. The random oligonucleotides contained in the enriched AAV particles were amplified by nested- polymerase chain reaction (PCR) and recloned into library plasmids for generating a secondary AAV peptide library for the next round of selection. In rounds 2–5, time to tissue harvest was extended to 6 days to better eliminate circulating unspecific particles. Recovered library genomes were analyzed by NGS after each round of selection (Figure 1). The complete NGS data have been deposited at the European Nucleotide Archive under Genbank accession no. PRJEB9646.
Figure 1.
Next-generation sequencing (NGS)-guided in vivo selection of a random X7-peptide adeno-associated virus (AAV) display library for identification of efficient and specific capsid targeting peptides. A random AAV display peptide library was injected intravenously in mice. Two (first selection round) or 6 (selection rounds 2–5) days after library administration, total DNA was isolated from the tissue of interest. The viral DNA containing the random library oligonucleotides of particles enriched in the target tissue was amplified by nested polymerase chain reaction (PCR) and cloned into library plasmid backbones to generate a secondary AAV display peptide library by transfection of AAV producer cells. The secondary library was used for a subsequent round of selection. NGS was used to analyze the selected oligonucleotide sequences after PCR-amplification from total DNA, isolated from murine lungs. Median of the changes in relative abundance of the 20 most frequent sequences (T20 Median) between the individual rounds was determined. As this median value sharply declined between rounds four and five, the selection process was stopped, and PCR-amplified virus DNA from the target and off-target organs was analyzed. Specificity and efficacy of dominant capsid variants was scored and AAV2-ESGHGYF was selected for further validation.
In the early phase of the selection, NGS revealed strong variability of the recovered library inserts, indicated by frequent changes of the most dominant peptides during the first four selection rounds. To better understand the kinetics of selection, we analyzed the change in relative frequency of the most dominant peptides during subsequent selection rounds. The median of the change in relative peptide frequency between subsequent selection rounds exceeded factor 6,000 among the 20 most dominant sequences (T20 Median). From round four–five, however, stabilization occurred, resulting in a decline of the T20 Median to 0.7. This was interpreted as an indicator for the beginning of the late selection phase in which well transducing capsid variants had been enriched sufficiently. Thus, the selection process was discontinued at this point and the recovered clones were analyzed regarding potential target transduction efficiency and specificity. To exclude irrelevant, less abundant sequences, only those were analyzed that individually accounted for >0.02% of all reads which combined made up >99% of all reads.
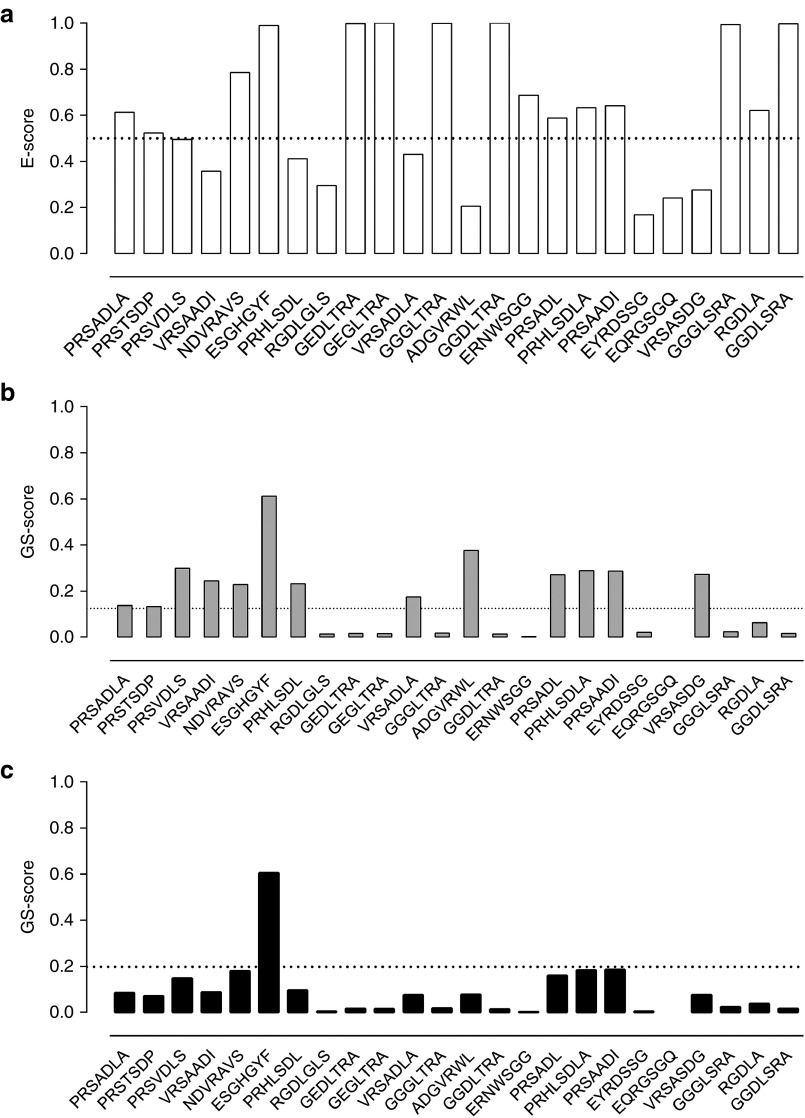
Our previous experience with in vivo screenings of AAV libraries as well as the NGS-based data presented here, indicate that even after five rounds of selection, a still very large diversity of recovered clones can make the identification of the most promising candidates very difficult. Therefore, we established three rating scores to allow such identification from the large total number of recovered candidates, based on relative frequency of a given clone in the target tissue and its specificity, i.e., its distribution in the target compared to control organs (Figure 2). At the time the selection was terminated, the relative frequency of the peptides might not have accurately reflected their transduction efficacies, as some less abundant sequences still increased in relative frequency. To evaluate this, we defined an enrichment score “E”, reflecting changes in relative abundance from before-last (Ry) to last (Rz) (i.e., fourth to fifth) selection round:
Figure 2.
Scoring of dominant capsid variants for specificity and efficacy. After five selection rounds, the targeting peptides accounting for >99% of all reads from the lung in the next-generation sequencing analysis were scored. Peptides appear in order of their relative abundance in the lung from left to right, ranging from PRSADLA (32.74%) to GGDLSRA (0.02%). Three of the displayed peptides deviate in length. (a) The enrichment score E describes for the target tissue the change in relative abundance from selections round four to five within a range from 0 to 1. Peptides with the same relative abundance in both rounds yield a value of 0.5 (dashed line), highly enriched sequences yield a value close to 1. (b) The general specificity score GS is a combination of the organ-specific S scores and describes the target specificity of a peptide. Peptides with the same relative abundance in lung and all three control organs score at 0.125 (dashed line). With increasing specificity, the score increases toward 1. (c) The combined score C is generated by multiplying E and GS scores and describes the peptide performance regarding specificity and efficacy with an ideal value of 1. A peptide that doubles its relative abundance from round four to five and shows only half of the relative abundance observed in the lung in all three off-target organs scores at 0.198 (dashed line). ESGHGYF was the only analyzed peptide exceeding these criteria.
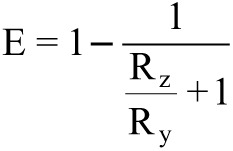 |
To evaluate tissue specificity, we performed deep sequencing of clones from three off-target organs (heart, liver, and kidney) after five selection rounds with the lung as target organ and compared their relative abundances (Ry) with those found within the lung as primary target (Rz). Therefore, we determined three organ-dependent specificity scores Sheart, Sliver, and Skidney using the same term as for the enrichment score E (see above). S values range from 0 to 1, with 0 representing the lowest and 1 the highest specificity. A general specificity score, GS was determined by multiplying the individual scores: GS = Sheart × Sliver × Skidney. This algorithm delineates target tissue specificity as it strongly “punishes” abundance in even a single off-target organ. To determine the most promising peptide regarding specificity and efficacy, a combined score “C”, was determined by multiplying GS and E (Figure 2).
Analysis of the enrichment score E indicated that the sequence motif G-G/E-G/D-L-T/S-R-A was strongly enriched, as was peptide ESGHGYF (Figure 2a). In GS- (Figure 2b) and C-score (Figure 2c), ESGHGYF dominated over all other sequences, suggesting it to be a highly specific and efficient lung targeting peptide. Therefore, the AAV capsids with ESGHGYF insert was chosen for further analyses.
AAV2 vectors displaying the ESGHGYF peptide mediate strong and specific gene expression in the lung
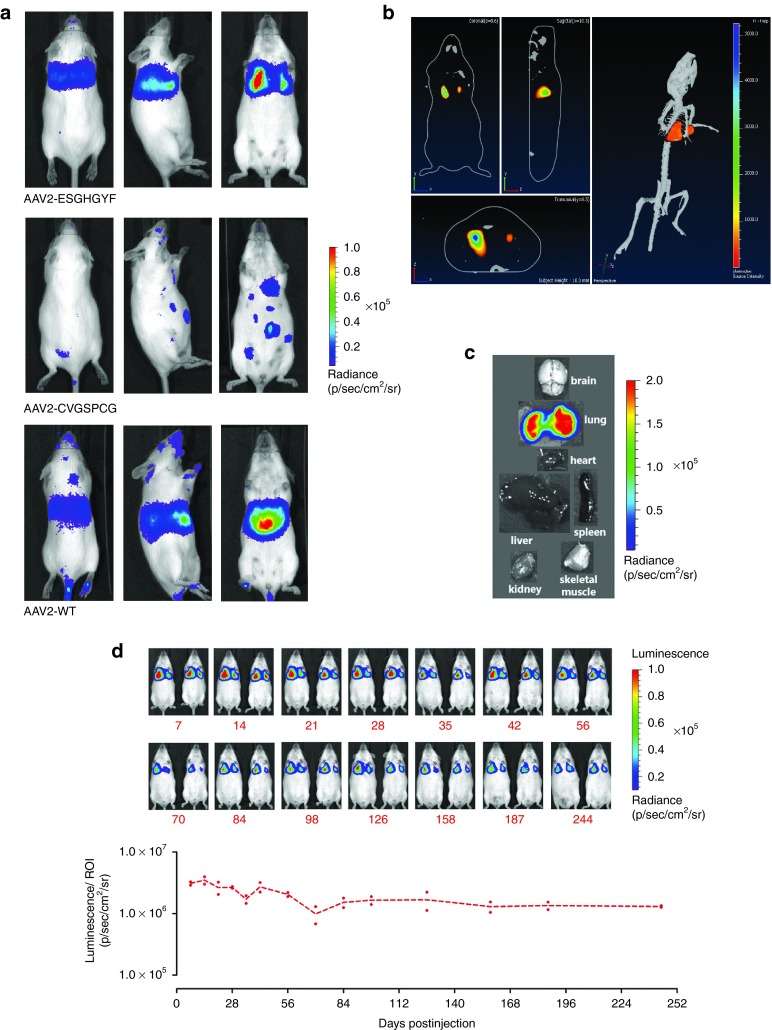
To analyze the in vivo tropism, targeting and control peptides were incorporated into the capsids of AAV vectors carrying a luciferase reporter gene. The peptide insertions appeared not to negatively influence capsid assembly or gene packaging as titers were comparable to unmodified capsid vectors. Vectors were injected intravenously into mice and bioluminescence was imaged in vivo after 14 days (Figure 3a). Indeed, the top-scoring peptide ESGHGYF mediated strong and lung-specific transgene expression. A random control clone from the unselected library (CVGSPCG) mediated weak gene expression mainly in the heart, and wild-type AAV2 mediated gene expression mainly in the liver. Both controls did not confer visible transgene expression in the lung. Almost no variability between repetitive experiments was observed (see Supplementary Figure S1), indicating a high degree of reproducibility. Three-dimensional reconstruction of bioluminescence images including virtual cross sections of a mouse injected with rAAV2-ESGHGYF vector confirmed lung specific expression (Figure 3b). In vivo data were also confirmed by imaging representative tissues ex vivo immediately after organ removal from ESGHGYF-injected animals (Figure 3c), whereas at the same rating scale, the weak luminescence signal mediated by control vectors was barely detectable ex vivo in any of the organs (data not shown).
Figure 3.
Luminescence imaging of mice after intravenous injection of AAV-vectors carrying a luciferase reporter gene. (a) Images were taken 14 days after i.v. administration of 5 × 1010 gp/mouse of vectors displaying the peptide ESGHGYF selected for lung tropism, the random control peptide CVGSPCG, or wild-type AAV2 capsids, respectively. Mice were imaged from the back (left panel), from the side (intermediate panel) and from the front (right panel). Images show representative examples of >3 animals per group. (b) Sagittal, coronal, and transaxial sections (left panel) and three-dimensional reconstruction (right panel) of the luminescence images of a mouse injected with AAV2-ESGHGYF vector (as in a), obtained by measuring different wavelengths of the emitted light, confirming the lung as source of luminescence. (c) Single organ of an ESGHGYF-injected mouse (as in a), measured ex vivo immediately after tissue explantation. (d) ESGHGYF-displaying vector was administered and long-term transgene expression was analyzed by bioluminescence imaging at 14-time points during a 244 days period (n = 2 animals). Upper panel: Imaged mice, rainbow scale as in a. lower panel: Values of luminescence/ region of interest.
The ESGHGYF-mediated, lung-targeted luciferase transgene expression was analyzed over 244 days after injection and quantified as emitted radiance from the lung as the defined region of interest (ROI) (Figure 3d). Throughout the whole period, transgene expression was stable at high levels and continued to be restricted to the lung. It peaked at day 14, and only showed a limited decrease until day 244 (Figure 3d; for original luminescence images with exact delineation of ROIs see Supplementary Figure S2). Of note, the specific transduction of lung tissue, that was observed after intravenous injection of rAAV2-ESGHGYF, was also seen after intraperitoneal vector administration (see Supplementary Figure S3), attesting to the specificity of the selected vector.
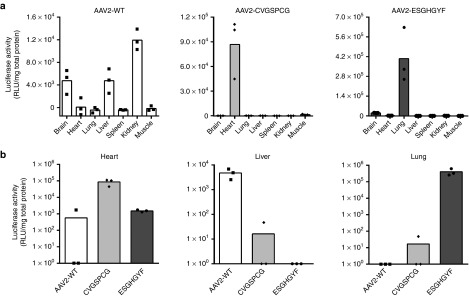
For additional quantitative accuracy, luciferase activity in tissue lysates of representative organs was determined 14 days after vector injection (Figure 4a). Here, wild-type AAV2 mediated weak gene expression in brain, liver and kidney and even less in all other tested organs. The random control peptide CVGSPCG mediated moderate gene expression in the heart (a common observation for capsids modified in the heparan sulfate-binding region47). In contrast, vectors displaying the peptide ESGHGYF mediated strong and specific gene expression in the lung (Figure 4a), confirming the intravital imaging results. In liver and heart—the most frequently transduced organs by AAV2—lung-targeted ESGHGYF vectors only mediated gene expression at background level. In the lung, in contrast, ESGHGYF conferred transgene expression >200× higher than wild-type AAV2 or CVGSPCG control vectors (Figure 4b).
Figure 4.
Determination of luciferase activity from tissue lysates. Luciferase transgene expression analysis in tissue lysates 14 days after vector administration. Data are shown as bars (mean) with plotted individual data points (n = 3 animals per group). (a) Organ distribution of luciferase transgene expression mediated by the three vectors as detailed in Figure 3a. (b) Direct comparison of transgene expression mediated by vectors carrying wild-type AAV2 capsids (AAV2-WT), CVGSPCG-displaying control capsids or ESGHGYF-displaying lung-targeted capsids in three organs in which transgene expression could be detected by bioluminescence imaging (heart, liver, and lung).
The impact of individual amino acids within the ESGHGYF peptide on the viral capsid was analyzed by alanine scanning mutagenesis. No abrogation of lung tropism was observed when replacing the first two amino acid positions of the inserted peptide by alanine, whereas replacements at all other positions either resulted in subtotal transduction deficiency (positions 3–4) or in retargeting toward heart and skeletal muscles (positions 5–7). These tissues were also transduced by capsids harboring a scrambled version (FHEYGSG) of the lung-homing peptide (see Supplementary Figure S4). Transduction of lung tissue by AAV2-ESGHGYF vector could not be blocked by soluble ESGHGYF peptide (see Supplementary Figure S5 and Supplementary Materials and Methods), indicating that the lung tropism is caused by the structural changes of the viral capsid (that are conferred by the ESGHGYF peptide) rather than the isolated peptide itself.
ESGHGYF-mediated targeting is based on lung-specific homing of circulating vectors
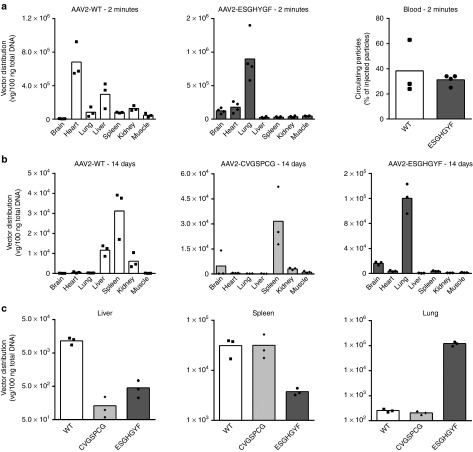
To investigate if the lung-specific transgene expression of intravenously administered ESGHGYF vectors is based on specific homing, we analyzed the vector distribution as early as 2 minutes after injection (Figure 5a). Quantification of vector genomes by real-time PCR revealed lung-specific homing of AAV2-ESGHGYF with vector copy numbers being 5–30 times higher in the lung than in any other organ, whereas wild-type AAV2 was recovered mainly from heart and liver. At this time point a considerable amount (~1/3) of the injected wild-type and ESGHGYF particles were still in the circulation and could be recovered from left ventricular blood (Figure 5a). The relative enrichment of ESGHGYF particles in the lung compared to off-target organs further increased and after 4 hours 6–100 times higher copy numbers were detected in the lung than in the control organs (see Supplementary Figure S6).
Figure 5.
Biodistribution of recombinant adeno-associated virus (AAV) vectors. Amounts of vectors were determined by quantitative real-time PCR of vector genomes. All experiments were performed after i.v. administration of 5 × 1010 gp/mouse. Data are shown as bars (mean) with plotted individual data points (n = 3–4 animals per group). (a) Distribution of wild-type AAV2 (left) and AAV2-ESGHGYF (middle) in seven analyzed organs, 2 minutes after vector administration, reflecting tissue homing of injected particles. Amount of circulating particles recovered from left ventricular blood (right). (b) Distribution of genomes delivered by AAV2-ESGHGYF or control vectors (wild-type AAV2 and AAV2-CVGSPCG), respectively, 14 days after vector administration. (c) Separate depiction of vector genome quantities in the three organs containing the highest amount of vector genomes (liver, spleen, and lung) comparing the three capsid variants used for gene delivery.
To analyze the correlation between vector homing and transgene expression, vector distribution was also measured 14 days after injection, the point at which transgene expression was analyzed in parallel experiments (see above). Genomes delivered by wild-type AAV2 could be recovered mainly from liver, kidney, and spleen. The spleen also contained most genomes of vectors displaying the random control peptide. Overall, the amount of vector genomes in the spleen was approximately equal with all tested capsid variants, suggesting nonspecific particle uptake in the reticular endothelial system, which is independent of transgene delivery and expression. In contrast, distribution of genomes delivered by the lung-targeted ESGHGYF-vectors strongly resembled the transgene expression data with highly specific enrichment in the lung. The amount of ESGHGYF-vector genomes in the lung was up to 250-fold higher than in the other organs and up to 470-fold higher than in lungs injected with wild-type or control capsid vectors (Figure 5b). The same vector genome distribution ratio between the organs was found 28 days after vector administration (data not shown). Direct comparison of the vector genome quantities between the three capsid variants is shown in Figure 5c for the three tissues in which relevant quantities of vector DNA could be detected. Taken together, these data suggest that ESGHGYF-mediated lung-specific transgene expression is brought about by tissue-specific homing of circulating vector particles. Albeit proving a strong influence on the viral tropism, the insertion of the ESGHGYF peptide did not seem to alter either reactivity with preexisting serum antibodies or the antibody response compared to wild-type AAV2. We found similar amounts of neutralizing antibodies against wild-type and the modified capsid in pooled human intravenous immunoglobulin and the sera of individual human subjects (see Supplementary Figure S7 and Supplementary Materials and Methods). Further, both capsid variants (wild-type or ESGHGYF) induced a cross-reactive neutralizing antibody response in mice (see Supplementary Figure S8 and Supplementary Materials and Methods).
AAV2-ESGHGYF delivers genes selectively to pulmonary endothelial cells
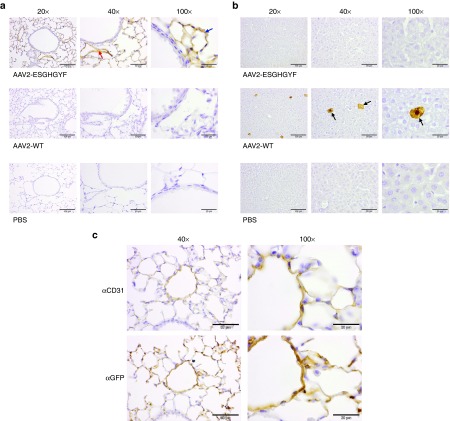
To identify the primary target of AAV2-ESGHGYF, we visualized transgene expression at the cellular level in the lung and a control organ 14 days after intravenous administration of AAV vectors (rAAV2) carrying the eGFP gene. In lungs of mice injected with rAAV2-ESGHGYF, immunohistochemistry revealed intense staining in endothelial cells throughout the entire pulmonary microvasculature and, slightly less intense, in larger pulmonary vessels (Figure 6a). In contrast, pulmonary tissue of mice injected with wild-type capsid rAAV2 showed no staining (Figure 6a). The liver (a tissue known to exhibit strong transgene expression after wild-type rAAV2 vector injection) was analyzed as control. Here, staining of hepatocytes was observed after wild-type rAAV2 but not after rAAV2-ESGHGYF administration (Figure 6b). The endothelial nature of the vector-transduced pulmonary cells was confirmed by CD31 staining, which exactly reproduced the eGFP staining pattern in serial sections of lungs of rAAV2-ESGHGYF injected mice (Figure 6c).
Figure 6.
AAV2-ESGHGYF-mediated specific gene delivery to pulmonary vasculature endothelial cells. Paraffin-embedded tissue sections of mouse lungs and control organs were stained by immunohistochemistry 14 days after i.v. vector administration of 1 × 1011 gp/mouse (AAV2-ESGHGYF or wild-type AAV2 as control). Each panel a and b displays a representative area of the tissue of interest, shown in different magnifications. Size of scale bars: 100 µm at 20× magnification; 50 µm at 40× magnification; and 20 µm at 100× magnification. (a) Representative area of the lung stained with an antibody against the transgene product GFP. Red arrow: Example of endothelial cells of larger pulmonary vessel; Blue arrow: Example of endothelial cells of pulmonary microvasculature. (b) Representative area of the liver stained with an antibody against GFP. Black arrows: examples of transduced hepatocytes (c) serial sections of a representative area of the lung of mice injected with rAAV2-ESGHGYF, stained with antibodies against the endothelial marker CD31 and against the transgene product GFP.
Discussion
Targeting vectors specifically to a tissue of interest after systemic administration has been an urgent but largely unmet need ever since the earliest days of gene therapy. Many targeting strategies have been developed but so far none of them could solve this problem completely.
To address this, we and others have introduced a vector evolution system enabling the selection of targeting ligands within the structural constraint of the viral capsid of AAV.35,36 While theoretically this system promises almost unlimited versatility in the development of specific vectors for any conceivable cell type of interest, so far its major use has been to select targeted vectors in vitro.35,36,37,38,39,40,41 Screening such libraries in vivo, however, mostly yielded vectors either with moderate transduction efficiency in the target tissue or low tissue specificity39,46 after systemic administration. Here, we present an improved system for in vivo selection of AAV display libraries and therefore introduce for the first time a tailored, truly tissue-specific, and very effective vector system.
Two determinants decide on the success of screening random AAV libraries in vivo: (i) the biological selection process itself with parameters such as amount of administered particles, circulation time, way of amplifying enriched clones from one selection round to the next, etc. and (ii) detection of the relevant clones within the large amount of recovered particles because the most abundant clones may not have the strongest transduction efficiencies and not the most specific homing properties to the tissue of interest. Besides optimizing the first point, we have addressed the second point by large-scale analysis of recovered clones and establishing a biomathematical algorithm that allows (i) to identify the suitable time point to discontinue the selection procedure and (ii) to select the capsid variant with the highest potential regarding transduction efficacy and target specificity for further evaluation. As early as in the first round of the selection, our NGS analyses revealed dominance of certain capsid variants. However, such dominance fluctuated from one round to the next, with different clones being prominent in each round. This variability could be due to the initially low copy numbers of each clone in the library. Therefore, some clones may not be evenly distributed despite their unselective tropism. Random recovery of such clones from the target organ and subsequent highly effective amplification can suggest dominance despite nonspecificity. Over several selection rounds, this effect diminishes as truly specific clones become steadily enriched in the target organ while others decrease in relative frequency due to nonspecific homing, resulting in dominant recovery, amplification, and visibility of capsid variants with superior target transduction. Analogous to phage display library screenings,48,49 the selection process was discontinued upon such decrease in variability. Nevertheless, the diversity of recovered clones was large at this point. NGS has repeatedly been used to identify targeting clones from phage and antibody display libraries, setting focus on the reduction of selection rounds more than on specificity.50,51,52 Here, we expanded the scoring analysis to the distribution of peptide variants in three off-target tissues, which was the key to identifying the enriched clone with target transduction efficiency and specificity. During preparation of the NGS probes, the crude tissue DNA was subjected to different PCR amplification steps to enrich the AAV library DNA and to add the necessary sequencing barcodes. Thus, the number of NGS reads from a given probe does not directly reflect the absolute number of library genomes in the respective organ. Our analysis was solely focused on the relative abundance of individual clones in each organ. The validity of our determined scores is confirmed by comparing them with functional data available for some of the recovered peptides. The most frequent peptides were PRSADLA and PRSTSDP, but both possess a low GS score. In congruence with this, we previously observed that both peptides mediate lung transduction but also strongly transduce liver, heart, and kidney.39 Similarly, EYRDSSG and NDVRAVS, both with a low GS score, have been shown to confer high transduction efficiency but only low organ specificity.35,39 The peptide ESGHGYF had by far the highest GS and C scores, and indeed showed uniquely strong transduction efficiency and specificity for the lung as the target tissue of this library selection. Of note, ESGHGYF represented only 6% of all reads in the lung after five rounds of selection and very likely would have been overlooked in a conventional analysis solely focusing on clone dominance.
The specificity of transgene expression in the pulmonary endothelium correlated with vector distribution, which indicates true specificity in gene delivery as opposed to a mere optimization of postentry vector processing. This unprecedented strength and specificity of transgene expression after systemic administration is composed of both, targeting to the lung and detargeting especially from the liver. Frequently, AAV2 capsid changes sterically close to the 588 site not only result in liver de-targeting but also augment gene expression in the heart.39,44,47 This was reconfirmed in our experiments with the control vectors displaying peptides CVGSPCG and FHEYGSG and by alanine scanning. In view of that it is remarkable, that the tropism of the ESGHGYF clone selected by using the technology described here did not show this pattern and was restricted to the lung.
The amino acid sequence of the ESGHGYF peptide does not show any striking similarities with one the different lung-homing peptides that have previously been identified by the phage display technique. The GFE-1 peptide (CGFECVRQCPER) was identified in vivo in murine lungs2 and has later been shown to bind the cell surface protein membrane dipeptidase,53 whereas the CGSPGWVRC peptide was identified in vitro on immortalized murine lung endothelial cells.54 Recently a tissue-penetrating homing peptide (CARSKNKDC) has been shown to confer selective homing to hypertensive pulmonary arteries but not to normal pulmonary vessels55 in a heparin sulfate-dependent manner.56 However, none of these peptides shares a common sequence motif with the ESGHGYF peptide. This is not surprising and can most likely be explained by the fundamental differences of the library systems that have been used to identify these peptides. The phage display technique allows the selection of well accessible peptides that are terminally fused to the phage's coat protein without being forced into major structural constraints. In an AAV display library, on the other hand, the peptides are embedded within a structurally very important area of the viral capsid, in which they may be forced into strong physicochemical constraints. Thus, in an AAV display library it is not necessarily the isolated peptide which is being selected for, more likely it is the modified viral capsid being structurally altered by insertion of this peptide. This also explains why the soluble ESGHGYF peptide did not show any effect in the competition assay with the AAV2-ESGHGYF vector.
Although the receptor targeted by the AAV2-ESGHGYF capsid has not been identified yet, the carbohydrates on the endothelial cell surface presumably are essential for specific targeting, since glycans are used as primary attachment receptors by all AAV serotypes analyzed so far.57 Our data indicate a specific receptor-mediated homing of AAV2-ESGHYGF to pulmonary endothelial cells rather than a global endothelial tropism with enrichment in the lung merely due to a first pass effect: (i) different injection routes (intravenous and intraperitoneal) led to specific enrichment in the lung and detargeting from the liver, although the majority of vector administered into the peritoneal cavity is supposed to enter the circulation via the portal vein, passing the hepatic sinusoids as first large vascular bed58; (ii) 2 minutes after intravenous injection (after ~10–50 circulation rounds59,60) as much as one-third of the injected vector copies could still be detected in the blood collected from the left ventricle, and—albeit being enriched in the lung—considerable particle numbers, likely still being in circulation, could be recovered from well perfused nontarget organs like brain, heart, and kidney. This would not have been the case if the lung-homing was solely explained by a first pass effect.
Although the insertion of the ESGHGYF peptide into the AAV2 capsid does not seem to have an positive influence on the antibody reactivity, the AAV2-ESGHGYF vector might very well be a promising candidate for gene therapy, as there are different potential options to circumvent unwanted immune responses in a clinical setting such as plasmapheresis61 or blocking of antibodies by applying an excess of empty particles.62 Taken together, this is the first report on the systematic selection of a gene therapy vector mediating strong and durable expression specifically in a target tissue other than the heart under circulation conditions in vivo. Thus, choosing the lung as therapeutically highly relevant target, vectors such as the one presented here might be used in gene therapy for disorders like pulmonary hypertension. Our NGS-guided approach of in vivo selection allows unprecedented exploitation of the high potential of random AAV display peptide libraries and could be used to obtain specific vectors for virtually any given target organ.
Materials and Methods
Preparation of the random AAV display peptide library. A random X7 AAV2 display peptide library with a theoretical diversity of 1 × 108 unique clones was produced using a two-step protocol as previously described.35,37 In short, the degenerate oligonucleotide encoding seven random amino acids (encoded by NNK) at nucleotide position 3967 in the AAV genome (amino acid position R588; VP1 numbering) was synthesized (Metabion, Martinsried, Germany) as follows: 5′-CAGTCGGCCAGAGAGGC(NNK)7GCCCAGGCGGCTGACGAG-3′. The second strand was synthesized by using sequenase (Amersham, Freiburg, Germany) and the primer 5′-CTCGTCAGCCGCCTGG-3′. The double-stranded insert was cleaved with BglI, purified with the QIAquick Nucleotide Removal Kit (Qiagen, Hilden, Germany) and ligated into the SfiI-digested pMT187-0–3 library plasmid.35 The diversity of the plasmid library was determined by the number of clones growing from a representative aliquot of transformed electro competent DH5α bacteria on agar containing 150 mg/ml ampicillin. Library plasmids were harvested and purified using Qiagen's Plasmid Preparation Kit. The AAV library genomes were packaged into chimeric wild-type and library AAV capsids (AAV transfer shuttles) by transfecting 2 × 108 293T-cells in ten 14.5-cm cell culture dishes at a 1:1:2 ratio of the plasmid pVP3cm (containing the wild-type cap gene with modified codon usage, without inverted terminal repeats),37 the library plasmids and the pXX6 helper plasmid.63 The resulting AAV library transfer shuttles were used to infect 2 × 108 293T-cells in ten 15-cm cell culture dishes at a multiplicity of infection (MOI) of 0.5 replicative units per cell. Cells were superinfected with Ad5 at an MOI of five plaque-forming units (pfu) per cell. The final random peptide AAV display library was harvested from the supernatant after 48 hours. The supernatant was concentrated using VivaSpin columns (Viva Science, Hannover, Germany), purified by iodixanol density gradient ultracentrifugation as previously described64 and titered by quantitative real-time PCR (qPCR) using the cap-specific primers 5′-GCAGTATGGTTCTGTATCTACCAACC-3′ and 5′-GCCTGGAAGAACGCCTTGTGTG-3′ with the LightCycler system (Roche Diagnostics, Mannheim, Germany).
In vivo screening of the random AAV display peptide library. 1 × 1011 genomic library particles were injected into the tail veins of healthy FVB/N mice (see below). After 48 hours, mice were sacrificed and organs of interest were removed. Total tissue DNA was extracted using the DNeasy tissue kit (Qiagen). The random oligonucleotides contained in AAV library particles which were enriched in the tissue of interest were amplified by nested PCR using the primers 5′-ATGGCAAGCCACAAGGACGATG-3′ and 5′-CGTGGAGTACTGTGTGATGAAG-3′ for the first PCR and 5′-GGTTCTCATCTTTGGGAAGCAAG-3′ as well as 5′-TGATGAGAATCTGTGGAGGAG-3′ for the second PCR. The PCR-amplified oligonucleotides were used to produce secondary libraries for four further rounds of selection (mice were sacrificed 6 days after library injection in round 2–5). Secondary libraries were produced like the primary library (as described earlier) but without the extra step of producing transfer shuttles. The secondary plasmid library was directly used to transfect 2 × 108 293T-cells in 15-cm cell culture dishes at a ratio of 25 library plasmids per cell, using Polyfect transfection reagent (Qiagen).
NGS of AAV peptide libraries. Amplicon libraries for illumina HiSeq2500 NGS were generated from corresponding peptide library DNA in two subsequent PCR reactions. In the first PCR, a forward primer with an Index sequence (ATCGATCG, TCGATCGA, CGATCGAT or GATCGATC; e.g., 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTATCGATCGCTGTATCTACCAACCTCCAGAGAGG-3′) was used in combination with reverse primer 5′-CAAGCAGAAGACGGCATACGAGATCTCTTCCGATCTCTGCCAGACCATGCCTGGAAGAACG-3′ to allow identification of individual samples. In the second PCR, the 5′ end of the amplicon was extended by forward primer 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ while the same reverse primer was used. For single read HiSeq2500 illumina sequencing 100 µl of a 2 nmol/l DNA solution were used (2 × 10−13 mol). Data analysis was performed by a custom script (see Supplementary Note 1).
Vector production and quantification. Recombinant AAV vectors were produced by triple transfection of HEK293T-cells. Cells were grown at 37 °C, 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA), supplemented with 1% penicillin/streptomycin (Invitrogen) and 10% fetal calf serum (Biochrom, Berlin, Germany). Plasmid DNA (see below) was transfected into 293T-cells with Polyfect transfection reagent (Qiagen). Four days after transfection, cells were harvested, lysed, and vectors were purified by iodixanol density-gradient ultracentrifugation as previously described.64 For transfections, we used pXX6 as adenoviral helper plasmid,63 the luciferase gene pUF2-CMV-luc,37 or the GFP gene pTR-CMV-GFP65 and a plasmid encoding the AAV capsid of interest. Plasmids encoding the AAV capsid mutants which had previously been selected from the AAV library and wild-type controls were modified pXX2-187 in ref. 38 and pXX2 in ref. 63, respectively. The oligonucleotide inserts encoding the modified peptides for the alanine scan were synthesized (Metabion) and further processed as the library inserts (described earlier).
For quantification of vector stocks, genomic titers were determined by qPCR using the CMV-specific primers 5′-GGCGGAGTTGTTACGACAT-3′ and 5′-GGGACTTTCCCTACTTGGCA-3′ using the LightCycler system (Roche), as previously described.66
Animals and vector administration. Initial in vivo selection with the random AAV2-peptide library, analysis of AAV luciferase expression vectors, and neutralizing antibody assay were performed in 8–12 weeks old FVB/N mice. The peptide competition experiment and GFP reporter gene experiments for immunohistochemistry were performed in 8–12 weeks old SCID mice. All experiments involving animals were conducted in accordance with the German Animal Protection Code. The protocol was approved by the responsible ethics review board and the local authorities. All experiments were performed under anesthesia with 2% isoflurane and oxygen. AAV vectors were injected into the tail vein or the peritoneal cavity at a total volume of 200 µl.
Assessment of luciferase reporter gene expression in vivo. AAV vectors were administered intravenously or intraperitoneally at a dose of 5 × 1010 vector genomes (vg) per mouse (n = 3 animals per injected AAV clone). At day 14, animals were anesthetized with isoflurane. Luciferase expression was analyzed using the Xenogen IVIS200 imaging system (Caliper Lifesciences, Hopkinton, MA) with the software Living Image 4.0 (Caliper) after intraperitoneal injection of 200 µl luciferin substrate (150 mg/kg, Xenogen) per mouse. Representative in vivo bioluminescence transgene expression images from different positions (ventral, dorsal, and lateral) were taken when luminescence in relative light units (photons/second/cm2) reached the highest intensity. Subsequently, animals were sacrificed, organs of interest were quickly removed and transgene expression images of single organs were taken immediately. Then, organs were snap frozen in liquid nitrogen and stored at −80 °C. Three-dimensional reconstructions of in vivo luminescence images were obtained by using the DLIT option of the software Living Image 4.0 (Caliper) and measuring the emitted light in five different wavelengths from 560 to 640 nm, for 3 minutes each.
To quantify luciferase expression, organs were homogenized in reporter lysis buffer (Promega, Madison, WI) using a Precelly's 24 tissue homogenizer (Peqlab, Erlangen, Germany) according to the manufacturer's instructions. Determination of luciferase reporter gene activity was performed in a luminometer (Mithras LB 940, Berthold Technologies, Bad Wildbad, Germany) over a 10-second interval after addition of 100 µl luciferase assay reagent (Promega) with 2 seconds delay between each measurement. Values were normalized to total protein levels in each sample with the Roti Nanoquant Protein Assay (Roth, Karlsruhe, Germany).
Analysis of vector distribution. For quantification of vector genome copy number in various organs, total tissue DNA from each organ was extracted using a tissue homogenizer (Precelly's 24 tissue homogenizer, Peqlab) and the DNeasy tissue kit (Qiagen) according to the manufacturer's instructions at different time points after administration of 5 × 1010 vg per mouse. DNA was quantified using a spectral photometer Nanodrop ND-2000C (Peqlab, Erlangen, Germany). Analysis of AAV vector DNA in tissues was performed by qPCR using CMV-specific primers as previously described,66 and normalized to 100 ng total DNA. For quantification of vector genome copy numbers in the circulation, blood was collected by left ventricular puncture and incubated for 10 minutes at room temperature. After centrifugation for 2 minutes at 10,000 ×g the serum was directly used as template for qPCR. PCR efficiency was controlled by a spiked-in plasmid. Total copy numbers in the blood were calculated, assuming a blood volume of 100 ml/kg body weight.67
Immunohistochemistry and histology. Lungs were fixed ex situ with 4% (w/v) paraformaldehyde via the trachea under a hydrostatic pressure of a 20 cm water column for 20 minutes, followed by 24 hours immersion in the same fixative. Lung tissues were embedded in paraffin. Two-micrometer sections were dewaxed, rehydrated, and used for immunohistochemistry. Immunohistochemistry was performed with polyclonal antibodies against GFP (A-11122; Invitrogen) or CD31 (ab28364; Abcam, Cambridge, MA). Endogenous peroxidase activity was inactivated with 1% H2O2 in methanol for 30 minutes. Prior to CD31 staining, sections were heated to 100 °C in citrate buffer, pH 6 for 20 minutes for antigen retrieval. After washing in phosphate-buffered saline, the sections were incubated for 30 minutes with phosphate-buffered saline, 10% goat serum (Vector Lab, Burlingame, CA), 2% powdered milk (Roth). Primary antibodies were allowed to bind for 1 hour at 37 °C. After washing in phosphate-buffered saline, the sections were incubated for 30 minutes with a secondary biotinylated goat antirabbit antibody (Vector Lab). Bound antibodies were visualized using the VECTASTAIN Elite ABC Kit (Vector Lab) and 3,3′-diaminobenzidine (Sigma-Aldrich, St. Louis, MO). Selected sections were counterstained with hemalum.
Photomicrographs were recorded by a digital camera (DP-25, Olympus, Tokyo, Japan) attached to a microscope (BX-51, Olympus) at 10- to 100-fold magnification using the software cell^A (Olympus).
SUPPLEMENTARY MATERIAL Figure S1. In vivo imaging of luminescence mediated by recombinant AAV2 vectors. Figure S2. Longterm transgene expression mediated by rAAV2-ESGHGYF. Figure S3. In vivo imaging of luminescence mediated by rAAV2-ESGHGYF after intraperitoneal injection. Figure S4. Functional consequences of alanine scan or sequence scrambling of the peptide ESGHGYF. Figure S5. Competition assay with soluble ESGHGYF peptide. Figure S6. Biodistribution of AAV2-ESGHGYF, four hours after administration. Figure S7. Prevalence of neutralizing antibodies against AAV2-WT and –ESGHGYF. Figure S8. Neutralizing antibody response after injection of mice with AAV2-WT and –ESGHGYF. Note S1. Bioinformatical data processing. Materials and Methods References
Author Contributions
Designed the experiments: J.K., S.M., T.S., K.R., O.J.M., M.T. Performed the experiments: J.K., S.M., T.S., L.L., E.S., A.H., K.R., D.I. Analyzed the data: J.K., S.M., T.S., M.A., K.R., O.J.M., R.P., W.A., M.T. Contributed material/technical equipment: L.L., O.J.M., J.A.K., M.T. Wrote the manuscript: J.K., T.S., K.R., O.J.M., M.T.
Acknowledgments
We thank Jude Samulski, University of North Carolina, Chapel Hill, NC for kindly providing the plasmids pXX2 and pXX6. The luciferase expression of vector-treated animals was monitored with kind permission of and support by Axel Leingärtner at the UCCH Core Facility for Optical in vivo Imaging, University Medical Center Hamburg-Eppendorf, Germany. We are grateful to Mascha Binder, University Medical Center Hamburg-Eppendorf for her support. This work was supported by the German Research Foundation (DFG, grant TR448/7-1 to M.T. and J.A.K.), the Else Kröner-Fresenius-Foundation (grant 2013_A83 to M.T.) and the Margarete Clemens Foundation (endowed professorship to M.T.). The University Medical Center Hamburg-Eppendorf (UKE) filed a patent application for the capsid-modified AAV vector AAV2-ESGHGYF on behalf of J.K., S.M., and M.T. The authors declare that there are no further competing financial interests.
Supplementary Material
References
- Pasqualini, R and Ruoslahti, E (1996). Organ targeting in vivo using phage display peptide libraries. Nature 380: 364–366. [DOI] [PubMed] [Google Scholar]
- Rajotte, D, Arap, W, Hagedorn, M, Koivunen, E, Pasqualini, R and Ruoslahti, E (1998). Molecular heterogeneity of the vascular endothelium revealed by in vivo phage display. J Clin Invest 102: 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arap, W, Haedicke, W, Bernasconi, M, Kain, R, Rajotte, D, Krajewski, S et al. (2002). Targeting the prostate for destruction through a vascular address. Proc Natl Acad Sci USA 99: 1527–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolonin, MG, Saha, PK, Chan, L, Pasqualini, R and Arap, W (2004). Reversal of obesity by targeted ablation of adipose tissue. Nat Med 10: 625–632. [DOI] [PubMed] [Google Scholar]
- Chen, YH, Chang, M and Davidson, BL (2009). Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat Med 15: 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Work, LM, Büning, H, Hunt, E, Nicklin, SA, Denby, L, Britton, N et al. (2006). Vascular bed-targeted in vivo gene delivery using tropism-modified adeno-associated viruses. Mol Ther 13: 683–693. [DOI] [PubMed] [Google Scholar]
- Trepel, M, Arap, W and Pasqualini, R (2001). Modulation of the immune response by systemic targeting of antigens to lymph nodes. Cancer Res 61: 8110–8112. [PubMed] [Google Scholar]
- Hajitou, A, Trepel, M, Lilley, CE, Soghomonyan, S, Alauddin, MM, Marini, FC 3rd et al. (2006). A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell 125: 385–398. [DOI] [PubMed] [Google Scholar]
- Wright, JF (2008). Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther 15: 840–848. [DOI] [PubMed] [Google Scholar]
- Mueller, C and Flotte, TR (2008). Clinical gene therapy using recombinant adeno-associated virus vectors. Gene Ther 15: 858–863. [DOI] [PubMed] [Google Scholar]
- Mingozzi, F and High, KA (2011). Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 12: 341–355. [DOI] [PubMed] [Google Scholar]
- Ylä-Herttuala, S (2012). Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol Ther 20: 1831–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindasamy, L, Padron, E, McKenna, R, Muzyczka, N, Kaludov, N, Chiorini, JA et al. (2006). Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol 80: 11556–11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki, K, Fuess, S, Storm, TA, Gibson, GA, Mctiernan, CF, Kay, MA et al. (2006). Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther 14: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S, Kapturczak, M, Loiler, SA, Zolotukhin, S, Glushakova, OY, Madsen, KM et al. (2005). Efficient transduction of vascular endothelial cells with recombinant adeno-associated virus serotype 1 and 5 vectors. Hum Gene Ther 16: 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taymans, JM, Vandenberghe, LH, Haute, CV, Thiry, I, Deroose, CM, Mortelmans, L et al. (2007). Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum Gene Ther 18: 195–206. [DOI] [PubMed] [Google Scholar]
- Zincarelli, C, Soltys, S, Rengo, G and Rabinowitz, JE (2008). Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 16: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Bowles, DE, Rabinowitz, JE and Samulski, RJ (2003). Marker rescue of adeno-associated virus (AAV) capsid mutants: a novel approach for chimeric AAV production. J Virol 77: 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshri, N, Koerber, JT, Kaspar, BK and Schaffer, DV (2006). Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat Biotechnol 24: 198–204. [DOI] [PubMed] [Google Scholar]
- Grimm, D, Lee, JS, Wang, L, Desai, T, Akache, B, Storm, TA et al. (2008). In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol 82: 5887–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber, JT, Jang, JH and Schaffer, DV (2008). DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol Ther 16: 1703–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, PA and Arnold, FH (2009). Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol 10: 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W, Zhang, L, Johnson, JS, Zhijian, W, Grieger, JC, Ping-Jie, X et al. (2009). Generation of novel AAV variants by directed evolution for improved CFTR delivery to human ciliated airway epithelium. Mol Ther 17: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon, KJ, Koerber, JT, Dickey, DD, Murtha, M, Keshavjee, S, Kaspar, BK et al. (2009). Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc Natl Acad Sci USA 106: 3865–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L, Jiang, J, Drouin, LM, Agbandje-McKenna, M, Chen, C, Qiao, C et al. (2009). A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc Natl Acad Sci USA 106: 3946–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, SJ, Blake, BL, Criswell, HE, Nicolson, SC, Samulski, RJ, McCown, TJ et al. (2010). Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB). Mol Ther 18: 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisowski, L, Dane, AP, Chu, K, Zhang, Y, Cunningham, SC, Wilson, EM et al. (2014). Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 506: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L, Li, J and Xiao, X (2011). Directed evolution of adeno-associated virus (AAV) as vector for muscle gene therapy. Methods Mol Biol 709: 127–139. [DOI] [PubMed] [Google Scholar]
- White, SJ, Nicklin, SA, Büning, H, Brosnan, MJ, Leike, K, Papadakis, ED et al. (2004). Targeted gene delivery to vascular tissue in vivo by tropism-modified adeno-associated virus vectors. Circulation 109: 513–519. [DOI] [PubMed] [Google Scholar]
- White, K, Büning, H, Kritz, A, Janicki, H, McVey, J, Perabo, L et al. (2008). Engineering adeno-associated virus 2 vectors for targeted gene delivery to atherosclerotic lesions. Gene Ther 15: 443–451. [DOI] [PubMed] [Google Scholar]
- Work, LM, Nicklin, SA, Brain, NJ, Dishart, KL, Von Seggern, DJ, Hallek, M et al. (2004). Development of efficient viral vectors selective for vascular smooth muscle cells. Mol Ther 9: 198–208. [DOI] [PubMed] [Google Scholar]
- Yu, CY, Yuan, Z, Cao, Z, Wang, B, Qiao, C, Li, J et al. (2009). A muscle-targeting peptide displayed on AAV2 improves muscle tropism on systemic delivery. Gene Ther 16: 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, W and Bartlett, JS (2003). RGD inclusion in VP3 provides adeno-associated virus type 2 (AAV2)-based vectors with a heparan sulfate-independent cell entry mechanism. Mol Ther 7: 515–525. [DOI] [PubMed] [Google Scholar]
- Münch, RC, Janicki, H, Völker, I, Rasbach, A, Hallek, M, Büning, H et al. (2013). Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol Ther 21: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, OJ, Kaul, F, Weitzman, MD, Pasqualini, R, Arap, W, Kleinschmidt, JA et al. (2003). Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat Biotechnol 21: 1040–1046. [DOI] [PubMed] [Google Scholar]
- Perabo, L, Büning, H, Kofler, DM, Ried, MU, Girod, A, Wendtner, CM et al. (2003). In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol Ther 8: 151–157. [DOI] [PubMed] [Google Scholar]
- Waterkamp, DA, Müller, OJ, Ying, Y, Trepel, M and Kleinschmidt, JA (2006). Isolation of targeted AAV2 vectors from novel virus display libraries. J Gene Med 8: 1307–1319. [DOI] [PubMed] [Google Scholar]
- Michelfelder, S, Lee, MK, deLima-Hahn, E, Wilmes, T, Kaul, F, Müller, O et al. (2007). Vectors selected from adeno-associated viral display peptide libraries for leukemia cell-targeted cytotoxic gene therapy. Exp Hematol 35: 1766–1776. [DOI] [PubMed] [Google Scholar]
- Michelfelder, S, Kohlschütter, J, Skorupa, A, Pfennings, S, Müller, O, Kleinschmidt, JA et al. (2009). Successful expansion but not complete restriction of tropism of adeno-associated virus by in vivo biopanning of random virus display peptide libraries. PLoS One 4: e5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi, K, Michelfelder, S, Korff, T, Hecker, M, Trepel, M, Katus, HA et al. (2012). Novel random peptide libraries displayed on AAV serotype 9 for selection of endothelial cell-directed gene transfer vectors. Gene Ther 19: 800–809. [DOI] [PubMed] [Google Scholar]
- Stiefelhagen, M, Sellner, L, Kleinschmidt, JA, Jauch, A, Laufs, S, Wenz, F et al. (2008). Application of a haematopoetic progenitor cell-targeted adeno-associated viral (AAV) vector established by selection of an AAV random peptide library on a leukaemia cell line. Genet Vaccines Ther 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber, JT, Klimczak, R, Jang, JH, Dalkara, D, Flannery, JG and Schaffer, DV (2009). Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Mol Ther 17: 2088–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelfelder, S, Varadi, K, Raupp, C, Hunger, A, Körbelin, J, Pahrmann, C et al. (2011). Peptide ligands incorporated into the threefold spike capsid domain to re-direct gene transduction of AAV8 and AAV9 in vivo. PLoS One 6: e23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, Y, Müller, OJ, Goehringer, C, Leuchs, B, Trepel, M, Katus, HA et al. (2010). Heart-targeted adeno-associated viral vectors selected by in vivo biopanning of a random viral display peptide library. Gene Ther 17: 980–990. [DOI] [PubMed] [Google Scholar]
- Dalkara, D, Byrne, LC, Klimczak, RR, Visel, M, Yin, L, Merigan, WH et al. (2013). In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci Transl Med 5: 189ra76. [DOI] [PubMed] [Google Scholar]
- Deverman, BE, Pravdo, PL, Simpson, BP, Kumar, SR, Chan, KY, Banerjee, A et al. (2016). Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol 34: 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, A, Schmidt, K, Leder, C, Müller, OJ, Wobus, CE, Bettinger, K et al. (2003). Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol 77: 11072–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodi, DJ, Soares, AS and Makowski, L (2002). Quantitative assessment of peptide sequence diversity in M13 combinatorial peptide phage display libraries. J Mol Biol 322: 1039–1052. [DOI] [PubMed] [Google Scholar]
- Derda, R, Tang, SK, Li, SC, Ng, S, Matochko, W and Jafari, MR (2011). Diversity of phage-displayed libraries of peptides during panning and amplification. Molecules 16: 1776–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias-Neto, E, Nunes, DN, Giordano, RJ, Sun, J, Botz, GH, Yang, K et al. (2009). Next-generation phage display: integrating and comparing available molecular tools to enable cost-effective high-throughput analysis. PLoS One 4: e8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravn, U, Gueneau, F, Baerlocher, L, Osteras, M, Desmurs, M, Malinge, P et al. (2010). By-passing in vitro screening–next generation sequencing technologies applied to antibody display and in silico candidate selection. Nucleic Acids Res 38: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- t Hoen, PA., Jirka, SM., Ten Broeke, BR., Schultes, EA., Aguilera, B, Pang, KH et al. (2012). Phage display screening without repetitious selection rounds. Anal Biochem 421: 622–631. [DOI] [PubMed] [Google Scholar]
- Rajotte, D and Ruoslahti, E (1999). Membrane dipeptidase is the receptor for a lung-targeting peptide identified by in vivo phage display. J Biol Chem 274: 11593–11598. [DOI] [PubMed] [Google Scholar]
- Giordano, RJ, Lahdenranta, J, Zhen, L, Chukwueke, U, Petrache, I, Langley, RR et al. (2008). Targeted induction of lung endothelial cell apoptosis causes emphysema-like changes in the mouse. J Biol Chem 283: 29447–29460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba, M, Alzoubi, A, O'Neill, K, Abe, K, Urakami, T, Komatsu, M et al. (2014). A novel vascular homing peptide strategy to selectively enhance pulmonary drug efficacy in pulmonary arterial hypertension. Am J Pathol 184: 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvinen, TA and Ruoslahti, E (2007). Molecular changes in the vasculature of injured tissues. Am J Pathol 171: 702–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietzsch, M, Broecker, F, Reinhardt, A, Seeberger, PH and Heilbronn, R (2014). Differential adeno-associated virus serotype-specific interaction patterns with synthetic heparins and other glycans. J Virol 88: 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas, G, Brindle, SD and Greengard, P (1971). The route of absorption of intraperitoneally administered compounds. J Pharmacol Exp Ther 178: 562–564. [PubMed] [Google Scholar]
- Tournoux, F, Petersen, B, Thibault, H, Zou, L, Raher, MJ, Kurtz, B et al. (2011). Validation of noninvasive measurements of cardiac output in mice using echocardiography. J Am Soc Echocardiogr 24: 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairez, O, Lonjaret, L, Ruiz, S, Marchal, P, Franchitto, N, Calise, D et al. (2013). Anesthetic regimen for cardiac function evaluation by echocardiography in mice: comparison between ketamine, etomidate and isoflurane versus conscious state. Lab Anim 47: 284–290. [DOI] [PubMed] [Google Scholar]
- Monteilhet, V, Saheb, S, Boutin, S, Leborgne, C, Veron, P, Montus, MF et al. (2011). A 10 patient case report on the impact of plasmapheresis upon neutralizing factors against adeno-associated virus (AAV) types 1, 2, 6, and 8. Mol Ther 19: 2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi, F, Anguela, XM, Pavani, G, Chen, Y, Davidson, RJ, Hui, DJ et al. (2013). Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci Transl Med 5: 194ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, X, Li, J and Samulski, RJ (1998). Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol 72: 2224–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin, S, Byrne, BJ, Mason, E, Zolotukhin, I, Potter, M, Chesnut, K et al. (1999). Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther 6: 973–985. [DOI] [PubMed] [Google Scholar]
- McCarty, DM, Monahan, PE and Samulski, RJ (2001). Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther 8: 1248–1254. [DOI] [PubMed] [Google Scholar]
- Rohr, UP, Heyd, F, Neukirchen, J, Wulf, MA, Queitsch, I, Kroener-Lux, G et al. (2005). Quantitative real-time PCR for titration of infectious recombinant AAV-2 particles. J Virol Methods 127: 40–45. [DOI] [PubMed] [Google Scholar]
- Riches, AC, Sharp, JG, Thomas, DB and Smith, SV (1973). Blood volume determination in the mouse. J Physiol 228: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.