Abstract
Virus filtration is a key clearance unit operation in the manufacture of recombinant protein, monoclonal antibody, and plasma-derived biopharmaceuticals. Recently, a consensus has developed among filter manufacturers and end users about the desirability of a common nomenclature and a standardized test for classifying and identifying virus-retentive filters. The Parenteral Drug Association virus filter task force has chosen PR772 as the model bacteriophage to standardize nomenclature for large-pore-size virus-retentive filters (filters designed to retain viruses larger than 50 to 60 nm in size). Previously, the coliphage PR772 (Tectiviridae family) has been used in some filtration studies as a surrogate for mammalian viruses of around 50 to 60 nm. In this report, we describe specific properties of PR772 critical to the support of its use for the standardization of virus filters. The complete genomic sequence of virulent phage PR772 was determined. Its genome contains 14,946 bp with an overall G+C content of 48.3 mol%, and 32 open reading frames of at least 40 codons. Comparison of the PR772 nucleotide sequence with the genome of Tectiviridae family prototype phage PRD1 revealed 97.2% identity at the DNA level. By dynamic light-scattering analysis, its hydrodynamic diameter was measured as 82 ± 6 nm, consistent with use in testing large-virus-retentive filters. Finally, dynamic light-scattering analysis of PR772 preparations purified on CsCl gradients showed that the phage preparations are largely monodispersed. In summary, PR772 appears to be an appropriate model bacteriophage for standardization of nomenclature for larger-pore-size virus-retentive filters.
Virus filtration is a critical operation during the manufacture of recombinant proteins, monoclonal antibodies, and plasma-derived biopharmaceuticals. Viruses are removed from process intermediates largely through a sieving-based mechanism, although protein absorptive effects occur depending on process conditions (13, 21).
There is no industry-wide uniform nomenclature or regulatory standard for applying a pore size or retention rating to virus-retentive filters. Virus filter manufacturers use different parameters for naming virus-retentive filters, including average pore size measurement, type of virus retained, size of virus retained, nominal (dextran or protein) molecular weight cutoff, and nominal molecular weight of proteins that can pass through the membrane (2). This lack of consensus among filter manufacturers makes the current nomenclature ambiguous; also, it is difficult for filter users to evaluate commercially available virus retention filters on a consistent basis.
One approach to advance to a more informative classification system is based on different pore sizes (34). Large-pore-size virus-retentive filters are defined as filters designed to remove retroviruses and other viruses of >50 nm by size exclusion but specifically not small viruses, e.g., >18 to 30 nm. Examples of these are the Pall Ultipor VF DV50, Asahi Planova 35N, and Millipore Viresolve NFR filters, which are designed to remove large viruses (>50 to 60 nm), e.g., endogenous retroviruses associated with continuous cell lines and relevant blood-borne pathogens, such as human immunodeficiency virus. Small-pore-size filters specifically target viruses in the 18- to 30-nm size range. The Pall Ultipor VF DV20, Asahi Planova 15N, and Millipore Viresolve NFP filters are designed to remove small viruses, e.g., murine minute virus, reported as a sporadic contaminant in raw material used in the manufacture of continuous cell line-derived products (18), hepatitis A virus, and human parvovirus B19, or other relevant blood-borne pathogens.
Bacteriophages are commonly used by filter manufacturers to evaluate the size exclusion properties of their virus removal filters and by some end users for preliminary evaluation of size-based filtration under process conditions (3, 4, 12, 19, 25-27). Because of its reported diameter (53 to 63 nm) (6, 15, 16) and its previous successful use in testing size exclusion properties of large-virus filters (4, 12, 24-26), PR772 has been chosen by the Parenteral Drug Association virus filter task force to be the model bacteriophage to standardize nomenclature for larger-pore-size virus filters (G. Sofer, unpublished data).
PR772 belongs to the Tectiviridae family of icosahedral double-stranded DNA bacteriophages (6, 7, 15, 16). The family Tectiviridae includes phages with a lipid membrane beneath the icosahedral shell (6). On the basis of the examination in the electron microscope of multiple phage preparations, the diameter of members of the family Tectiviridae recognized by the International Committee on Taxonomy of Viruses is about 63 nm (6). The prototype phage PRD1 has been extensively characterized (7-10, 29, 30, 33). Recently, combined electron microscopy (EM) and X-ray imaging yielded a quasiatomic model of PRD1 and the average dimensions of the PRD1 particle were calculated to be as follows: vertex to vertex, 698 Å; edge to edge, 655 Å; facet to facet, 637 Å (29). Its genome has been sequenced (14,927 bp) and contains 34 open reading frames (ORFs) (9). The 5′ end of the PRD1 genomic DNA is covalently attached to a terminal protein that serves to prime DNA replication. Phages of the Tectiviridae family can be separated into two groups based the complementary inverted terminal repeat (ITR) sequences located at their 5′ and 3′ termini (31). Group 1 includes phages PRD1 and PR5 isolated in North America, and group 2 comprises phages PR4, PR772, and L17 isolated in Europe, Africa, and Australia, respectively (31). Interestingly, these enterobacteriophages of the Tectiviridae family have several properties in common with other, seemingly unrelated, viruses that infect evolutionarily distant hosts. These viruses include mammalian adenoviruses (10), the Bacillus bacteriophage Bam35 (28, 35), and the Paramecium virus PCBV-1 (23). The similarities between members of the family Tectiviridae and adenoviruses include genome replication, capsid architecture, fold of the major coat protein, and vertex organization (10, 33). These elements of similarity suggest that Tectiviridae family phages may be particularly good model viruses for these mammalian viruses in virus filter performance studies.
Coliphage PR772 was originally isolated from the wastewater treatment system in Pretoria, South Africa (15, 16). While less is known about the structure of PR772 than about that of PRD1, it can be surmised to be highly similar on the basis of the overall similarity of Tectiviridae family phages (15, 16). Initial estimates of the PR772 capsid size of 53 nm were based on transmission EM (TEM) of phosphotungstate-stained and dried phages (15), a procedure that may shrink particles (17). Given the consensus size of members of the family Tectiviridae (63 nm), the precise atomic resolution sizing data from PRD1 and the overall DNA homology between members of the family Tectiviridae (6, 7, 29, 31, 32), it can be presumed that PR772's natural hydrated size is more likely to be closer to 63 nm. To our knowledge, beyond the ITRs and some 5′ regions, the PR772 genome has not been sequenced.
Phage PR772 has proven useful in the past to evaluate the size exclusion properties of large-pore-size virus filters under various operating conditions (4, 12, 24-26). Further, routine filter testing with PR772 is preferable because of its nonpathogenic host compared to the usual host of PRD1 (Salmonella enterica serovar Typhimurium), which is a pathogen (risk group 2 organism) that must be handled in a level 2 containment laboratory. In this study, specific properties of the phage (e.g., hydrodynamic size, genome sequence, and aggregation potential) were analyzed to support its use to standardize the nomenclature of larger-pore-size virus filters.
MATERIALS AND METHODS
Phage propagation and preparation.
PR772 and its bacterial host, Escherichia coli K-12 J-53, were obtained from the Félix d'Hérelle Reference Center for Bacterial Viruses (Université Laval, Québec City, Québec, Canada). Bacteriophages were grown on the surface of agar plates by the overlay method. For each of the 10 to 20 tryptic soy agar (Difco, Detroit, Mich.) plates, 105 PFU of PR772 were added to 2 ml of mid-log-phase bacteria and incubated for 5 min at 20 to 25°C. Nine milliliters of 50°C soft agar (0.7% agarose in tryptic soy broth) was mixed and poured onto the surface of a 150-mm-diameter tryptic soy agar plate. After solidification, the plates were incubated at 37°C for 16 h to allow bacterial growth and semiconfluent lysis of the bacterial lawn by PR772. Plaques produced by PR772 are translucent in appearance, indicating incomplete lysis of the surrounding bacteria. Phages were recovered from each plate by washing with 10 ml of phosphate-buffered saline (PBS) for 4 h at 20 to 25°C or overnight at 2 to 8°C with gentle agitation on an orbital shaker. The crude washes were centrifuged for 15 min at 10,000 × g and then filtered through a 0.45-μm-pore-size filter for direct use or further purification by CsCl density gradient ultracentrifugation (15). Each plate typically yielded between 4 × 1010 and 4 × 1011 PFU. The filtered supernatants intended for purification were centrifuged at 90,000 × g for 2 h. The phage pellet was resuspended overnight at 2 to 8°C in 7 ml of PBS. CsCl (∼3 g) was then added to bring the density of the phage suspension to 1.3 g/ml, and the mixture was ultracentrifuged for 16 h at 300,000 × g. Phage bands were visible if sufficient phage existed in the starting material (>1011 PFU). The CsCl gradient ultracentrifugation yielded two or more bands, with the top translucent white band usually containing the most viable phages. The phage band was drawn out of the ultracentrifuge tube with a needle and syringe and dialyzed for 24 h against three changes of PBS. The CsCl gradient purification method resulted in 11- to 59-fold concentration of the phages over crude lysates.
DNA preparation, subcloning, and sequencing.
About 7 × 1011 PFU of CsCl-purified PR772 were digested overnight at 50°C with 0.1 mg of proteinase K per ml in 5 ml of 100 mM NaCl-10 mM Tris (pH 8.0)-25 mM EDTA-0.5% sodium dodecyl sulfate. The digestion mixture was extracted for 1 h at 20 to 25°C with phenol-chloroform-isoamyl alcohol (25:24:1 volume ratio). After extraction, the phage DNA was precipitated from the aqueous phase by the addition of sodium acetate (0.3 M) and 2 volumes of ethanol. After centrifugation for 5 min at 10,000 × g, the DNA pellet was resuspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). The PR772 DNA was digested with RsaI (5′ GTAC 3′) or HaeIII (5′ GGCC 3′) and randomly cloned into the SmaI site of pUC18. All restriction enzymes were purchased from New England Biolabs (Beverly, Mass.). RsaI produces 20 visible fragments on an agarose gel (3,051, 1,880, 1,284, and 1,000 bp, a 931- and 949-bp doublet; 827, 760, and 493 bp, a 463- and 468-bp doublet; 410, 377, and 311 bp; a 261- and 279-bp doublet; and 210 bp, a triplet spanning 174 to 192 bp; data not shown) and an undeterminable number of bands of less than 150 bp (five according to sequence analysis). Similarly, HaeIII generates 12 small fragments (1,103, 839, 755, 723, 696, and 609 bp; a 512- and 523-bp doublet; a 447- and 456-bp doublet, and a 380- and 394-bp doublet; data not shown) and an undeterminable number of bands of less than 350 bp (57 by sequence analysis). The ligation mixtures were transformed into MAX efficiency competent E. coli DH5α cells (Gibco BRL) in accordance with the manufacturer's instructions and then plated on Luria-Bertani plates containing ampicillin (50 μg/ml) and grown overnight. Plasmid DNA minipreparations from single colonies grown overnight were made by a boiling lysis-isopropanol precipitation method. Clones were analyzed by restriction enzyme analysis (EcoRI/HindIII double digest) on a 1% Ultrapure (Gibco BRL, Gaithersburg, Md.) agarose gel. Plasmid clones with PR772 DNA inserts (29 RsaI subclones and 18 HaeIII subclones) were sequenced with pUC18-M13 forward, pUC18-M13 reverse, and internal primers from Amplicon Express (Pullman, Wash.) or Bioserve (Laurel, Md.). Assembly of the subclone sequences was done with the AssemblyLign software package included with MacVector (Accelrys, San Diego, Calif.). Sequencing of the subclone DNA generated a double-stranded sequence from ∼37% of the PR772 genome after assembly of overlapping fragments. Finally, gaps (nonsubcloned areas) in the genome sequence were completed with synthetic primers (Amplicon Express) and the PR772 genome as the template. The entire phage genome was sequenced on both DNA strands.
Bioinformatic analysis.
The sequence analysis were performed with the Genetic Computer Group (Madison, Wis.) sequence analysis software package, version 10.3, including GenBank release 138.0, EMBL (abridged) release 76.0, PIR-Protein release 77.08, NRL_3D release 28.0, SWISS-PROT release 42.0, and Pfam release 10.0. BLAST searches were performed with the database search programs available at http://www.ncbi.nlm.nih.gov/BLAST/ (1). The isoelectric point and molecular weight were calculated with tools available at http://us.expasy.org/tools/pi_tool.html.
TEM.
To confirm the morphology of PR772, phages were observed under an electron microscope as described previously (22). Phages from the crude lysate were sedimented at 25,000 × g for 60 min in a Beckman (Palo Alto, Calif.) ultracentrifuge (model J2-21, rotor JA-18.1), and pellets were washed twice in buffer (10 mM Tris-HCl, 1 mM MgSO4, 10 mM sodium azide, pH 7). A drop of phage suspension was then deposited on a carbon-coated Formvar grid and mixed with an equal volume of uranyl acetate (2%, pH 4.0). Excess liquid was withdrawn with filter paper. Specimens were studied with a Philips EM 300 electron microscope operated at 60 kV. Magnification was monitored with catalase crystals (Worthington, Freehold, N.J.) (20). Similar preparations of PR772 were also fixed with 2.5% glutaraldehyde, stained with osmium tetroxide, sectioned, and examined by TEM by JFE Enterprises (Brookville, Md.).
Filtration.
PR772 was diluted to ∼108 PFU/ml in PBS containing 15 mg of bovine serum albumin (Sigma, St. Louis, Mo.) per ml, 5 mM EDTA, and 0.1% Triton X-100. About 5 ml of phage was passed through 100-nm-rated membranes, either film-cast or track-etched membrane filters. The membrane manufacturing processes for film-cast and track-etched membranes differ; for detailed information, see reference 5. The film-cast membranes used were Millex-VV Durapore polyvinylidene difluoride (Millipore Corporation, Bedford, Mass.), Acrodisc Supor PES, and AcroPak 20 Fluorodyne II polyvinylidene difluoride (Pall Corporation, East Hills, N.Y.); the track-etched Isopore VCTP polycarbonate (Millipore Corporation) filter was also tested. The agar overlay method was used to enumerate phage before and after filtration through the 100-nm-pore-size-rated filters.
LLS analysis.
To perform laser light-scattering (LLS) analysis, three samples of CsCl-purified PR772, 1.0 × 10−5, 2.0 × 10−5, and 3.0 × 10−5 g/ml in PBS, were prepared by diluting the stock solution (A280 of about 0.77, assumed to correspond to a concentration of about 3.85 × 10−4 g/ml) to known concentrations in volumetric flasks. Each of the diluted solutions was filtered through a 0.2 μm-pore-size filter and directly used to fill a dust-free light-scattering cell for LLS experiments. A standard laboratory-built LLS spectrometer equipped with a BI-9000 AT digital correlator and a solid-state laser (200 mW, 532 nm; DPSS; Coherent Inc., Santa Clara, Calif.) was used to perform LLS studies over a scattering angular range of 20 to 120°C.
DLS analysis.
In dynamic light-scattering (DLS) analysis, the intensity-intensity time correlation function G(2)(t) in the self-beating mode was measured according to G(2)(t) = A[1+ β|g(1)(t)|2], where A is the measured baseline, β is a coherence factor, t is the delay time, and g(1)(t) is the normalized first-order electric field time correlation function. g(1)(t) is related to the line width distribu-tion G(Γ) by g(1)(t) = ∫∞0G(Γ)e−ΓtdΓ. By using a Laplace inversion program, CONTIN, the normalized distribution function of the characteristic line width, G(Γ), was obtained. The average line width, Γ̄, was calculated according to Γ̄ = ∫ΓG(Γ)dΓ. The polydispersity index, PDI, was defined as PDI = μ2/Γ̄2 with μ2 = ∫(Γ − Γ̄)2G(Γ)dΓ, where Γ̄ is a function of both C and q, which can be expressed as Γ̄/q2 = D(1 + kdC)[1 + f(Rgq)2] with D, kd, f, and C being the translational diffusive coefficient, the diffusion second virial coefficient, a dimensionless constant, and the concentration, respectively. The scattering vector, q, is defined as q = (4π n/λ)sin(θ/2) with n, λ, and θ being the solvent refractive index, the wavelength of light in a vacuum, and the scattering angle, respectively. When the concentration is extremely dilute and Rgq ≪ 1, Γ/q2 is approximately equal to D. D can be further converted into the hydrodynamic radius, Rh, by using the Stokes-Einstein equation, D = kBT/6πηRh, where kB, T, and η are the Boltzmann constant, the absolute temperature, and the viscosity of the solvent, respectively.
LLS calibration.
An aqueous suspension of latex spheres (Polybead Carboxylate Microsphere; Polyscience Inc., Warrington, Pa.) was used to calibrate the LLS setup. According to vendor claims, the latex spheres have a nominal diameter of 48.1 ± 5.2 nm. The size distribution of the latex spheres measured by DLS (the average Rh was 24 ± 2 nm) was consistent with the vendor claims, demonstrating that our setup was accurate, precise, and reliable. The size distribution of latex spheres was very narrow, with the PDI being on the order of less than 0.05.
Nucleotide sequence accession number.
The complete genome sequence (14,946 bp) of PR772 is available under GenBank accession number AY441783.
RESULTS
Comparative genome analysis.
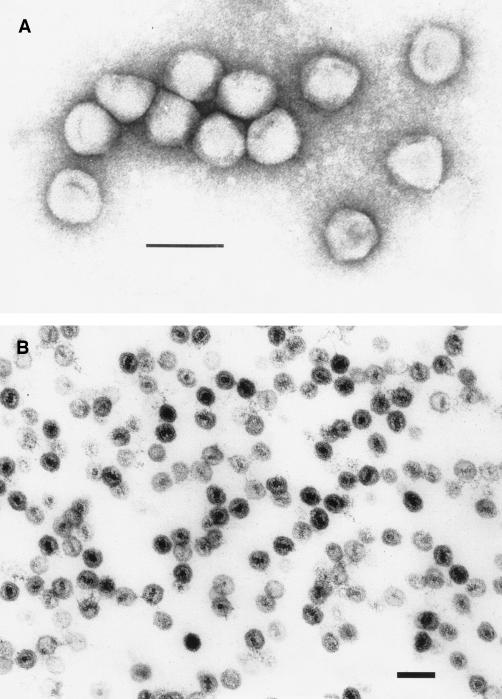
The entire genomic sequence of coliphage PR772 was resolved. Its linear DNA contains 14,946 bp with an overall GC content of 48.3 mol% G+C, which is very close to the 50.8 mol% G+C of E. coli strain K-12 (11). The PR772 5′ and 3′ ITR sequences are identical to each other and to the previously reported sequence (31). Five common endonucleases (BamHI, BglII, EcoRI, HindIII, and Sau3AI) do not cut the phage DNA. It has been previously noted that phages lack restriction enzyme sites, presumably as an evolutionary response to pressures from their host restriction enzymes, particularly members of the Tectiviridae family, which also have a broad host range (6, 7, 9). Bioinformatic analysis showed 32 ORFs of at least 40 codons (Table 1). The gene organization was very compact, resulting in only very few short intergenic regions. All of the ORFs except two (orf31 and orf32) were coded in the same orientation. Comparison of the PR772 and PRD1 genomes revealed that their DNA sequences are 97.2% identical (9). TEM of PR772 confirmed that it belongs to the Tectiviridae family and is structurally similar to PRD1 (Fig. 1). Unique restriction endonuclease fragments present in PR772 but not PRD1 include 3,051-, 1,284-, 493-, and 410-bp RsaI fragments and 1,103- and 839-bp HaeIII fragments (data not shown). The largest difference between the two genomes appears to be an insertion of 12 nucleotides (orf26-orf27, coordinates 12,729 to 12,740) accounting for most of the size difference (PR772, 14,946 bp; PRD1, 14,925 bp). According to the standard BLAST protocol, all 32 of the ORFs showed homology with the previously characterized gene products of PRD1. Moreover, 11 of the ORFs displayed 100% identity to PRD1 ORFs. They included the following: ORF1, terminal protein; ORF6, penton protein; ORF13, DNA-packaging ATPase; ORF14, unknown; ORF15, DNA packaging; ORF16, major capsid protein; ORF18, DNA packaging; ORF19, unknown; ORF20, DNA delivery; ORF21, DNA delivery; and ORF22, membrane protein of unknown function.
TABLE 1.
Comparison between gene products of PR772 and PRD1
| PR772
|
Descriptiona,c | PRD1
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | Coordinates | Length (amino acids) | Molecular weight | pI | Genea | ORFa,b | Coordinates | Protein | Molecular weight | pI | % amino acid identity | |
| 1 | 236. . .1015 | 259 | 29,514.32 | 10.15 | Genome terminal protein | VIII | 233. . .1012 | P8 | 29,514.23 | 10.79 | 100 | |
| 2 | 1019. . .2680 | 553 | 63,295.11 | 6.06 | DNA polymerase (N) | I | 1016. . .2677 | P1 | 63,336.03 | 6.68 | 99 | |
| 3 | 2637. . .3131 | 164 | 18,827.46 | 9.04 | Muramidase | XV | 2679. . .3128 | P15 | 17,268.62 | 8.75 | 99 | |
| 4 | 3131. . .4906 | 591 | 63,653.68 | 4.39 | Receptor binding | II | 3128. . .4903 | P2 | 63,821.56 | 4.19 | 95 | |
| 5 | 3456. . .3590 | 44 | 5,182.38 | 12.55 | b | 3453. . .3587 | 5,113.17 | 12.98 | 91 | |||
| 6 | 4910. . .5290 | 126 | 13,746.45 | 4.99 | Penton protein | XXXI | c | 4907. . .5287 | P31 | 13,746.41 | 4.75 | 100 |
| 7 | 5103. . .5297 | 64 | 7,297.95 | 12.02 | d | 5103. . .5294 | 7,096.00 | 12.38 | 97 | |||
| 8 | 5290. . .6312 | 340 | 34,465.13 | 4.51 | Spike protein, vertex | V | 5287. . .6309 | P5 | 34,449.03 | 4.40 | 99 | |
| 9 | 6331. . .6591 | 86 | 9,494.68 | 4.43 | Assembly (N) | XVII | 6328. . .6588 | P17 | 9,537.67 | 4.20 | 98 | |
| 10 | 6581. . .6787 | 68 | 7,436.65 | 4.40 | Assembly (N) | XXXIII | f | 6578. . .6784 | P33 | 7,450.66 | 4.10 | 96 |
| 11 | 6787. . .7287 | 166 | 17,401.25 | 3.98 | Minor capsid protein | VI | 6784. . .7284 | P6 | 17,573.38 | 3.64 | 91 | |
| 12 | 7032. . .7643 | 203 | 20,687.30 | 7.80 | Assembly (N) | X | 7029. . .7640 | P10 | 20,688.19 | 8.44 | 95 | |
| 13 | 7640. . .8323 | 227 | 25,786.90 | 9.00 | DNA packaging ATPase | IX | 7637. . .8320 | P9 | 25,786.82 | 9.34 | 100 | |
| 14 | 8335. . .8463 | 42 | 4,541.35 | 5.90 | i | 8332. . .8460 | 4,541.33 | 6.53 | 100 | |||
| 15 | 8463. . .8591 | 42 | 4,670.63 | 6.53 | DNA packaging (M) | XX | j | 8460. . .8588 | P20 | 4,670.61 | 7.36 | 100 |
| 16 | 8598. . .9785 | 395 | 43,474.91 | 5.30 | Major capsid protein | III | 8595. . .9782 | P3 | 43,447.62 | 4.98 | 100 | |
| 17 | 9430. . .9684 | 84 | 9,271.11 | 12.30 | h | 9427. . .9681 | 9,295.16 | 12.37 | 96 | |||
| 18 | 9804. . .9947 | 47 | 5,492.63 | 5.00 | DNA packaging (M) | XXII | k | 9801. . .9944 | P22 | 5,492.61 | 4.54 | 100 |
| 19 | 10047. . .10169 | 40 | 4,399.20 | 9.52 | l | 10044. . .10166 | 4,399.18 | 9.78 | 100 | |||
| 20 | 10171. . .10443 | 90 | 9,788.88 | 9.77 | DNA delivery (M) | XVIII | m | 10168. . .10440 | P18 | 9,788.85 | 10.39 | 100 |
| 21 | 10443. . .10607 | 54 | 5,457.50 | 9.99 | DNA delivery (M) | XXXII | n | 10440. . .10604 | P32 | 5,457.48 | 10.79 | 100 |
| 22 | 10620. . .10826 | 68 | 6,694.74 | 8.34 | (M) | XXXIV | o | 10617. . .10823 | P34 | 6,694.72 | 9.30 | 100 |
| 23 | 10836. . .11090 | 84 | 9,270.63 | 5.61 | Minor capsid protein | XXX | p | 10833. . .11087 | P30 | 9,186.53 | 7.69 | 97 |
| 24 | 11205. . .11828 | 207 | 22,282.11 | 7.83 | DNA delivery | XI | 11202. . .11825 | P11 | 22,209.02 | 8.71 | 71 | |
| 25 | 11839. . .12192 | 117 | 12,608.34 | 8.64 | Infectivity (M) | XVI | s | 11836. . .12189 | P16 | 12,551.21 | 8.68 | 94 |
| 26 | 12193. . .13002 | 269 | 27,454.92 | 4.69 | Transglycosylase (M) | VII | 12190. . .12987 | P7 | 27,073.50 | 4.92 | 96 | |
| 27 | 12538. . .13002 | 154 | 15,457.43 | 4.22 | DNA delivery (M) | XIV | 12535. . .12987 | P14 | 15,014.02 | 4.31 | 96 | |
| 28 | 12999. . .13352 | 117 | 12,763.80 | 5.73 | t | 12984. . .13337 | 12,777.79 | 5.80 | 99 | |||
| 29 | 13321. . .13707 | 128 | 13,583.77 | 9.52 | u | 13390. . .13692 | 10,588.01 | 6.85 | 77 | |||
| 30 | 13631. . .13903 | 90 | 10,283.11 | 8.86 | v | 13616. . .13888 | 10,230.03 | 8.32 | 99 | |||
| 31 | 14147. . .13863 | 94 | 10,430.90 | 5.24 | Single-stranded DNA-binding protein (N) | XIX | 14132. . .13848 | P19 | 10,460.89 | 5.00 | 98 | |
| 32 | 14702. . .14220 | 160 | 16,664.08 | 6.62 | Single-stranded DNA-binding protein (N) | XII | 14687. . .14205 | P12 | 16,650.00 | 7.52 | 99 | |
For ORF h, the molecular weight and pI were calculated with http://us.expasy.org/tools/pi_tool.html.
M, integral membrane protein; N, nonstructural protein.
FIG. 1.
TEM of PR772. (A) Phage PR772 stained with 2% uranyl acetate. Original magnification, ×297,000; bar, 100 nm. (B) Sectioned PR772 fixed with 2.5% glutaraldehyde and stained with osmium tetroxide. Original magnification, ×150,000; bar, 100 nm.
The most divergent deduced protein was ORF24, which was 71% identical to P11 of PRD1, an integral membrane protein responsible for viral infectivity and DNA delivery (8). Most of the differences were due to three frame shifts in the 5′ region of orf24 (data not shown). Another significant difference was in ORF29 (128 amino acids) of PR772, which was larger by 28 amino acids than its homologous protein (ORF u, 100 amino acids) in PRD1 (Table 1). This increase was due to the insertion of one nucleotide (coordinate 13,382) within the gene that changed the reading frame.
Filtration.
As an initial test of monodispersion of crude and CsCl gradient-purified PR772 preparations, phage titer losses were tracked before and after filtration through 100-nm-pore-size film-cast and track-etched filters. As shown in Table 2, most PR772 phages passed through 100-nm-pore-size filters from various manufacturers. Additionally, there is no significant difference in titer reduction with either crude or CsCl gradient-purified preparations of PR772. Moreover, the three separate CsCl gradient preparations of PR772 did not differ significantly in their filtration properties. Thus, prefiltration of PR772 through 100-nm-pore-size filters is feasible prior to use in virus-retentive filter rating studies.
TABLE 2.
Filter sizing of PR772 preparation with 100-nm-pore-size filters
| Filter type and name | Phage prepn | Phage titer
|
% Passage | |
|---|---|---|---|---|
| Before filtration | After filtration | |||
| 100-nm film cast | ||||
| Millex-VV Durapore PVDF | A (plate lysate) | 6.1 × 107 ± 0.4 × 107 | 8.9 × 107 ± 2.1 × 107 | 145 |
| B (CsCl gradient) | 4.8 × 108 | 4.7 × 108 | 97 | |
| C (CsCl gradient) | 8.1 × 108 ± 5.9 × 108 | 6.0 × 108 ± 1.8 × 108 | 74 | |
| D (CsCl gradient) | 7.0 × 108 ± 2.3 × 108 | 3.1 × 108 ± 1.6 × 108 | 45 | |
| Acrodisc Supor | C | 8.1 × 108 ± 5.9 × 108 | 4.2 × 108 ± 0.8 × 108 | 51 |
| D | 7.0 × 108 ± 2.3 × 108 | 3.7 × 108 ± 1.4 × 108 | 53 | |
| AcroPak 20 Fluorodyne II | C | 8.1 × 108 ± 5.9 × 108 | 3.3 × 108 ± 2.6 × 108 | 41 |
| D | 7.0 × 108 ± 2.3 × 108 | 7.0 × 108 ± 4.4 × 108 | 100 | |
| 100-nm track etched, Isopore VCTP | C | 8.1 × 108 ± 5.9 × 108 | 4.2 × 108 ± 1.6 × 108 | 51 |
| D | 7.0 × 108 ± 2.3 × 108 | 2.9 × 108 ± 0.6 × 108 | 41 | |
DLS.
The suitability of PR772 for virus filter testing depends on two physical properties, i.e., diameter and size uniformity. On the basis of multiple EM studies, members of the family Tectiviridae are generally accepted by the International Committee on Taxonomy of Viruses to have a physical diameter of 63 nm. However, it is of interest to demonstrate if additional physical techniques can be used to measure the size of PR772, as well as other properties. DLS is a technique that measures temporal intensity fluctuations in scattered light due to the Brownian motion of particles (14). With the Stokes-Einstein equation [D = kBT/(6πηRh)], the particle diffusion coefficient (D) measured by DLS can be processed to yield the Rh and then the hydrodynamic diameter. For non-solvent-interacting, solid, spherical particles, Rh represents the physical size. For solvent-interacting, irregularly shaped, and nondraining particles (e.g., most proteins and viruses), Rh depends on the conformation and hydration state. However, while the hydrodynamic diameter of a phage measured by DLS may be different from the diameter measured by EM, it is highly reflective of the actual particle translational behavior in the fluid.
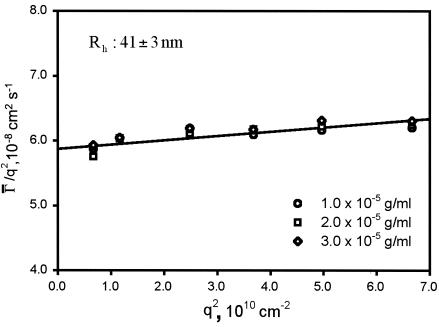
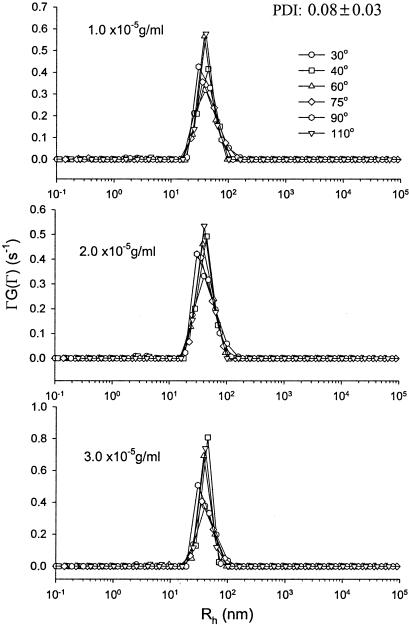
DLS measurements of phage PR772 were performed within 24 h after sample preparation. Each sample was measured at six angles at least three times. Besides the PR772 samples, the solvents (PBS buffer and benzene) were also scanned as references. During DLS measurements, the difference between the measured baseline and the calculated baseline was less than 0.1% and the measured baseline was accumulated to at least 107 cps. Table 3 shows the gamma (Γ̄/s−1) values obtained by CONTIN analysis of all the three PR772 samples measured at six different scattering angles. Figure 2 shows plots of Γ̄/q2 versus q2. No concentration dependence was observed. The translational diffusion coefficient, D, was (5.9 ± 0.4) × 10−8 cm2 s−1. The Rh, calculated accordingly from the Stokes-Einstein equation, was 41 ± 3 nm. Thus, the estimated hydrodynamic diameter of PR772 is 82 ± 6 nm. Figure 3 shows the size distribution of phage PR722 at different scattering angles for all three samples. Only a unimodal size distribution was observed in the system, and its distribution was relatively narrow, with a PDI of 0.08 ± 0.03. The low degree of polydispersity of the phage PR772 preparation, 0.08, was comparable to that of the latex bead standards used for calibration (<0.05). In summary, phage PR772 preparations made by CsCl gradient banding are highly monodispersed suspensions of particles with a hydrodynamic diameter of ∼82 nm.
TABLE 3.
Gamma (Γ̄/102 s−1) values measured at different angles
| Concn (g/ml) and measurement no. | Value at angle of:
|
|||||
|---|---|---|---|---|---|---|
| 30° | 40° | 60° | 75° | 90° | 110° | |
| 1.0 × 10−5 | ||||||
| 1 | 3.92 | 6.91 | 15.8 | 22.4 | 30.3 | 41.8 |
| 2 | 3.94 | 7.10 | 15.2 | 22.6 | 31.0 | 40.8 |
| 3 | 3.89 | 7.05 | 15.2 | 22.3 | 30.6 | 41.4 |
| Avg | 3.91 | 7.02 | 15.4 | 22.4 | 30.6 | 41.4 |
| 2.0 × 10−5 | ||||||
| 1 | 3.96 | 7.10 | 15.3 | 22.0 | 31.3 | 41.2 |
| 2 | 3.72 | 6.91 | 15.1 | 22.9 | 30.7 | 42.4 |
| 3 | 3.82 | 7.05 | 15.2 | 23.4 | 30.7 | 41.3 |
| Avg | 3.83 | 7.02 | 15.2 | 22.8 | 30.9 | 41.6 |
| 3.0 × 10−5 | ||||||
| 1 | 3.96 | 6.91 | 15.7 | 22.8 | 31.6 | 41.4 |
| 2 | 3.86 | 7.14 | 14.8 | 22.6 | 31.2 | 42.4 |
| 3 | 4.03 | 7.05 | 15.7 | 22.7 | 31.3 | 42.4 |
| Avg | 3.95 | 7.03 | 15.4 | 22.7 | 31.4 | 42.1 |
FIG. 2.
Plots of Γ̄/q2 versus q2 for PR772 at estimated concentrations of 1.0 × 10−5, 2.0 × 10−5, and 3.0 × 10−5 g/ml.
FIG. 3.
CONTIN results for the size distribution of PR772 at estimated concentrations of 1.0 × 10−5, 2.0 × 10−5, and 3.0 × 10−5 g/ml.
DISCUSSION
Filter manufacturers currently use different parameters to name virus retention filters (2), making the filter nomenclature ambiguous and confusing to the end user. Recently, a consensus has emerged among filter manufacturers and end users in the biopharmaceutical industry about the desirability of a common nomenclature system to facilitate performance comparisons between virus removal filters from different manufacturers. Bacteriophages have been used for decades in medical size-based removal applications (3); more recently, filter manufacturers have used phages to evaluate the size exclusion properties of their viral removal filters (3, 4, 12, 19, 25-27). On the basis of its reported 53- to 63-nm diameter (6, 15, 16) and its previous successful use in testing size exclusion properties of large-virus filters (4, 12, 24-26), PR772 has been suggested by the Parenteral Drug Association virus filter task force to be a potential model bacteriophage to standardize nomenclature for large-pore-size virus filters (G. Sofer, unpublished data). This study was designed to conduct additional work and expand on the previous characterization of PR772 published several decades ago. Other phages in the Tectiviridae family (e.g., PRD1) have been extensively characterized, and a similar characterization of PR772 is important to justify its use in filter studies. In this report, we have characterized the size, genome, structure, and other properties of PR772 important for its use in filter testing.
This phage is easy to handle and stable, and high phage titers can be obtained readily. Unlike other members of the family Tectiviridae (e.g., PRD1, which is usually propagated on S. enterica serovar Typhimurium), production of PR772 does not involve the handling of pathogenic host bacteria. Phage PR772 titers can be rapidly and easily enumerated by plaque-counting techniques. Thus, low levels of contaminating nucleic acids, a concern for quantitative PCR assay of mammalian viruses (36), are unlikely to interfere with measurements of PR772 titers. Analysis of the complete genomic sequence of PR772 revealed a high level of identity (97%) with the genome of the well-characterized tectivirus PRD1. It contains a similar set of genes and presumed gene products, with minimal substitutions and additional nucleotides. In fact, the overall gene order of the PRD1 and PR772 genomes is so conserved that putative functions were assigned to almost all of the gene products. The differences between these two phage genomes are mainly due to point mutations and to a single insertion-deletion of 12 nucleotides in one ORF. Detailed bioinformatic analysis did not reveal undesired DNA sequences (e.g., virulence factor, antibiotic resistance), indicating that this phage-host system appears to be suitable for routine laboratory work. The availability of its complete nucleotide sequence also provides a powerful tool for quality control of the phage preparations.
The hydrodynamic diameter of phage PR772 measured by light scattering (82 nm) differs modestly from the generally accepted physical diameter of members of the family Tectiviridae (63 nm) (6). Unlike EM, DLS measures the hydrodynamic size of hydrated particles in a fluid (14). Thus, it could be argued that the size of PR772 measured in this way should be more reflective of the actual hydrated size of PR772 particles during an actual filtration process than the dry phage size determined by EM. Both measurements of the diameter of members of the family Tectiviridae are somewhat smaller than that of endogenous rodent retroviruses or human immunodeficiency virus (80 to 100 nm) but still large enough to be considered nonfilterable through the large-pore-size virus-retentive filters that are designed to retain viruses larger than 50 to 60 nm in size (e.g., Pall Ultipor VF DV50, Millipore Viresolve NFR, and Asahi Planova 35N filters). Thus, testing with PR772 represents a worst-case condition for large-pore-size filters. Therefore, PR772's size appears to be appropriate for testing of these virus filters. Both CsCl gradient-purified and plate lysate PR772 preparations were similarly filterable by 100-nm-pore-size filters; thus, these filters can be used in a PR772 prefiltration step prior to use in virus-retentive filter rating. In addition, our light-scattering experiments indicated that live PR772 phages prepared on CsCl gradients are almost entirely monodispersed, a critical attribute for their use as model phages in virus-retentive filter testing.
In summary, the structural and molecular properties of PR772 favor its use as an appropriate model bacteriophage for nomenclature standardization of larger-pore-size virus-retentive filters. In addition, our data are consistent with the use of PR772 as a potential model virus for preliminary evaluation of viral clearance provided by virus-retentive filters in scaled-down studies (conducted under process conditions) designed to demonstrate size-based filtration clearance.
Acknowledgments
J. Siegel and K. Epstein assisted in preparing and screening PR772 subclones for sequence analysis. J. Carter provided Millipore 100-nm filter samples. J. Martin offered support and advice for this project and provided Pall 100-nm filter samples. J. Higbee (Amplicon Express) and S. Fitzsimmons (CDER/FDA) advised us in DNA sequencing and sequence assembly. J. Brown, P. Krause, and S. Kozlowski critically reviewed the manuscript.
This work was supported in part by a grant from the Natural Sciences and Engineering Research Council of Canada (S.M.).
The opinions expressed in this report are those of the authors and not necessarily those of the Food and Drug Administration or the U.S. Government.
REFERENCES
- 1.Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aranha, H. 2001. Viral clearance strategies for biopharmaceutical safety. Part 2: filtration for viral clearance. Biopharm 14:1-8. [Google Scholar]
- 3.Aranha-Creado, H., and H. Brandwein. 1999. Application of bacteriophages as surrogates for mammalian viruses: a case for use in filter validation based on precedents and current practices in medical and environmental virology. PDA J. Pharm. Sci. Technol. 53:75-82. [PubMed] [Google Scholar]
- 4.Aranha-Creado, H., K. Oshima, S. Jafari, G. Howard, Jr., and H. Brandwein. 1997. Virus retention by a hydrophilic triple-layer PVDF microporous membrane filter. PDA J. Pharm. Sci. Technol. 51:119-124. [PubMed] [Google Scholar]
- 5.Baker, R. W. 1991. Membrane separation systems—recent developments and future directions, vol. 2. Noyes/William Andrew Publishing, Park Ridge, N.J.
- 6.Bamford, D. H., and H.-W. Ackermann. 2000. Family Tectiviridae, p. 111-116. In M. Van Regenmortel, C. Fauquet, D. Bishop, E. Carstens, M. Estes, S. Lemon, J. Maniloff, M. Mayo, D. McGeoch, C. Pringle, and R. Wickner (ed.), Virus taxonomy, classification and nomenclature of viruses: 7th report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif..
- 7.Bamford, D. H., J. Caldentey, and J. K. Bamford. 1995. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45:281-319. [DOI] [PubMed] [Google Scholar]
- 8.Bamford, J. K., J. J. Cockburn, J. Diprose, J. M. Grimes, G. Sutton, D. I. Stuart, and D. H. Bamford. 2002. Diffraction quality crystals of PRD1, a 66-MDa dsDNA virus with an internal membrane. J. Struct. Biol. 139:103-112. [DOI] [PubMed] [Google Scholar]
- 9.Bamford, J. K., A. L. Hanninen, T. M. Pakula, P. M. Ojala, N. Kalkkinen, M. Frilander, and D. H. Bamford. 1991. Genome organization of membrane-containing bacteriophage PRD1. Virology 183:658-676. [DOI] [PubMed] [Google Scholar]
- 10.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 11.Blattner, F., G. Plunkett, C. Bloch, N. Perna, V. Burland, M. Riley, J. Collado-Vides, J. Glasner, C. Rode, G. Mayhew, J. Gregor, N. Davis, H. Kirkpatrick, M. Goeden, D. Rose, B. Mau, and Y. Shao. 1997. The complete genome of Escherichia coli K-12. Science 5331:1453-1474. [DOI] [PubMed] [Google Scholar]
- 12.Brough, H., C. Antoniou, J. Carter, J. Jakubik, Y. Xu, and H. Lutz. 2002. Performance of a novel Viresolve NFR virus filter. Biotechnol. Prog. 18:782-795. [DOI] [PubMed] [Google Scholar]
- 13.Burns, D. B., and A. L. Zydney. 1999. Effect of solution pH on protein transport through ultrafiltration membranes. Biotechnol. Bioeng. 64:27-37. [DOI] [PubMed] [Google Scholar]
- 14.Chu, B. 1991. Laser light scattering, 2nd ed. Academic Press, Inc., New York, N.Y.
- 15.Coetzee, J. N., G. Lecatsas, W. F. Coetzee, and R. W. Hedges. 1979. Properties of R plasmid R772 and the corresponding pilus-specific phage PR772. J. Gen. Microbiol. 110:263-273. [DOI] [PubMed] [Google Scholar]
- 16.Coetzee, W. F., and P. J. Bekker. 1979. Pilus-specific, lipid-containing bacteriophages PR4 and PR772: comparison of physical characteristics of genomes. J. Gen. Virol. 45:195-200. [DOI] [PubMed] [Google Scholar]
- 17.Earnshaw, W. C., J. King, and F. A. Eiserling. 1978. The size of the bacteriophage T4 head in solution with comments about the dimension of virus particles as visualized by electron microscopy. J. Mol. Biol. 122:247-253. [DOI] [PubMed] [Google Scholar]
- 18.Garnick, R. 1998. Raw materials as a source of contamination in large-scale cell culture. Dev. Biol. Stand. 93:21-29. [PubMed] [Google Scholar]
- 19.Graf, E. G., E. Jander, A. West, H. Pora, and H. Aranha-Creado. 1999. Virus removal by filtration. Dev. Biol. Stand. 99:89-94. [PubMed] [Google Scholar]
- 20.Luftig, R. 1967. An accurate measurement of the catalase crystal period and its use as an internal marker for electron microscopy. J. Ultrastruct. Res. 20:91-102. [DOI] [PubMed] [Google Scholar]
- 21.Menon, M. K., and A. L. Zydney. 1999. Effect of ion binding on protein transport through ultrafiltration membranes. Biotechnol. Bioeng. 63:298-307. [PubMed] [Google Scholar]
- 22.Moineau, S., J. Fortier, H.-W. Ackermann, and S. Pandian. 1992. Characterization of lactococcal bacteriophages from Quebec cheese plants. Can. J. Microbiol. 38:875-882. [Google Scholar]
- 23.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshima, K. H., T. Evans-Strickfaden, and A. Highsmith. 1998. Comparison of filtration properties of hepatitis B virus, hepatitis C virus and simian virus 40 using a polyvinylidene fluoride membrane filter. Vox Sang. 75:181-188. [PubMed] [Google Scholar]
- 25.Oshima, K. H., T. T. Evans-Strickfaden, A. K. Highsmith, and E. W. Ades. 1995. The removal of phages T1 and PP7, and poliovirus from fluids with hollow-fiber ultrafilters with molecular weight cut-offs of 50,000, 13,000, and 6,000. Can. J. Microbiol. 41:316-322. [DOI] [PubMed] [Google Scholar]
- 26.Oshima, K. H., T. T. Evans-Strickfaden, A. K. Highsmith, and E. W. Ades. 1996. The use of a microporous polyvinylidene fluoride (PVDF) membrane filter to separate contaminating viral particles from biologically important proteins. Biologicals 24:137-145. [DOI] [PubMed] [Google Scholar]
- 27.Phillips, M. W., and A. J. DiLeo. 1996. A validatible porosimetric technique for verifying the integrity of virus-retentive membranes. Biologicals 24:243-253. [DOI] [PubMed] [Google Scholar]
- 28.Ravantti, J., A. Gaidelyte, D. H. Bamford, and J. K. Bamford. 2003. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrate a viral lineage. Virology 313:401-414. [DOI] [PubMed] [Google Scholar]
- 29.San Martin, C., R. M. Burnett, F. de Haas, R. Heinkel, T. Rutten, S. D. Fuller, S. J. Butcher, and D. H. Bamford. 2001. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure 9:917-930. [DOI] [PubMed] [Google Scholar]
- 30.San Martin, C., J. T. Huiskonen, J. K. Bamford, S. J. Butcher, S. D. Fuller, D. H. Bamford, and R. M. Burnett. 2002. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nat. Struct. Biol. 9:756-763. [DOI] [PubMed] [Google Scholar]
- 31.Savilahti, H., and D. H. Bamford. 1986. Linear DNA replication: inverted terminal repeats of five closely related Escherichia coli bacteriophages. Gene 49:199-205. [DOI] [PubMed] [Google Scholar]
- 32.Savilahti, H., and D. H. Bamford. 1993. Protein-primed DNA replication: role of inverted terminal repeats in the Escherichia coli bacteriophage PRD1 life cycle. J. Virol. 67:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokolova, A., M. Malfois, J. Caldentey, D. I. Svergun, M. H. Koch, D. H. Bamford, and R. Tuma. 2001. Solution structure of bacteriophage PRD1 vertex complex. J. Biol. Chem. 276:46187-46195. [DOI] [PubMed] [Google Scholar]
- 34.van Reis, R., and A. Zydney. 2001. Membrane separations in biotechnology. Curr. Opin. Biotechnol. 12:208-211. [DOI] [PubMed] [Google Scholar]
- 35.Verheust, C., G. Jensen, and J. Mahillon. 2003. pGIL01, a linear tectiviral plasmid prophage originating from Bacillus thuringiensis serovar israelensis. Microbiology (Reading) 149:2083-2092. [DOI] [PubMed] [Google Scholar]
- 36.Xu, Y., and K. Brorson. 2003. An overview of quantitative PCR assays for biologics quality and safety evaluations. Dev. Biol. 113:89-98. [PubMed] [Google Scholar]