Abstract
Repetitive element anchored PCR was used to evaluate the genetic profiles of Escherichia coli isolated from surface water contaminated with urban stormwater, sanitary sewage, and gull feces to determine if strains found in environmental samples reflect the strain composition of E. coli obtained from host sources. Overall, there was less diversity in isolates collected from river and beach sites than with isolates obtained from human and nonhuman sources. Unique strain types comprised 28.8, 29.2, and 15.0% of the isolate data sets recovered from stormwater, river water, and beach water, respectively. In contrast, 50.4% of gull isolates and 41.2% of sewage isolates were unique strain types. River water, which is expected to contain E. coli strains from many diffuse sources of nonpoint source pollution, contained strains most closely associated with other river water isolates that were collected at different sites or on different days. However, river sites impacted by sewage discharge had approximately 20% more strains similar to sewage isolates than did sites impacted by stormwater alone. Beach sites with known gull fecal contamination contained E. coli most similar to other beach isolates rather than gull isolates collected at these same sites, indicating underrepresentation of possible gull strains. These results suggest large numbers of strains are needed to represent contributing host sources within a geographical location. Additionally, environmental survival may influence the composition of strains that can be recovered from contaminated waters. Understanding the ecology of indicator bacteria is important when interpreting fecal pollution assessments and developing source detection methodology.
Contamination of surface waters by fecal pollution constitutes a serious environmental and public health threat. In large complex systems, such as the urbanized coastal areas of the Great Lakes, fecal pollution can be introduced from multiple sources, including sewage overflows, agricultural runoff, and urban stormwater. Identifying and eliminating the source of contamination is not straightforward because assessment of fecal pollution generally relies on a limited number of surface water samples to measure fecal indicator organism densities. Escherichia coli is one of the U.S. Environmental Protection Agency (EPA) recommended indicator organisms for freshwater systems and is a sensitive measure of fecal pollution since it is common to almost all warm-blooded animals, including humans (35). Detection of E. coli by standard microbiological methods provides no information as to the originating host source. However, identifying the source of fecal pollution is a high priority in order to better understand the potential health risk and to mitigate the source of pollution. Human sources of fecal pollution can contain human pathogens such as Salmonella spp., Shigella spp., pathogenic E. coli, and enteroviruses, including hepatitis A (25). Agricultural animals can also serve as a vector for important pathogens including Cryptosporidium parvum and E. coli 0157:H7 (13, 25). The health risk from diffuse, nonpoint runoff is relatively unknown, but some studies have noted increased reports of illness following swimming near stormwater outfalls (18, 26). Generally, the pathogens that could be expected to occur in contaminated waters are dependent on the host source reservoir from which they are derived.
Methods to determine sources of fecal pollution have included phenotypic and genetic characterization of fecal indicator bacteria (32). Studies have employed DNA fingerprinting to discern characteristic patterns and associations among E. coli strains in order to classify the strains according to the host from which they were derived (7, 11, 17, 23, 27, 31). Overall, studies that have assessed the relationships among strains by either cluster analysis (17) or discriminant analysis using banding patterns (7, 27) have found that strains from a host source do not form an exclusive group of genetically similar strains but rather can display a wide range of diversity. However, strains that are highly similar based on repetitive element anchored PCR (rep-PCR) fingerprints have been found to be from the same host source group. For example, isolates from humans, geese, ducks, sheep, pigs, chickens, and cows were correctly classified into host groups for >80.0% of isolates by using Jackknife analysis, which matches one isolate against all other isolates in a data set to determine which host group is most similar to the isolate being evaluated (11). Further, in a study using 440 isolates from humans, gulls, cattle, and dogs, it was found that strains with >85% similarity in rep-PCR banding patterns were from the same host group, with rare exceptions (23). Correct classification into a host group with similarity measures of ribotype patterns and amplified fragment length polymorphism patterns has also been reported (7, 17). It is unknown if these associations among strains from specific host types are a function of geographical proximity of the hosts, unspecified selection processes, or actual host specificity traits of the strains.
One important consideration for source-tracking studies is that nonpoint source runoff, e.g., urban stormwater, is increasingly recognized as a major source of fecal pollution (1, 36). Urban stormwater is expected to carry E. coli from numerous types of host animals that contribute only a minor amount when considered as individual host groups but, when combined, comprise the major portion of fecal pollution in urban rivers. Many of these minor host sources are often not specifically characterized as part of source-tracking studies or are characterized in low numbers relative to other major host sources of pollution. It is essentially unknown how genetic characterizations of strains from hosts such as humans, agricultural animals, or wildlife species that are included in data sets can be extrapolated to the uncharacterized host E. coli populations that may occur in contaminated systems.
Most studies describing genetic relationships among strains have focused on E. coli taken from host animals (7, 8, 11) with a few exceptions (27). E. coli strains found in contaminated surface waters may not be directly comparable to E. coli that is isolated directly from host sources, thereby complicating the utility of source tracking using data sets of host source characteristics. Differences in survivorship may impact comparisons of host and environmental isolates; for example, only a small subset of E. coli from a host may survive in the environment, but these E. coli comprise the majority of strains that are isolated from contaminated waters. Previous studies provide evidence that E. coli can persist in the benthos environment and subsequently be detected in overlying surface waters (6). Residual populations were reported in one study, where fecal coliform levels in wastewater subjected to low temperatures decrease rapidly but then stabilize to 1 to 10% of the initial population size (34). In addition, E. coli that has been isolated from septic tanks has been found to be less diverse and genetically distinct than strains of E. coli from the inhabitants of the households served by those systems (15).
In this study, we evaluated the genetic profiles of E. coli strains found in stormwater, where fecal pollution is derived from multiple uncharacterized host sources, and compared these profiles to known host sources of pollution. We have also compared E. coli strains recovered from surface water with known sources of contamination with E. coli strains collected directly from these same sources to determine if host source comparisons were relevant to investigating fecal pollution in environmental samples. These environmental samples included river water subjected to stormwater discharges, e.g., primarily impacted by nonpoint sources of fecal pollution, river water contaminated with sewage overflows, and beach water samples that were highly impacted by roosting ring-billed gulls.
MATERIALS AND METHODS
Isolation of E. coli.
Host and environmental E. coli strains were isolated over a three-and-a-half-year period during 2000-2003. Primary isolation of E. coli from all samples followed the EPA method for E. coli enumeration (37). In short, fecal samples or aliquots of sewage treatment plant influent were inoculated onto m-TEC (Hardy Diagnostics, Santa Maria, Calif.) agar plates by using a 20-μl loop, and environmental water samples were filtered through 0.45-μm-pore-size filters (Millipore, Bedford, Mass.) and placed on m-TEC agar plates. All samples were incubated for 2 h at 35°C and 18 h at 44.5°C.
E. coli strains from sewage were collected from two separate wastewater treatment plants; one plant serves a combined sewer area and receives both stormwater and sanitary sewage during wet weather flows and, therefore, we did not obtain samples from this plant unless there had been no rainfall in the preceding 48 h. The second treatment plant receives flow from the separated sewer system that serves residential areas and the influent is primarily sanitary sewage. For isolation of E. coli, 500 ml of 24-h flow-weighted influent samples from the treatment plants were mixed well, and 20-μl aliquots of each sample were inoculated onto m-TEC agar plates, with mixing of the sample between inoculations, for a total of 15 to 20 individual plates for each wastewater treatment plant sample. In order to maximize sample distribution, one isolate per plate was subcultured to 250 μl of EC media containing 4-methylumbelliferyl-β-d-glucuronide (MUG media; Remel, Lenexa, Kans.) to confirm E. coli identification as described previously (23).
Fecal samples from host animals, including gulls, cattle, dogs, and bison, were obtained using a cotton-tipped swap to collect newly deposited fecal material at various sites. E. coli isolates from the primary m-TEC agar plates were subcultured into MUG media for confirmation. For the final strain data set, one isolate per fecal sample for gull and other host animals was used for comparison of DNA fingerprint patterns. Gull isolates were primarily obtained at three beaches within a 5-mile radius; all of these sites harbor a large ring-billed gull population, and beach water samples were collected at two of these sites. Collection sites and isolate numbers are shown in Table 1.
TABLE 1.
E. coli isolates from host and environmental samples
| Isolate types | No. of isolates (n = 2,315) | Source
|
Reference | |
|---|---|---|---|---|
| Type of samplea | Geographical and temporal sample distribution | |||
| Sewage | 490 | Wastewater treatment plant influent from two treatment plants | 25 flow-weighted samples from metropolitan Milwaukee over 27 monthsb | 23, this study |
| Gull | 230 | Fecal samples collected from beach sites | 7 beach sites on Lake Michigan Southwestern shore, 35 collection days over 27 monthsc | 23, this study |
| Cattle | 103 | 100-ml samples from feedlot detention systems | 4 farm sites in Southwestern Wisconsin, two collection days per site | 23 |
| Other hosts | 53 | Fecal samples from dogs and raccoons | Individual samples of animals in Milwaukee River Basin watershed over 12 mo | This study |
| Other hosts outgroup | 48 | Pelican and bison fecal samples | Isolates obtained from Florida and Utah, respectively | This study |
| Stormwater | 295 | In-line, flow-weighted samples from stormwater conveyance system | Stormwater system in metropolitan Milwaukee, discharges to two major rivers that drain to Lake Michigan | This study |
| River water-stormwaterd | 513 | Flow-weighted samples during storm events (two sites), or 1-liter grab samples in triplicate for each site (transect of 10 sites) | 12 river sites on two major tributaries that drain to Lake Michigan, 15 collection days over 24 mo | This study |
| River water CSOe | 134 | Flow-weighted samples during storm events (one site), or 1-liter grab samples in triplicate for each site (transect of four sites) | 4 sites within the combined sewer system collected during 3 separate CSO events over a 24-mo period | This study |
| Beach water | 353 | 1-liter grab sample in triplicate for each site | Two Lake Michigan beaches in metropolitan Milwaukee, 5 to 10 sites per beach, 15 collection days | This study |
| Gull pond | 23 | 1-liter grab sample in triplicate | Stormwater detention pond located near a landfill site in metropolitan Milwaukee | This study |
One isolate per fecal sample was used for individual host animals; multiple samples were taken at each cattle feedlot detention system.
Twenty sewage isolates collected in the United Kingdom sanitary sewage conveyance system were also analyzed.
Approximately 75% of the gull isolates were collected from three of the sites within a 5-mile radius in the metropolitan Milwaukee area; two of these sites were used to collect beach water samples.
Isolates were collected from river water that received stormwater discharges and no reported sanitary discharges.
Isolates were collected from river water sites during a CSO.
Stormwater samples were collected from an in-line monitoring system in metropolitan Milwaukee (prior to discharge to the rivers) and were diluted 1:100 prior to filtering 1 ml onto m-TEC agar for isolation of E. coli. River water samples were collected in duplicate or triplicate using a 1-liter grab sampler. Twelve sites were sampled that included a suburban and highly urbanized 15-km transect of the Menomonee River and the confluence of the Menomonee and Milwaukee River, which are two major tributaries that discharge to Lake Michigan in downtown Milwaukee. The majority of samples were collected following rain events, with matching base flow samples collected for the river sites. Flow-weighted samples were also collected for five rain events at two U.S. Geological Survey gauging stations that contained automated samplers: one site upstream in the watershed and one site in the estuary. Postrain event sampling at the river sites included sampling during two combined sewage overflow (CSO) events, which discharge a combination of stormwater and sanitary sewage into the rivers at some of the sites. Isolates obtained from river sites receiving CSO discharge were designated as CSO isolates. All other isolates from river samples were impacted by only urban stormwater discharges. Beach water samples were collected at seven sites on Lake Michigan in metropolitan Milwaukee during base flow conditions and following rain events. All environmental water samples were placed on ice following collection and handled according to the EPA guidelines for the analysis of water samples for E. coli (38). Isolates were obtained from the primary m-TEC agar plates and further processed as described for the handling of isolates from fecal samples. All isolates were cataloged and stored in 26% glycerol solution at −70°C until analyzed.
DNA fingerprinting.
PCR was used to amplify target DNA from bacterial isolates to generate fingerprint patterns. The target DNA was the repetitive extragenic palindrome sequence, a noncoding region found repeatedly interspersed in bacterial genomes (39). Approximately 2 μl of whole-cell preparations at an optical density of 1 provided templates for each 25-μl PCR. Primers employed to generate amplified fragments included REP1R and REP2I primers (39). PCR and cycling parameters were as described by Rademaker and de Bruijn (28). Reactions were run for 30 cycles with a 42°C annealing temperature on a PTC-225 thermocycler (MJ Research, Waltham, Mass.) to perform the amplifications. One sample of E. coli strain K-12 and three wild-type E. coli isolates from a previous run were included in every PCR setup as controls to determine if variability in PCR amplification occurred. The banding patterns of the control strain K-12 and the repeated wild-type strains were analyzed concurrently with the test samples (described below) to assure consistency in reaction products. rep-PCR runs that did not demonstrate reproducibility of patterns in the control strains were discarded.
Separation of amplified genomic fragments was accomplished via gel electrophoresis using 1% agarose gels made with 1× Tris-ascetate-EDTA and run at 70 V for 16 h at 4°C. A 1-kb molecular weight marker (Invitrogen, Grand Island, N.Y.) was run in three to four lanes of each gel as an external reference standard in order to allow for the correction of gel irregularities due to electrophoresis. Gels were stained with 0.6 μg of ethidium bromide/ml in 1× Tris-acetate-EDTA and visualized under UV light. Banding patterns were digitally photodocumented by using an EpiChemi II darkroom bioimaging system (UVP, Inc., Uplands, Calif.).
Cluster analysis, diversity indices, and group statistics of DNA fingerprint patterns.
Digital images of gels were entered into a genomic fingerprint analysis program, Bionumerics v. 3.0 (Applied Maths, Kortrijk, Belgium). Banding patterns were compared using a densitometric curve-based method that evaluates the intensity as well as the position of the bands to generate pairwise similarity scores (Pearson coefficient) that were subsequently used for cluster analysis. The Pearson coefficient proved more accurate for rep-PCR comparisons than the other methods that account only for band position based on visual inspection of identical strain K-12 banding patterns and correspondence of the similarity scores. For comparisons, a 1.0% optimization setting was found to give the highest similarity recognition among multiple samples of strain K-12 while excluding dissimilar strains.
Cluster analysis was carried out by constructing dendrograms using the unweighted pair-group method using arithmetic means tree-building method. Strain types were designated based on clonal characteristics, and clades were defined as essentially clonal rep-PCR patterns with no more than one band difference between patterns (40). A similarity score value of 85% was used as a cutoff for designating strain types; this value was based upon comparison of patterns generated by repeated analysis of strain K-12 (n = 115), where identical patterns were found to be >90% similar based on comparison of banding patterns. Wild-type strains with one band difference produced similarity scores above 85% and were considered the same strain type. Diversity indices were calculated on the basis of rep-PCR patterns using the Shannon diversity index and equitability (2). For diversity calculations, strain groups were normalized to a sample size of 230 by randomly removing strains with a randomized data set generated in Excel (Microsoft, 2000). To construct rarefaction curves, each strain type data set was randomly sampled using EcoSim 7.0 (16) for 1,000 iterations. Data were plotted using SigmaPlot 8.02 (SPSS, Chicago, Ill.). The average slope was calculated to determine strain types per 100 isolates sampled.
Jackknife analysis was used to assess the robustness of isolate host or environmental group assignment based on maximum similarity coefficients. Isolates from stormwater were designated as one group on the assumption that they are from nonhuman sources. All isolates were manually assigned to a host or environmental group and then matched pairwise to all other isolates in the data set. The percentage of isolates correctly identified to their original group was then calculated, as well as the percentage of misclassification into other groups, e.g., the percentage of isolates that had DNA fingerprints more similar to an isolate from another group.
RESULTS
Diversity of E. coli from hosts compared with environmental water samples.
E. coli isolates from two potential sources of fecal pollution, gulls and sewage, were characterized by using rep-PCR DNA fingerprinting and compared with E. coli isolates from in-line stormwater, river water, and beach water samples. The strains from environmental water samples were collected from multiple samples over time and across several sites within the watershed to achieve a broad representation of E. coli in the watershed that discharges to Lake Michigan (Table 1). The diversity among strains isolated directly from environmental samples was less than what was found in strains isolated from a particular host source (Table 2). For example, 50.4% of the isolates from gulls and 41.2% of the isolates from sewage were shown to be unique strains in the data set. In contrast, only 28.8% of the stormwater isolates and 29.2% of river water isolates were found as unique strains. Beach water samples yielded the highest frequency of isolation of nonunique strains, where only 15% of the strain isolates were found to be unique fingerprints and all other isolates matched one or more isolates in the data set. Diversity indices for each of these groups are shown in Table 3. In the case of stormwater, river, and beach isolates, cluster analysis using a global dendrogram constructed with all strains demonstrated that identical strains were typically recovered from different sites or from samples collected on different days.
TABLE 2.
Diversity and relative abundance of E. coli strains found in hosts and environmental water samples
| Strain type or clade size (no. of isolates in clade) | Sewage isolates (n = 490)
|
Gull isolates (n = 230)
|
Stormwater isolates (n = 295)
|
River isolates (n = 647)
|
Beach isolates (n = 353)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of clades | Proportion of isolates in category | No. of clades | Proportion of isolates in category | No. of clades | Proportion of isolates in category | No. of clades | Proportion of isolates in category | No. of clades | Proportion of isolates in category | |
| Unique strainsa | 202 | 41.2 | 116 | 50.4 | 85 | 28.8 | 189 | 29.2 | 53 | 15.0 |
| 2a | 41 | 16.7 | 27 | 23.4 | 26 | 17.6 | 53 | 16.9 | 14 | 7.9 |
| 3 | 11 | 6.7 | 14 | 18.3 | 10 | 10.2 | 12 | 5.6 | 3 | 2.5 |
| 4 | 10 | 6.1 | 3 | 5.2 | 2 | 2.7 | 13 | 8.0 | 6 | 6.8 |
| 5 | 6 | 6.1 | 7 | 11.9 | 8 | 6.2 | 3 | 4.2 | ||
| 6 | 1 | 2.6 | 3 | 6.1 | 6 | 5.6 | 2 | 3.4 | ||
| 7 | 1 | 1.4 | 4 | 9.5 | 1 | 1.1 | 2 | 4.0 | ||
| 8 | 4 | 6.5 | 2 | 5.4 | 1 | 2.3 | ||||
| 9 | 1 | 1.8 | 2 | 5.1 | ||||||
| 10 | 2 | 5.7 | ||||||||
| 11 | 1 | 2.2 | 1 | 1.7 | ||||||
| 12 | 1 | 2.4 | ||||||||
| 13 | 1 | 3.7 | ||||||||
| 14 | 1 | 2.2 | ||||||||
| 15 | 2 | 4.6 | 1 | 4.2 | ||||||
| >15 | 2 (16) | 6.5 | 1 (23) | 7.8% | 1 (18) | 2.8 | 1 (18) | 5.1 | ||
| 1 (20) | 3.1 | 1 (23) | 6.5 | |||||||
| 1 (22) | 3.4 | 1 (30) | 8.5 | |||||||
| 1 (29) | 4.5 | 1 (53) | 15.0 | |||||||
| 1 (37) | 5.7 | |||||||||
| Total no. of strain typesb | 280 | 161 | 140 | 291 | 94 | |||||
Only one isolate found with a given rep-PCR fingerprint pattern.
A strain type was defined as isolates that were >85% similar based on comparison of rep-PCR banding patterns using the Pearson coefficient with no more than one band difference between isolates.
TABLE 3.
Genotypic diversity of E. coli based on rep-PCR DNA fingerprinting patterns of strains recovered from host animals and contaminated water samples
| Source of strains | Parameter
|
||
|---|---|---|---|
| Total no. of strain typesa | Shannon diversity index (H)b | Equitability (J)c | |
| Sewage | 147 | 4.747 | 0.9511 |
| Gulls | 160 | 4.932 | 0.972 |
| Stormwater | 119 | 4.474 | 0.936 |
| River water | 138 | 4.558 | 0.925 |
| Beach water | 73 | 3.613 | 0.842 |
Total number of strain types in a normalized sample size of 230.
Shannon diversity index calculated as H = −Σ (Pi ln Pi).
Equitability calculated from H using the equation J = H/Hmax, where Hmax is the theoretical maximum Shannon diversity index.
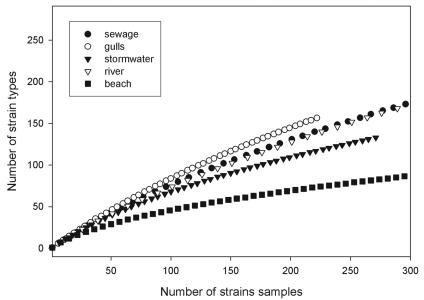
The sampling saturation was assessed by using rep-PCR DNA fingerprints to differentiate among strain types (Fig. 1). rep-PCR fingerprints that were >85% similar were considered a single strain type, or clonal line. In general, this equated to either a difference of one band between strains or a difference in the intensity of the amplified bands with essentially the same pattern between strains. Cattle isolates were not assessed due to the low number of farms represented in the data set. Likewise, the other host sources such as raccoons, dogs, pelicans, and bison were not included due to the low number of strains that were characterized. The sewage and gull host groups did not appear to be near sampling saturation for possible strains, even within the limited geographical region that was the subject of this study. The average slope of the line was the highest for gull isolates and was approximately 0.7 (e.g., 70 unique strains per 100 sampled); the average slope of the plotted data for sewage isolates was 0.57; for stormwater, 0.47; and for river isolates, 0.45. Beach water isolates were the only group that appeared to have a good representation of possible strains; the average slope was found to be 0.26, where 50% sampling saturation of the total strains types found was reached after 100 isolates were sampled.
FIG. 1.
Assessment of sampling saturation of possible E. coli strains from host and environmental groups. A rarefaction curve was generated from iterative sampling using EcoSim software (16) to determine the abundance of strain types found in each group (gulls, sewage, stormwater, river, or beach) for the number of strains sampled. Third-order regression lines calculated using all data points were r2 = >0.99 for all five series.
Cluster analysis and group statistics of E. coli rep-PCR fingerprints.
Cluster analysis of all rep-PCR fingerprints (n = 2,315) did not reveal distinct grouping of strains by host type; major divisions in the dendrogram at 55, 65, and 75% similarity produced groupings that contained sewage and gull isolates, as well as isolates from one or more of the environmental samples. However, the dendrogram produced small groupings of highly similar strains that were primarily from a single type of host or environmental sample. Jackknife analysis was performed to assess the extent in which strains within a host or environmental group were more similar to each other when compared to all other strains in the data set. Jackknife analysis removes one isolate from the dendrogram, reconstructs the pairwise similarity matrix and dendrogram, and then “matches” the isolate back to the data set to determine which user-defined group it belongs to, in this case, by host or environmental sample type. Matching of isolates to the entire data set demonstrated that strains from a type of sample (e.g., gull, sewage, stormwater, river water, beach water) were most similar to other strains from the same host or environmental source (Table 4). These findings may be a function of geographic distribution rather than host source specificity, since both stormwater and river water are expected to contain E. coli from a broad range of host sources and therefore are not expected to be similar. Yet, stormwater and river isolates were most similar to the other isolates in each respective environmental sample type and gave a classification rate of 73.1% for stormwater and 64.4% for river water isolates. These results were considerably higher than random associations, since stormwater comprises approximately 10% of the data set and river water 28% of the data set.
TABLE 4.
Rates of correct grouping of strains within specific host or environmental groups based on maximum similarity of rep-PCR fingerprints using Jackknife analysis
| Host or environmental isolate group | % of isolates classified asa:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sewage | Gull | Stormwater | River (stormwater) | River (CSO) | Beach | Gull pond | Cow | Other hosts (watershed) | Other hosts (outgroup) | |
| Sewage | 67.4 | 4.5 | 3.6 | 12.7 | 4.7 | 4.5 | — | 2.1 | 0.2 | 0.2 |
| Gull | 11.9 | 39.9 | 8.6 | 16.9 | 6.5 | 11.5 | 0.7 | 2.2 | 1.1 | 0.7 |
| Stormwater | 3.7 | 4.8 | 73.1 | 9.9 | 4.8 | 2.7 | — | 0.7 | — | 0.3 |
| River water (stormwater) | 7.9 | 5.0 | 8.8 | 64.4 | 6.9 | 5.2 | — | 0.9 | 0.7 | 0.2 |
| River water (CSO) | 17.1 | 4.8 | 11.6 | 20.5 | 37.7 | 4.8 | — | 1.4 | — | 2.1 |
| Beach | 3.6 | 8.7 | 3.2 | 7.8 | 3.6 | 72.8 | — | — | — | 0.3 |
| Gull pond | — | 4.3 | 4.3 | — | 4.3 | 21.7 | 65.2 | — | — | — |
| Cow | 3.9 | 2.9 | 2.9 | 5.8 | 3.9 | 1.9 | — | 76.8 | — | — |
| Other hosts (watershed) | 1.9 | — | 1.9 | 9.4 | — | — | — | — | 86.8 | — |
| Other hosts (outgroup) | 4.2 | 2.1 | 2.1 | 2.1 | 2.1 | 2.1 | — | — | — | 85.4 |
Bold indicates values of correct classification; values given across the remainder of the row are the misclassification rates; —, no isolates were classified into this group.
Assessment of data set performance for source identification using environmental samples with known sewage or gull contamination.
E. coli isolates that were collected from river sites during sewage overflow events (e.g., CSO isolates) were compared to the data set to assess the discriminatory power of data set comparisons for identifying human sources. E. coli in these samples is expected to be derived from human sources as well as stormwater containing nonpoint source urban runoff. When comparing the CSO isolates to all isolates, 37.7% of the E. coli obtained from CSO-contaminated river water were most similar to other isolates within the CSO group; however, there was a high rate of classification into the sewage group (17.1% of isolates) (Table 4). The CSO isolates were also matched against the data set containing only isolates from known hosts and stormwater, where 38% of CSO isolates were identified as sewage, 43% as stormwater, and 19% were matched to gull, cow, or other host categories. River water isolates recovered from sites that were not impacted by CSO discharge were matched against these same groups, where it was found that 57% of the isolates matched most closely to stormwater isolates, 19% to sewage isolates, and the remaining proportion of isolates (24%) to gull, cow, or other hosts.
Similar comparisons were carried out using E. coli collected at beach sites and from a detention pond adjacent to a landfill; all of the sites harbored a large ring-billed gull population and no other sources of contamination were evident. Almost 73% of the beach isolates were most similar to each other, which is consistent with what was found with the other environmental group isolates (Table 4). Matching the beach isolates to host or stormwater isolates demonstrated that 23% matched most closely to gulls, 51% to stormwater, 17% to sewage, and the remainder to other host categories.
Comparison of outgroups and geographically distinct strains.
Three groups of strains were used to evaluate the representatives of the data set and assess trends in associations between the host and environmental groups. Dog and raccoon isolates were expected to be present in stormwater samples; these isolates were found to be most closely related to river and stormwater isolates, but there were no identical matches to any other isolate in the data set. Bison and pelican strains were expected to be distinct from the data set since they were collected from distinct geographic areas; however, four of the bison strains resembled environmental strains from one beach site with the exception of a one band difference. Likewise, two pelican strains matched gull isolates exactly, and one pelican strain matched a sewage isolate exactly. Interestingly, 18 of 20 sewage strains from the United Kingdom were identical or near identical to Milwaukee sewage strains. The United Kingdom sewage strains were not similar to any of the environmental isolates.
DISCUSSION
Fecal pollution in urban coastal systems is expected to originate from an array of human and nonhuman sources, and multiple pollution control measures may be necessary to meet the requirements of the Clean Water Act and its amendments (32). Identifying the major contributing sources of contamination is the critical component for accurate assessment and successful control measures. Source identification approaches have included methods independent of indicator organisms such as F+ RNA coliphages (5, 9), detection of human-specific viruses (4, 20), and detection of polymorphisms in the ribosomal genes of Bacteroides-Prevotella (3). There are numerous source detection methods based on indicator organism phenotypic or genotypic characteristics, including evaluating percentages of fecal streptococci (10), assessing antibiotic resistance (19, 41), and DNA fingerprinting of fecal indicator bacteria (11, 17, 27, 29). Findings from these studies suggest that no single parameter will be diagnostic for simultaneously determining sources of fecal pollution and quantifying relative contributions. Most likely, a combination of complementary approaches will be necessary to compensate for deficiencies or uncertainty factors in chosen methodologies.
Incorporating indicator bacteria characteristics into source-tracking studies is attractive since source identification would be intricately tied to the same biological indicator of the fecal contamination. Utilizing genetic traits in E. coli for fecal pollution source tracking is based on the hypothesis that there is host-specific structure within the E. coli population. Most studies that have described the population genetics of E. coli have focused on subsets of pathogenic strains or have employed multilocus enzyme electrophoresis to assess clonality within the natural population (24, 30, 33). Overall, the amount of genetic variability that can be explained from host specificity of strains remains controversial (14). In studies that focus on using rep-PCR and ribotyping analysis of E. coli for source tracking, the results have been comparable (8), and overall trends in associations between E. coli isolates and the hosts from which they were derived are consistent for independent studies in different geographical regions (7, 11, 27).
We were interested in determining how the composition of strains collected from environmental samples would compare with strains collected from host sources. Previous studies have focused on characterizing E. coli recovered directly from host sources, either fecal samples or composite samples of feedlot detention ponds and sewage influent (8, 11, 23). All of these studies have demonstrated that there is a high amount of diversity within the E. coli population; therefore, accurate strain representation is necessary to reflect what might be expected in surface water contaminated with fecal pollution. For example, sampling more than one isolate per animal may overestimate the frequency of that strain in the natural population (or in a particular host group) given that animals, including humans, can be colonized with one predominant strain of E. coli (23). Overall, the high diversity in the natural population demonstrates that broad sampling of the host population is necessary to determine possible strains that might be introduced into the environment.
While we have gained some insight into what may be required to characterize host source E. coli, few studies have investigated whether or not the same strain types can be recovered from the environment. We have found that strains in contaminated surface waters had lower diversity than what was represented in a broad sampling of host sources. The strategy for collection of the water samples is expected to have a major effect on the profile of strains recovered, and a broad sampling of contaminated waters is needed to produce good representation of possible strains. In this study, duplicate or triplicate samples were collected at multiple sites. The diversity assessments were based on the assumption that adequate random sampling had occurred to represent strains found in stormwater, river water, or beach water in the watershed. This assumption was supported by performing cluster analysis to determine if patterns could be observed that demonstrate the association of identical strains to samples or sites on the same day. The identical, or nonunique, strains recovered, which in turn decrease the diversity assessments, were not associated with a particular sample or set of duplicate or triplicate samples but did correspond with the general sample type (stormwater, beach water, or river water). For example, 15 identical strains collected from river water samples (Table 2) were recovered from a total of 12 different sites or different days at the same site (data not shown). Similar to sampling considerations that may bias a host isolate data set, isolation of the same strain type from a single sample or site on the same day would in effect bias the overall diversity calculation and disproportionately contribute to the low diversity in the environmental strain data set. In addition, the diversity indices should be interpreted in light of possible underrepresentation of possible strains in each of these groups given the lack of sampling saturation (Fig. 1).
In this study, surface waters contaminated with sewage discharge from a CSO contained a higher percentage of strains that were similar to strains obtained from sewage influent (primarily human sources) than did river water contaminated with stormwater only. Taken together, these results would indicate that at least some of the genetic variability among E. coli could be explained by host niche. A recent study by Scott et al. also suggests that DNA fingerprinting may be useful for differentiating human and nonhuman sources, even when applied across a large geographical region (29). Their conclusions are further supported by our study, in which 20 sewage strains collected from the United Kingdom were most similar to sewage strains collected in the Milwaukee River Basin, even when compared to >2,300 total strains, where sewage isolates comprised <25% of the total data set.
Beach isolates resembled gull isolates at a relatively low frequency, despite documentation that gulls contribute to fecal pollution at some beach sites used in this study (22) and have been implicated in other studies to have impacted water quality (12, 21). The high amount of diversity noted in isolates collected from gull feces may indicate that more extensive sampling would be necessary to adequately describe the possible E. coli strains that are derived from gulls. In addition, the beach isolates showed less diversity than what would be expected from multiple birds roosting near a beach site. These results may suggest that only a subset of E. coli survive in the environment, decreasing the reliability of comparing beach isolates to a data set of gull isolates. Alternatively, a limited number of animals, e.g., gulls, may be contributing to the fecal pollution since gulls have been found to be colonized with one predominate strain of E. coli (23).
We anticipated finding broadly diverse rep-PCR fingerprint profiles for the stormwater E. coli isolates, which would reflect the diffuse nature of the bacterial contamination in urban runoff. However, the diversity among strains isolated directly from stormwater was considerably less than what was found in strains isolated from a particular host source. This may indicate that interrelationships among strains are not primarily host dependent since there was a high amount of similarity among strains from stormwater, which is expected to carry fecal pollution from many different sources. Identical strains may indicate possible clonal propagation, and this could account for some of the low diversity. However, many of the identical strains were found at different sites or on different days. Alternatively, the strains that were detected may be a product of selective die-off, which presents itself as a limited range of persistent strains that can be isolated in the environment.
DNA fingerprinting may not be a cost-feasible methodology to identify and quantify fecal pollution sources given the extensive diversity and undercharacterized genetic structure of the natural E. coli population. However, this approach is useful in understanding the ecology of E. coli in the secondary environment (e.g., surface waters) outside the host. Replication of cells outside the host or persistence of residual strain types have implications for recreational water testing, where limited survival and lack of environmental growth are necessary characteristics for bacteria that are to be used as indicator organisms. These types of analyses offer valuable insight into the potential to create persistent residual populations that may confound recreational water testing. There does not appear to be a proportional relationship between fecal indicator bacteria from a host and what is actually detected in the environment, which will be an important consideration when developing methods for fecal pollution source tracking.
Acknowledgments
This work was funded by the Milwaukee Metropolitan Sewerage District and the Wisconsin Department of Natural Resources.
I thank the EPA laboratories in Gulf Breeze for providing pelican isolates and the Robens Centre for Public and Environmental Health for providing sewage isolates. I also thank Alissa Salmore, Erika Jensen, Hilary Street, Josh Harris, and Magnolia Tulod for technical assistance.
REFERENCES
- 1.Bannerman, R. T., D. W. Owens, R. B. Dodds, and N. J. Hornewer. 1993. Sources of pollutants in Wisconsin stormwater. Water Sci. Technol. 28:241-259. [Google Scholar]
- 2.Begon, M., J. L. Harper, and C. R. Townsend. 1986. Ecology: individuals, populations, and communities. Sinaure Associates, Inc., Sunderland, Mass.
- 3.Bernhard, A. E., and K. G. Fields. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehm, A. B., J. A. Fuhrman, R. D. Mrse, and S. B. Grant. 2003. Tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon Bay, Catalina Island, California. Environ. Sci. Technol. 37:673-680. [DOI] [PubMed] [Google Scholar]
- 5.Brion, G. M., J. S. Meschke, and M. D. Sobsey. 2002. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res. 36:2419-2425. [DOI] [PubMed] [Google Scholar]
- 6.Byappanahalli, M., M. Fowler, D. Shively, and R. Whitman. 2003. Ubiquity and persistence of Escherichia coli in a Midwestern coastal stream. Appl. Environ. Microbiol. 69:4549-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carson, C. A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carson, C. A., B. L. Shear, M. R. Ellersieck, and J. D. Schnell. 2003. Comparison of ribotying and repetitive extragenic palindromic-PCR for identification of fecal Escherichia coli from humans and animals. Appl. Environ. Microbiol. 69:1836-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, D., S. C. Long, and M. D. Sobsey. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 69:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devriese, L. A., B. Pot, and M. D. Collins. 1993. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J. Appl. Bacteriol. 75:399-408. [DOI] [PubMed] [Google Scholar]
- 11.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogary, L. R., S. K. Haack, M. J. Wolcott, and R. L. Whitman. 2003. Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull feces. Appl. Environ. Microbiol. 94:865-878. [DOI] [PubMed] [Google Scholar]
- 13.Fujioka, R. S. 2002. Microbial indicators of water quality, p. 234-243. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 14.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 15.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 16.Gotelli, N. J., and G. L. Entsminger. 2004. EcoSim: null models software for ecology. Version 7.0. Acquired Intelligence Inc. & Kesey-Bear, Burlington, Vt. http://homepages.together.net/∼gentsmin/ecosim.htm.
- 17.Guan, S., R. Xu, S. Chen, J. Odumeru, and C. Gyles. 2002. Development of a procedure for discriminating among Escherichia coli isolates from animal and human sources. Appl. Environ. Microbiol. 68:2690-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haile, R. W., J. S. Witte, M. Gold, R. Cressey, C. McGee, R. C. Millikan, A. Glasser, N. Harawa, C. Ervin, P. Harmon, J. Harper, J. Dermend, J. Alamillo, K. Barrett, M. Nided, and G. Wang. 1999. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology 10:355-363. [PubMed] [Google Scholar]
- 19.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, S., R. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levesque, B., P. Brousseau, P. Simard, E. Dewailly, M. Meisels, D. Ramsay, and J. Joly. 1993. Impact of the ring-billed gull (Larus delawarensis) on the microbiological quality of recreational water. Appl. Environ. Microbiol. 59:1228-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLellan, S. L., and A. K. Salmore. 2003. Evidence for localized bacterial loading as the cause of chronic beach closings in a freshwater marina. Water Res. 37:2700-2708. [DOI] [PubMed] [Google Scholar]
- 23.McLellan, S. L., A. D. Daniels, and A. K. Salmore. 2003. Genetic characterization of Escherichia coli populations of host sources of fecal pollution by using DNA fingerprinting. Appl. Environ. Microbiol. 69:2587-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milkman, R. 1997. Recombination and population structure in Escherichia coli. Genetics 46:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moe, C. 2002. Waterborne transmission of infectious agents, p. 184-204. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 26.O'Shea, M. L., and R. Field. 1992. Detection and disinfection of pathogens in storm-generated flow. Can. J. Microbiol. 38:267-276. [DOI] [PubMed] [Google Scholar]
- 27.Parveen, S., K. M. Portier, K. Robinson, L. Edmiston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rademaker, J. L. W., and F. J. de Bruijn. 1997. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer assisted pattern analysis, p. 151-171. In G. Gaetano-Anolles and P. M. Gresshoff (ed.), DNA markers: protocols, applications, and overviews. J. Wiley & Sons, New York, N.Y.
- 29.Scott, T. M., S. Parveen, K. M. Portier, J. B. Rose, M. L. Tamplin, S. R. Farrah, A. Koo, and J. Lukasik. 2003. Geographical variation in ribotype profiles of Escherichia coli isolates from humans, swine, poultry, beef, and dairy cattle in Florida. Appl. Environ. Microbiol. 69:1089-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selender, R. K., D. A. Caugant, and T. S. Whittam. 1987. Genetic structure and variation in natural populations of Escherichia coli, p. 1625-1648. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 31.Seurinck, S., W. Verstraete, and S. D. Siciliano. 2003. Use of 16S-23S rRNA intergenic spacer region PCR and repetitive extragenic palindromic PCR analyses of Escherichia coli isolates to identify nonpoint fecal sources. Appl. Environ. Microbiol. 69:4942-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 33.Souza, V., M. Rocha, A. Valera, and L. E. Eguiarte. 1999. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65:3373-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torrella, F., J. P. Lopez, and C. J. Banks. 2003. Survival of indicators of bacterial and viral contamination in wastewater subjected to low temperatures and freezing: application to cold climate waste stabilization ponds. Water Sci. Technol. 48:105-112. [PubMed] [Google Scholar]
- 35.U.S. Environmental Protection Agency. 1986. Ambient water quality criteria for bacteria-1986. EPA-440/5-84/002. Office of Water Regulation and Standards, Criteria and Standards Division, Washington, D.C.
- 36.U.S. Environmental Protection Agency. 1998. The quality of our nation's waters: 1996. EPA841-S-97-001. Office of Water, Washington, D.C.
- 37.U.S. Environmental Protection Agency. 2000. Improved enumeration methods for recreational water quality indicators: enterococci and Escherichia coli. EPA/821/R-97/004. Office of Science and Technology, Washington, D.C.
- 38.U.S. Environmental Protection Agency. 2002. Method 1603: Escherichia coli (E. coli) in water by membrane filtration using modified membrane-thermotolerant Escherichia coli agar (modified mTEC). EPA-821-R-02-023. Office of Water, Washington, D.C.
- 39.Versalvoic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittam, T. M. 1995. Genetic population structure and pathogenicity in enteric bacteria, p. 217-245. In S. Baumberg, J. P. W. Young, E. M. H. Wellington, and J. R. Saunders (ed.), Population genetics of bacteria (Symposium 52). Cambridge University Press, Cambridge, United Kingdom.
- 41.Wiggins, B. A., R. W. Andrews, R. A. Conway, C. L. Corr, E. J. Dobratz, D. P. Dougherty, J. R. Eppard, S. R. Knupp, M. C. Limjoco, J. M. Mettenburg, J. M. Rinehardt, J. Sonsino, R. L. Torrijos, and M. E. Zimmerman. 1999. Use of antibiotic resistance analysis to identify nonpoint sources of fecal pollution. Appl. Environ. Microbiol. 65:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]