Abstract
Verticillium wilt is a vascular disease of cotton. The causal fungus, Verticillium dahliae, secretes elicitors in culture. We have generated ∼1,000 5′-terminal expressed sequence tags (ESTs) from a cultured mycelium of V. dahliae. A number of ESTs were found to encode proteins harboring putative signal peptides for secretion, and their cDNAs were isolated. Heterologous expression led to the identification of a protein with elicitor activities. This protein, named V. dahliae necrosis- and ethylene-inducing protein (VdNEP), is composed of 233 amino acids and has high sequence identities with fungal necrosis- and ethylene-inducing proteins. Infiltration of the bacterially expressed His-VdNEP into Nicotiana benthamiana leaves resulted in necrotic lesion formation. In Arabidopsis thaliana, the fusion protein also triggered production of reactive oxygen species and induced the expression of PR genes. When added into suspension cultured cells of cotton (Gossypium arboreum), the fusion protein elicited the biosynthesis of gossypol and related sesquiterpene phytoalexins at low concentrations, and it induced cell death at higher concentrations. On cotton cotyledons and leaves, His-VdNEP induced dehydration and wilting, similar to symptoms caused by a crude preparation of V. dahliae elicitors. Northern blotting showed a low level of VdNEP expression in the mycelium during culture. These data suggest that VdNEP is a wilt-inducing factor and that it participates in cotton-V. dahliae interactions.
Recognition plays a central role in interactions between plants and their pathogens. Successful pathogens must be able to recognize and overcome host-plant defense responses. Plants also have evolved sophisticated mechanisms to detect a multitude of potential pathogens in their natural environments and to activate diverse defense mechanisms (15). Elicitors are molecules that trigger defense responses in plants, and they play significant roles in the signal exchange between plants and pathogens (33). Diverse plant defense responses induced by elicitors involve de novo synthesis and accumulation of antimicrobial phytoalexins (11, 16), induction of cell death (hypersensitive response) (6, 17, 44), production of activated oxygen species (oxidative burst) (4, 13), and modification of plant cell walls by deposition of callose (41). The structural and cultivar specificity of elicitors and their ability to trigger plant defense responses at very low concentrations strongly suggest the existence of receptors at the plant plasma membrane and a downstream signal transduction cascade (12).
Verticillium dahliae Kleb. is a phytopathogenic fungus that causes wilt disease in a wide range of crops (34), including cotton. The fungus is widespread in most cotton-growing areas and is a major threat to cotton production. Many reports have shown that V. dahliae produces both low- and high-molecular-weight phytotoxic metabolites in culture fluids; these secreted substances behave as elicitors inducing phytoalexin formation, as well as the formation of phytotoxins triggering wilt symptoms associated with disease development. Buchner et al. (8) found that a potato isolate of V. dahliae produced a high-molecular-mass protein-lipopolysaccharide complex (PLPC) which was associated with pathogenicity. A low-molecular-mass (<3-kDa) phytotoxic polypeptide fraction was partially purified from this PLPC, and it was found that the chlorosis-inducing activity of the PLPC was due mainly to this small peptide (29). From cotton isolate, a PLPC of 197 kDa was found to induce necrosis and wilting in cotton seedlings and also stimulated phenylalanine ammonia-lyase activity and phytoalexin biosynthesis in cotton suspension cells. The PLPC was partially purified from the culture filtrate, and it consisted of five protein-containing components (28). Davis et al. (10) purified a 65-kDa glycoprotein from V. dahliae culture fluids, which acted as an elicitor of phytoalexin formation. Chu et al. (9) reported a secreted glycoprotein of 26 kDa which induced cotton phytoalexin production and leaf wilting. Thus, there appears to be a broad spectrum of elicitors and/or phytotoxins that are related to the pathogenicity of V. dahliae. To further study the interactions between V. dahliae and cotton plants and the mechanisms of wilt development, pure elicitor molecules are preferred to rule out effects of contaminating proteins and carbohydrates in the crude preparation, and cloning of the elicitor genes should be a great help to this approach.
Here, we report the isolation of a V. dahliae cDNA that encodes a necrosis- and ethylene-inducing protein (VdNEP). We present evidence that VdNEP has a high activity in triggering plant defense responses in Nicotiana benthamiana, Arabidopsis thaliana, and cotton and that it also induces wilting of cotton leaves and cotyledons. This protein may play an important role in promoting wilting symptoms of cotton plants.
MATERIALS AND METHODS
Materials.
V. dahliae strain Vd-8, a cotton strain isolated from Gossypium hirsutum, was kindly provided by the Jiangsu Academy of Agricultural Science. The fungus was cultured in liquid Czapek medium (each liter contained 2 g of NaNO3, 1 g of K2HPO3, 0.5 g of MgSO4 · 7H2O, 0.5 g of KCl, 0.01g of FeSO4 · 7H2O, and 30 g of sucrose) at 25°C by shaking at 150 rpm. Plants of N. benthamiana and G. hirsutum L. cv. Zhong-12 were grown at 28°C in a greenhouse with a photoperiod of 16 h of light and 8 h of dark. Plants of A. thaliana (Col-0) were grown at 22°C. The G. arboreum suspension cells were cultured as previously reported (23). Most of the chemicals were purchased from Sigma (St. Louis, Mo.).
DNA and RNA isolation and analysis.
Genomic DNA of V. dahliae was isolated as described previously (22). After complete digestion, about 20 μg of DNA per lane was separated by electrophoresis on an agarose gel and transferred onto a nitrocellulose membrane. The probe for VdNEP was obtained by 32P labeling of the PCR product amplified from the VdNEP cDNA, with primers NEPEcoR (5′-GAATTCATGCTTCCCTCCGCAGTCT-3′) and NEPNot (5′-GCGGCCGCTTAAAACGCGGCGCGCATG-3′). After hybridization overnight, the blot was washed twice in 2× SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) for 15 min at room temperature and 42°C and twice in 0.2× SSC-0.1% SDS at 55°C for 15 min, and then the blot was exposed to X-ray film. Total RNA was isolated as previously described (25). For Northern blotting, a total of 15 μg of RNA was loaded per lane. The membrane was hybridized and washed as described above for DNA blotting. For reverse transcription-PCR (RT-PCR), the first strand of cDNA was synthesized with 1 μg of total RNA in a 20-μl RNA PCR mixture (TaKaRa, Dalian, China). After a 10-fold dilution, 1 μl of the RT products was used as a template in PCR amplification. Primers used in RT-PCR were PR1F (5′-GTAGGTGCTCTTGTTCTTCCC-3′) and PR1R (5′-CACATAATTCCCACGAGGATC-3′) for A. thaliana PR1 (AtPR1) (M59196), ACS6F (5′-CATAAGTGTTGCGGAAGTAA-3′) and ACS6R (5′-GGCAATGGAACGAACC-3′) for AtACS6 (U73786), PDFF (5′-AAGTTTGCTTCCATCATCACC-3′) and PDFR (5′-ATACACACGATTTAGCACCAA-3′) for AtPDF1.2 (AY063779), and ACTF (5′-TTCCTCAATCTCATCTTCTTCC-3′) and ACTR (5′-GACCTGCCTCATCATACTCG-3′) for AtACT (ACTIN2; U37281).
Expressed sequence tag (EST) generation and analysis.
A cDNA library of V. dahliae was constructed with ZAP Express (Stratagene, La Jolla, Calif.) from the mycelium cultured in liquid Czapek medium for 10 to 14 days. After excision, individual clones were picked and cDNA inserts were sequenced from the 5′ end. Automated sequence similarity searches were done for each EST using the BLASTX algorithm to identify putative homologues in the database. ESTs were classified with a system based on the Munich Information Center for Protein Sequences yeast functional class catalogue (http://mips.gsf.de/proj/yeast/catalogues/funcat/index.html), with the cell wall protein category added.
Expression of VdNEP in plants with pSfinx.
The open reading frames (ORFs) of the cDNA candidates were PCR amplified with SfiI sites introduced, and the products were subcloned into the potato virus X-based binary expression vector pSfinx (40). The constructs were subsequently transferred into Agrobacterium tumefaciens strain Mog101, containing the helper plasmid pIC-SArep. The transformed Agrobacterium cells were grown as described previously (40), and different concentrations of Agrobacterium cells in a 10 mM MgCl2 solution (105 to 107 CFU/ml) were infiltrated into N. benthamiana leaves with 1-ml plastic syringes without needles.
Expression of VdNEP in Escherichia coli.
The cDNA of VdNEP without signal peptide was amplified with primers NEPEcoRs (5′-GAATTCCAGCAGCCCCCCAAGGTT-3′) and NEPNot. After digestion, the PCR product was inserted into pET-32a(+) (Novagen, Madison, Wis.). E. coli BL21(DE3) was transformed with the construct, and VdNEP was expressed as a His tag fusion protein (His-VdNEP) by induction with 1.0 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). Proteins were examined by SDS- polyacrylamide gel electrophoresis. Protein purification was performed with the Ni- nitrilotriacetic acid resin (QIAGEN, Valencia, Calif.), and the purified proteins were dialyzed against a phosphate-buffered saline (PBS) buffer (pH 7.4) at 4°C. The protein concentrations were determined with the Bio-Rad (Hercules, Calif.) protein assay kit.
Assay of necrosis- and wilt-inducing activity.
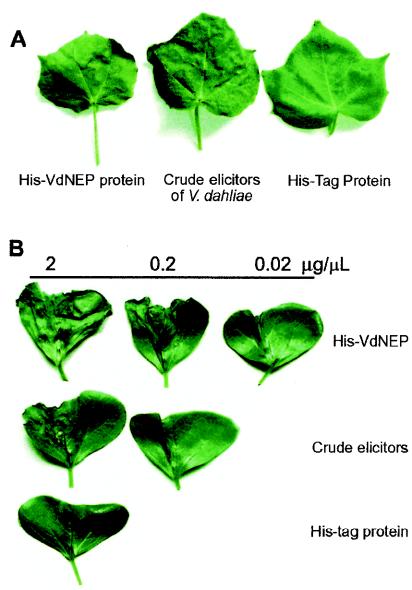
The proteins in PBS buffer were infiltrated into leaves of 3- to 4-week-old plants (for A. thaliana and N. benthamiana) or into cotyledons of 2-week-old cotton seedlings (G. hirsutum L. cv. Zhong-12). For each assay, a 10-μl protein solution at the concentrations noted in Fig. 5 was used for infiltration, which was performed by using 1-ml plastic syringes without needles. Cotton leaves with freshly cut petioles were dipped into a 50-μl protein solution. After the solution was absorbed, the leaves were dipped into water. Lesion formation or wilting symptoms were viewed and photographed each day after treatment.
FIG. 5.
Wilting of cotton (G. hirsutum L. cv. Zhong-12) leaves and cotyledons induced by His-VdNEP and the crude elicitors of V. dahliae 24 h posttreatment. (A) Leaves dipped into 50-μl protein solutions (0.5 μg/μl); (B) cotyledons infiltrated with 10-μl protein solutions at the concentrations indicated.
Examination of cell death, DNA laddering, and H2O2 production.
Treated G. arboreum suspension cells were stained with Evans blue, followed by several washings with double-distilled water, and the optical density of the supernatant was determined as described previously (3). After being stained, the cells were mounted and observed under a light microscope. Dead cells were stained blue.
Treated leaves were collected, boiled for 1 min in lactophenol (10 ml of lactic acid, 10 ml of glycerol, 10 ml of liquid phenol, and 10 ml of double-distilled water) containing 10 mg of trypan blue, and then decolorized in chloral hydrate (2.5 g of chloral hydrate dissolved in 1 ml of distilled water) for at least 30 min. The leaves were then mounted on a slide with chloral hydrate and photographed.
Genomic DNA was isolated from G. arboreum suspension cells collected at 36 h after treatment with an apoptosis DNA extraction kit (Watson Biotechnologies, Inc., Shanghai, People's Republic of China). DNA was run on a 2% agarose gel containing ethidium bromide. DNA laddering was observed and photographed under UV light.
To view H2O2 produced by plant leaves, treated leaf samples were incubated for 8 h with 1 mg of 3,3′-diaminobenzidine (DAB) solution (pH 3.8)/ml and then decolorized in 96% ethanol. The sample was mounted on a slide with 60% glycerol and examined under a light microscope. H2O2 was detected as reddish-brown coloration.
Quantitative determination of cotton sesquiterpene aldehydes.
Suspension-cultured cells of G. arboreum were treated with His-VdNEP or other elicitors, and the treated cells were harvested 36 h after treatment. Gossypol and related sesquiterpene aldehydes were quantitatively determined by a phloroglucinol-HCl reagent, as previously described (27).
Preparation of a crude elicitor mixture of V. dahliae.
Total elicitors secreted by V. dahliae during culture were prepared as previously described (9). Briefly, (NH4)2SO4 was added to the culture fluid to 80% saturation, mixed well, and left to stand at 4°C for another 2 h. The solution was centrifuged at 10,000 × g for 15 min, and the pellet was dissolved in PBS buffer and dialyzed at 4°C.
Nucleotide sequence accession numbers.
The five cDNA sequences of V. dahliae have been deposited in the GenBank database under accession no. AY524789 (VdNEP), AY524790 (03c01), AY524791 (01f09), AY524792 (10g12), and AY524793 (06h05).
RESULTS
Analysis of V. dahliae ESTs.
Generation of ESTs offers a low-cost strategy to identify substantial gene inventories in the absence of gene information (39). We generated a set of ESTs from V. dahliae strain Vd-8, a cotton isolate, to identify potential elicitor genes. High-quality sequences were obtained from 1,053 cDNA clones with sequenced tags longer than 250 bp. A BLASTX search revealed that a high proportion (∼60%) of the ESTs showed no or low similarities (E value, ≥10−2) to protein sequences deposited in the NCBI database, and those sequences were not discussed here. The remaining ESTs were assigned to 17 major functional groups (Table 1), and as expected, most of them show higher similarities to fungal genes. Recently Neumann and Dobinson (31) obtained over 1,000 ESTs from each of two cDNA libraries of a tomato isolate of V. dahliae, and they also found a high proportion of sequences with unknown functions.
TABLE 1.
Classification of ESTs from V. dahliae
| Functional classification of ESTsa | No. of ESTs |
|---|---|
| Metabolism | 41 |
| Cell cycle and DNA processing | 16 |
| Transcription | 7 |
| Protein synthesis | 49 |
| Protein fate | 22 |
| Cellular transport and transport mechanism | 6 |
| Cellular communication and/or cellular transduction mechanism | 9 |
| Cell rescue, defense, and virulence | 34 |
| Regulation of and/or interaction with cellular environment | 17 |
| Subcellular localization | 19 |
| Protein activity regulation | 2 |
| Protein with binding function and cofactor requirement | 1 |
| Transport facilitation | 14 |
| Cell wall proteins | 10 |
| Energy | 61 |
| Unclassified proteins | 32 |
| Hypothetical proteins | 62 |
| Total | 402 |
ESTs were classified based on the Munich Information Center for Protein Sequences Saccharomyces cerevisiae functional catalogue (http://mips.gsf.de/proj/yeast/catalogues/funcat/index.html), with the cell wall protein category added.
From the functional group “cell rescue, defense, and virulence,” five ESTs were selected for further analysis (Table 2). They were considered elicitor candidates because they showed similarities to putative microbial elicitors (necrosis-inducing proteins, allergens that were glycoproteins, and proteinases) and because each was predicted to have a eukaryotic signal peptide for protein secretion (http://www.cbs.dtu.dk/services/SignalP-2.0/). For example, ESTs 01f09 and 03c01 are similar to allergens from Aspergillus fumigatus (5), and EST 07g01 shows sequence similarity to an NEP from Fusarium oxysporum (19).
TABLE 2.
Five ESTs of V. dahliae selected for analysis of elicitor activities
| Code no. | Function annotation | E value (BLASTX) |
|---|---|---|
| SPVD07g01 | Necrosis- and ethylene-inducing peptide, AAC97382, F. oxysporum | 8e-56 |
| SPVD01f09 | Allergen Asp F 2, AAC69357, A. fumigatus | 7e-65 |
| SPVD03c01 | Allergen rAsp f 9, CAA11266, A. fumigatus | 9e-47 |
| SPVD06h05 | Subtilisin-like serine protease PR1C, CAD11898, Metarhizium anisopliae | 6e-12 |
| SPVD10g12 | Aspartic proteinase, T47207, Neurospora crassa | 2e-45 |
Necrosis-inducing activity of VdNEP.
cDNAs corresponding to the five ESTs were isolated and sequenced. To determine if they induce plant defense reactions, ORFs of the five candidates were inserted into pSfinx for Agrobacterium-mediated plant expression (40). Necrotic lesions were clearly observed on N. benthamiana leaves 10 days postinfiltration with Agrobacterium harboring pSfinx-07g01 (Fig. 1) but not on leaves with Agrobacterium harboring other candidate ORFs or with the empty pSfinx vector. Thus, among the five candidate cDNAs, 07g01 encoded a product that was able to induce necrosis in N. benthamiana. However, these results do not exclude the possibility that the other four candidates could also have elicitor activities, since such factors as expression level and protein modification may obscure the activity.
FIG. 1.
Induction of necrotic lesions induced by expressing VdNEP in N. benthamiana leaves. The leaf had been infiltrated with A. tumefaciens strain Mog101 (105 to 107CFU/ml) containing pSfinx-VdNEP or the empty pSfinx vector for 10 days.
The cDNA of 07g01 codes for a protein of 233 amino acid residues with a predicted molecular mass of 25.9 kDa and an isoelectric point of 8.84. This protein was predicted to have an N-terminal signal peptide of 18 amino acid residues and two transmembrane domains, with no putative glycosyl residues. A BLASTX search of the GenBank database showed that the deduced protein was most similar to NEPs from other fungi and oomycetes. Comparison of the amino acid sequences revealed that it had sequence identities of 62, 38, 36, and 33% with the NEPs from F. oxysporum f. sp. erythroxyli (AAC97382) (2), Pythium monospermum (AAQ89593), Pythium aphanidermatum (AAD53944) (41), and Phytophthora sojae (AAM48171) (35), respectively. The V. dahliae protein reported here is thus named VdNEP.
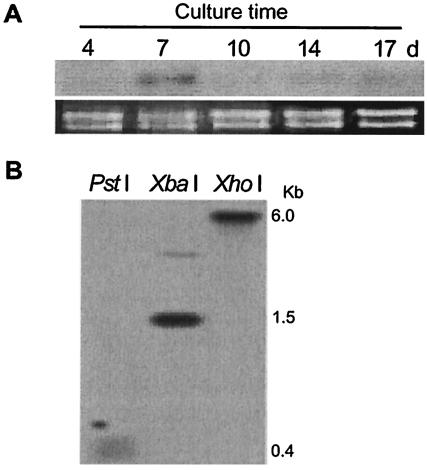
Northern blotting indicated that the expression level of VdNEP was generally low during culture, but a slightly higher expression level appeared in the mycelium cultured for 7 days (Fig. 2A). Southern hybridization revealed a single copy of VdNEP in the V. dahliae genome, with possibly a distantly related gene, since there was an additional weak hybridization band (Fig. 2B).
FIG. 2.
(A) Northern analysis of VdNEP expression in suspension cultures of V. dahliae; (B) Southern blot of V. dahliae genomic DNA hybridized with a VdNEP probe. d, days.
Defense responses of N. benthamiana and Arabidopsis plants triggered by VdNEP.
In order to assay the elicitor activity more directly, VdNEP was expressed in E. coli as a His tag fusion protein (His-VdNEP). A protein band of about 43 kDa appeared after induction with IPTG (data not shown), which was in good agreement with the calculated size of the fusion protein. After infiltration of the purified His-VdNEP into N. benthamiana leaves at a concentration as low as 0.02 μg/μl, necrosis appeared around the infiltrated sites. The necrotic lesions were visually apparent within 2 days postinfiltration, and then the lesions dried out (data not shown).
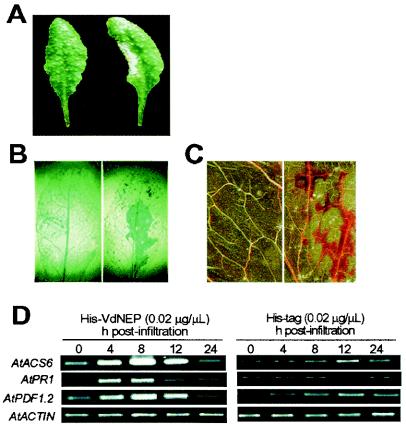
To further examine the activity of VdNEP, the fusion protein was applied to a well-defined Arabidopsis system. When infiltrated into leaves of A. thaliana, His-VdNEP fusion protein at a concentration of 0.02 μg/μl again induced necrotic lesions. Symptoms included water-soaked brownish lesions, which were eventually dried out after 48 h (Fig. 3A). Trypan blue staining showed that treated leaf cells died within 12 h (Fig. 3B). In addition, His-VdNEP triggered the production of reactive oxygen species (such as H2O2) in leaves within 6 h of infiltration (Fig. 3C), preceding the onset of lesion formation.
FIG. 3.
Defense responses of A. thaliana induced by His-VdNEP. (A) Necrotic lesion on a treated leaf 48 h postinfiltration; (B) cell death in a treated leaf revealed by trypan blue staining 12 h postinfiltration; (C) H2O2 production in a treated leaf revealed by 3′,3′-diaminobenzidine staining 6 h postinfiltration; (D) expression of PR genes in treated leaves. In the experiments shown in panels A to C, the right leaf was infiltrated with His-VdNEP, and the left was infiltrated with the His tag. For each infiltration, 10 μl of the protein solution (0.02 μg/μl) was applied. Transcripts of ACTIN2 were amplified by 26 cycles of PCR, and those of other genes were amplified by 30 cycles.
The expression of genes encoding pathogenesis-related proteins of treated Arabidopsis leaves was then investigated. ACS6 encodes 1-aminocyclopropane-1-carboxylate synthase for ethylene biosynthesis (19, 37), and PR1 and PDF1.2 are the marker genes of salicylic acid- and jasmonate acid-dependent defense signaling pathways, respectively (14, 16). RT-PCR showed that the infiltration of His-VdNEP rapidly induced the expression of all three genes (Fig. 3D). Thus, in Arabidopsis leaves both SA- and ethylene-JA-dependent defense pathways of plants were activated by His-VdNEP.
Cotton defense responses to His-VdNEP.
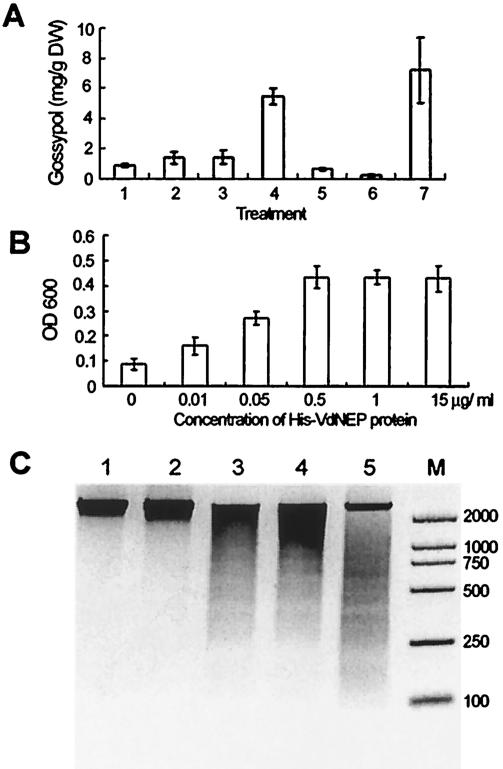
Previous investigation showed that transcription levels of genes encoding enzymes of the gossypol pathway were greatly increased upon elicitation with a crude preparation of V. dahliae elicitors or upon pathogen inoculation, followed by increased accumulation of gossypol and related sesquiterpene phytoalexins (7, 23, 24, 25). We found that, when it was applied to cotton suspension-cultured cells, His-VdNEP was highly active in eliciting the formation of sesquiterpene aldehydes (Fig. 4A). The elicitation of phytoalexin accumulation was dose dependent, with doses ranging from 0.01 to 0.1 μg of the fusion protein/ml. At higher concentrations (0.5 μg/ml or above), however, His-VdNEP suppressed phytoalexin formation. We then examined the viability of the treated cells by Evans blue staining. At 36 h posttreatment, the portion of dead cells was positively correlated with His-VdNEP concentrations below 0.5 μg/ml. At a low concentration of His-VdNEP (0.05 μg/ml) only a small fraction (about 30%) of cotton cells died, whereas at higher concentrations (0.5 μg/ml or above) all cells died (Fig. 4B), and as a consequence, the formation of phytoalexins ceased.
FIG. 4.
Responses of G. arboreum suspension-cultured cells to His-VdNEP. (A) Induced formation of sesquiterpene aldehydes (gossypol equivalents) in cells treated for 36 h. Lanes: 1, His tag protein, 0.1 μg/ml; 2 to 6, His-VdNEP at 0.01, 0.05, 0.1, 0.5 and 1 μg/ml, respectively; 7, crude elicitors of V. dahliae (1 μg protein/ml). DW, dry weight. (B) Relative increase in cell death estimated by monitoring Evans blue retention by determining the absorbance at 600 nm. (C) DNA laddering induced by His-VdNEP. Lanes: 1, PBS buffer; 2, His tag protein at 0.5 μg/ml; 3 to 5, His-VdNEP at 0.05, 0.1 and 0.5 μg/ml, respectively; M, DL-2000 marker (Takara Biotechnology, Dalian, China).
Fragmentation of DNA is one of the best-established criterions for confirming an elicitor-dependent programmed cell death during hypersensitive responses of plants (36, 41). We examined the nuclear DNA of His-VdNEP-treated cotton cells. After cells were treated for 36 h with different concentrations of His-VdNEP, DNA laddering of 180-bp fragments was clearly observed. The His tag alone did not induce DNA laddering of cotton cells, and neither did PBS buffer (Fig. 4C). This indicates that programmed cell death did occur in cells treated with His-VdNEP.
Leaf wilt is the key symptom of cotton plants infected with V. dahliae. Previous investigations showed that various components of glycoproteins or of PLPC secreted by V. dahliae could cause cotton leaf wilting. It is therefore interesting to test if VdNEP induces wilt on cotton plants. Due to difficulties of infiltrating solutions into cotton leaves, we instead conducted the test by dipping cotton leaves with fresh cut petioles into a 50-μl protein solution (0.5 μg/μl). We found that the leaves were wilting 24 h after being dipped into the His-VdNEP solution, and the whole leaf dried out after 48 h. Similar symptoms were also observed on leaves dipped into a solution of crude elicitors. Wilting did not occur on leaves dipped into a control His tag solution (Fig. 5A).
The effects of His-VdNEP and the crude elicitor preparation on cotton cotyledons of seedlings was also investigated by infiltration. Both solutions induced dehydration and wilting in cotyledons within 24 h postinfiltration. The severity of wilting was positively related to the protein concentrations applied, and His-VdNEP was more effective than the crude elicitors in inducing cotyledon wilting, with a specific activity approximately 10 times higher (Fig. 5B).
DISCUSSION
Cotton plants infected by V. dahliae show symptoms of leaf vein browning and chlorosis, wilting, premature defoliation, and, most severely, plant death. Control of V. dahliae is difficult because the pathogen can survive for years in the soil (38). Previous studies suggested that the formation of wilting symptoms in infected plants was due mainly to the effect of toxins produced by V. dahliae, and the phytotoxic factors included PLPC, glycoproteins, and small peptides. Here, we report cDNA cloning of a protein component, VdNEP, that acted as a wilt-inducing factor on excised cotton leaves and cotyledons and as an elicitor in inducing phytoalexin production and programmed cell death of cotton suspension-cultured cells. The bacterially expressed fusion protein of His-VdNEP also induced necrotic lesions in N. benthamiana leaves and triggered a complex of defense responses in Arabidopsis plants. The wilting symptom observed in His-VdNEP-treated cotton leaves was similar to that evoked by a mixture of elicitors secreted by V. dahliae. Furthermore, His-VdNEP exhibited a higher specific activity of wilt induction than the crude elicitors prepared from V. dahliae culture fluids. These results strongly suggest that VdNEP is associated with the production of vascular wilt symptoms in V. dahliae-infected cotton plants.
The V. dahliae strain used in this investigation is a cotton isolate. Interestingly, although VdNEP from this strain exerted elicitor activities on all three dicot plants examined here, the responses of host and nonhost plants were not identical. VdNEP induced wilting on cotton leaves and cotyledons, whereas on leaves of Arabidopsis and N. benthamiana, which are nonhost plants of this strain, it caused necrosis formation but not wilting. The wilt-inducing activity has not been reported for NEPs before, and wilting development triggered by VdNEP seemed to be specific to susceptible host plants. The tomato Ve genes, which encode cell surface-like receptors, are implicated in race-specific resistance to infection by V. dahliae (21). The race 1 isolates caused little or no damage on tomato carrying the Ve gene, but they produced severe symptoms on plants lacking the Ve gene. Phytotoxic peptide produced by race 1 of V. dahliae was associated with the production of wilt symptoms in susceptible hosts, including tomato plants lacking the Ve gene (30). Similarly, bacterial Avr proteins expressed in plants carrying the cognate R protein generally induce a hypersensitive response that is linked to resistance, but they cause disease-like symptoms in plants lacking the R protein (32).
The NEPs and their homologues have been found in many fungal and oomycete species, including Phytophthora (13, 35), Fusarium (2), Pythium (41), and Verticillium (this report), as well as in some eubacteria (13, 35). The genomic DNA sequences of Magnaporthe grisea also contain NEP homologues (EAA46838 and EAA49539). The NEPs of F. oxysporum, P. aphanidermatum, and Phytophthora parasitica were all isolated from culture fluids, indicating that they are secretory proteins. The presence in a broad spectrum of microorganisms, the absence in host organisms, and the activity of activating defense responses in hosts are the characteristics of pathogenesis-associated molecular patterns (1, 13). Members of this elicitor family from phytopathogenic microorganisms are likely key molecules in causing necrosis or wilting in plants, but the molecular mechanisms remain elusive. Our understanding of wilt- and necrosis-inducing signals triggered by NEPs awaits isolation of the plant receptors, and this will certainly help to design new approaches to enhancing cotton resistance to, or tolerance of, V. dahliae.
Many reports showed that V. dahliae infection, or treatment with the fungal elicitor, activated the expression of defense response and PR genes in cotton (18, 26, 28, 45). We found that His-VdNEP protein could induce PR1 and PDF1.2 gene expression in Arabidopsis. A previous report (13) showed that NPP1 from P. parasitica activated PR1 gene expression soon after infiltration into Arabidopsis leaves, similar to our findings. On the other hand, when V. dahliae isolated from cauliflower was inoculated into Arabidopsis plants, infection did not appear to trigger the accumulation of either PR1 or PDF1.2 transcripts, although anthocyanin accumulation was enhanced and stunting, leaf chlorosis, and acceleration of flowering were induced (42). One of the possible explanations of this discrepancy is that the VdNEP gene was not expressed when this V. dahliae strain was inoculated into Arabidopsis plants or that the VdNEP level was too low to activate PR gene expression.
Finally, infiltration of His-VdNEP into Arabidopsis leaves increased the expression level of ACS6, which encodes a key enzyme for ethylene biosynthesis (37). The plant hormone ethylene is involved in many aspects of plant growth and development, including seed germination, root hair development, root nodulation, flower senescence, abscission, and fruit ripening (20, 43). Whether ethylene plays a role in triggering or promoting such symptoms as wilting and necrosis in V. dahliae-plant interactions needs further investigation.
Acknowledgments
We thank B. Han and Y. J. Zhang for their help in EST sequencing. We thank David Baulcombe at the Sainsbury Laboratory, Mathieu Joosten at Wageningen University, and Plant Bioscience Limited (Norwich, United Kingdom) for providing the binary PVX vector pSfinx.
This research was supported by the Science and Technology Committee of Shanghai, the National 863 Program of China, and the National Natural Science Foundation of China.
REFERENCES
- 1.Aderem, A., and R. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, J., C. Jennings, and J. D. Anderson. 1997. The 24-KDa protein from Fusarium oxysporum f. sp. erythroxyli: occurrence in related fungi and the effect of growth medium on its production. Can. J. Microbiol. 43:45-55. [DOI] [PubMed] [Google Scholar]
- 3.Baker, C. J., and N. M. Mock. 1994. An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell Tissue Organ Cult. 39:7-12. [Google Scholar]
- 4.Baker, C. J., E. W. Orlandi, and N. M. Mock. 1993. Harpin, an elicitor of the hypersensitive response in tobacco caused by Erwinia amylovora, elicits active oxygen production in suspension cells. Plant Physiol. 102:1341-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee, B., P. A. Greenberger, J. N. Fink, and V. P. Kurup. 1998. Immunological characterization of Asp f 2, a major allergen from Aspergillus fumigatus associated with allergic bronchopulmonary aspergillosis. Infect. Immun. 66:5175-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer, D. W., Z. M. Wei, S. V. Beer, and A. Collmer. 1995. Erwinia chrysanthemi HarpinEch: an elicitor of the hypersensitive response that contributes to soft-rot pathogenesis. Mol. Plant-Microbe Interact. 8:484-491. [DOI] [PubMed] [Google Scholar]
- 7.Bianchini, G. M., R. D. Stipanovic, and A. A. Bell. 1999. Induction of δ-cadinene synthase and sesquiterpenoid phytoalexins in cotton by Verticillium dahliae. J. Agric. Food Chem. 47:4403-4406. [DOI] [PubMed] [Google Scholar]
- 8.Buchner, V., A. Nashmias, and Y. Burstein. 1982. Isolation and partial characterization of a phytotoxic glycopeptide from a protein-lipopolysaccharide complex produced by a potato isolate of Verticillium dahliae. FEBS Lett. 138:261-264. [Google Scholar]
- 9.Chu, Z. Q., J. W. Jia, X. J. Zhou, and X. Y. Chen. 1999. Isolation of glycoproteins from Verticillium dahliae and their phytotoxicity. Acta Bot. Sin. 41:972-976. [Google Scholar]
- 10.Davis, D. A., P. S. Low, and P. Heinstein. 1998. Purification of a glycoprotein elicitor of phytoalexin formation from Verticillium dahliae. Physiol. Mol. Plant Pathol. 52:259-273. [Google Scholar]
- 11.Ebel, J. 1986. Phytoalexin synthesis: the biochemical analysis of the induction process. Annu. Rev. Phytopathol. 24:235-264. [Google Scholar]
- 12.Ebel, J., and E. G. Cosio. 1994. Elicitor of plant defense responses. Int. Rev. Cytol. 148:1-36. [Google Scholar]
- 13.Fellbrich, G., A. Romanski, A. Varet, B. Blume, F. Brunner, S. Englehardt, G. Felix, B. Kemmerling, M. Krzymowska, and T. Nürnberger. 2002. NPP1, a Phytophthora-associated trigger of plant defense in Arabidopsis. Plant J. 32:375-390. [DOI] [PubMed] [Google Scholar]
- 14.Glazebrook, J. 2001. Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 4:301-308. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, M. G. 1996. Microbial elicitors and their receptors in plants. Annu. Rev. Phytopathol. 34:387-412. [DOI] [PubMed] [Google Scholar]
- 16.Hammond-Kosack, K. E., and J. D. G. Jones. 1996. Resistance gene-dependent plant defense response. Plant Cell 8:1773-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, S. Y., H. C. Huang, and A. Collmer. 1993. Pseudomonas syringae harpinpss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73:1255-1266. [DOI] [PubMed] [Google Scholar]
- 18.Hill, M. K., K. J. Lyon, and B. R. Lyon. 1999. Identification of disease response genes expressed in Gossypium hirsutum upon infection with the wilt pathogen Verticillium dahliae. Plant Mol. Biol. 40:289-296. [DOI] [PubMed] [Google Scholar]
- 19.Jennings, J. C., P. C. Apel-Birkhold, N. M. Mock, C. J. Baker, J. D. Anderson, and B. A. Bailey. 2001. Induction of defense responses in tobacco by the protein Nep1 from Fusarium oxysporum. Plant Sci. 161:891-899. [Google Scholar]
- 20.Johnson, P. R., and J. R. Ecker. 1998. The ethylene gas signal transduction pathway: a molecular perspective. Annu. Rev. Genet. 32:227-254. [DOI] [PubMed] [Google Scholar]
- 21.Kawchuk, L. M., J. Hachey, D. R. Lynch, F. Kulcsar, G. van Rooijen, D. R. Waterer, A. Robertson, E. Kokko, R. Byers, R. J. Howard, R. Fischer, and D. Prufer. 2001. Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA 98:6511-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, K. N., D. I. Rouse, and T. L. German. 1994. PCR primers that allow intergeneric differentiation of ascomycetes and their application to Verticillium spp. Appl. Environ. Microbiol. 60:4324-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C. J., P. Heinstein, and X. Y. Chen. 1999. Expression pattern of genes encoding farnesyl diphosphate synthase and sesquiterpene cyclase in cotton suspension-cultured cells treated with fungal elicitors. Mol. Plant-Microbe Interact. 12:1095-1104. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., C. R. Benedict, R. D. Stipanovic, and A. A. Bell. 1999. Purification and characterization of S-adenosyl-l-methionine: desoxyhemigossypol-6-O-methyltransferase from cotton plants. An enzyme capable of methylating the defense terpenoids of cotton. Plant Physiol. 121:1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, P., Y. H. Wang, G. D. Wang, M. Essenberg, and X. Y. Chen. 2001. Molecular cloning and functional identification of (+)-δ-cadinene-8-hydroxylase, a cytochrome P450 mono-oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J. 28:95-104. [DOI] [PubMed] [Google Scholar]
- 26.McFadden, H. G., R. Chapple, R. De Feyter, and E. Dennis. 2001. Expression of pathogenesis-related genes in cotton stems in response to infection by Verticillium dahliae. Physiol. Mol. Plant Pathol. 58:119-131. [Google Scholar]
- 27.Meng, Y. L., J. W. Jia, C. J. Liu, W. Q. Liang, P. Heinstein, and X. Y. Chen. 1999. Coordinated accumulation of (+)-δ-cadinene synthase mRNAs and gossypol in developing seeds of Gossypium hirsutum and a new member of the cad1 family from G. arboreum. J. Nat. Prod. 62:248-252. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, R., V. Slater, and I. A. Dubery. 1994. A phytotoxic protein-lipopolysaccharide complex produced by Verticillium dahliae. Phytochemistry 35:1449-1453. [Google Scholar]
- 29.Nachmias, A., V. Buchner, and Y. Burstein. 1985. Biological and immunochemical characterization of a low molecular weight phytotoxin isolated from a protein-lipopolysaccharide complex produced by a potato isolate of Verticillium dahliae Kleb. Physiol. Plant Pathol. 26:43-55. [Google Scholar]
- 30.Nachmias, A., V. Buchner, L. Tsror, Y. Burstein, and N. Keen. 1987. Differential phytotoxicity of peptides from culture fluids of Verticillium dahliae races 1 and 2 and their relationship to pathogenicity of the fungi on tomato. Phytopathology 77:506-510. [Google Scholar]
- 31.Neumann, M. J., and K. F. Dobinson. 2003. Sequence tag analysis of gene expression during pathogenic growth and microsclerotia development in the vascular wilt pathogen Verticillium dahliae. Fungal Genet. Biol. 38:54-62. [DOI] [PubMed] [Google Scholar]
- 32.Nimchuk, Z., L. Rohmer, J. H. Chang, and J. L. Dangl. 2001. Knowing the dancer from the dance: R-gene products and their interactions with other proteins from host and pathogen. Curr. Opin. Plant Biol. 4:288-294. [DOI] [PubMed] [Google Scholar]
- 33.Nürnberger, T. 1999. Signal perception in plant pathogen defense. Cell Mol. Life Sci. 55:167-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pegg G. F. 1989. Pathogenesis in vascular disease of plants, p. 51-94. In E. C. Tjamos and C. Beckman (ed.), Vascular wilt diseases of plants. Springer, Berlin, Germany.
- 35.Qutob, D., S. Kamoun, and M. Gijzen. 2002. Expression of Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32:361-373. [DOI] [PubMed] [Google Scholar]
- 36.Ryerson, D. E., and M. C. Heath. 1996. Cleavage of nuclear DNA into oligonucleosomal fragments during cell death induced by fungal infection or by abiotic treatments. Plant Cell 8:393-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato, T., and A. Theologis. 1989. Cloning the mRNA encoding 1-aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc. Natl. Acad. Sci. USA 86:6621-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnathorst, W. C. 1981. Life cycle and epidemiology of Verticillium, p. 81-111. In M. A. Mace, A. A. Bell, and C. H. Beckman (ed.), Fungal wilt diseases of plants. Academic Press, New York, N.Y.
- 39.Soanes, D. M., W. Skinner, J. Keon, J. Hargreaves, and N. J. Talbot. 2002. Genomics of phytopathogenic fungi and the development of bioinformatics resources. Mol. Plant-Microbe Interact. 15:421-427. [DOI] [PubMed] [Google Scholar]
- 40.Takken, L. W., R. Luderer, S. H. E. J. Gabriëls, N. Westerink, R. Lu, P. J. G. M. de Wit, and M. H. A. J. Joosten. 2000. A functional cloning strategy, based on a binary PVX-expression vector, to isolate HR-inducing cDNAs of plant pathogens. Plant J. 24:275-283. [DOI] [PubMed] [Google Scholar]
- 41.Veit, S., J. M. Wörle, T. Nürnberger, W. Koch, and H. U. Seitz. 2001. A novel protein elicitor (PaNie) from Pythium aphanidermatum induces multiple defense responses in carrot, Arabidopsis, and tobacco. Plant Physiol. 127:832-841. [PMC free article] [PubMed] [Google Scholar]
- 42.Veronese, P., M. L. Narasimhan, R. A. Stevenson, J. K. Zhu, S. C. Weller, K. V. Subbarao, and R. A. Bressan. 2003. Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana. Plant J. 35:574-587. [DOI] [PubMed] [Google Scholar]
- 43.Wang, K. L.-C., H. Li, and J. R. Ecker. 2002. Ethylene biosynthesis and signaling networks. Plant Cell 14:S131-S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, L. M. 1995. Elicitins from Phytophthora and basic resistance in tobacco. Proc. Natl. Acad. Sci. USA 92:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, X. J., S. Lu, Y. H. Xu, J. W. Wang, and X. Y. Chen. 2002. A cotton cDNA encoding a pathgenesis-related 10 protein with in vitro ribonuclease activity. Plant Sci. 162:629-636. [Google Scholar]