Abstract
Objective(s):
This study addresses feature selection for breast cancer diagnosis. The present process uses a wrapper approach using GA-based on feature selection and PS-classifier. The results of experiment show that the proposed model is comparable to the other models on Wisconsin breast cancer datasets.
Materials and Methods:
To evaluate effectiveness of proposed feature selection method, we employed three different classifiers artificial neural network (ANN) and PS-classifier and genetic algorithm based classifier (GA-classifier) on Wisconsin breast cancer datasets include Wisconsin breast cancer dataset (WBC), Wisconsin diagnosis breast cancer (WDBC), and Wisconsin prognosis breast cancer (WPBC).
Results:
For WBC dataset, it is observed that feature selection improved the accuracy of all classifiers expect of ANN and the best accuracy with feature selection achieved by PS-classifier. For WDBC and WPBC, results show feature selection improved accuracy of all three classifiers and the best accuracy with feature selection achieved by ANN. Also specificity and sensitivity improved after feature selection.
Conclusion:
The results show that feature selection can improve accuracy, specificity and sensitivity of classifiers. Result of this study is comparable with the other studies on Wisconsin breast cancer datasets.
Keywords: Breast cancer, Classification feature, Selection data mining
Introduction
A major class of problems in medical science involves the diagnosis of disease, based on a number of tests done on the patients. Because of welter of data, the ultimate diagnosis may be difficult to obtain, even for a medical expert.
Improvements in facilities caused very large databases can be collected in medicine which needs to discover relationships buried in data. Data mining approaches in medical domain are using intensively for these purposes (1, 2). One of the application areas of analysing database is automated diagnostic systems. These systems can help doctors in their decision making. Another application is finding ways to improve patient outcome, reduce cost and enhance clinical studies. In addition, need for automated diagnosis has been most acute in case of deadly disease like cancer where early detection can greatly enhance the chances of long-term survival and reduce the costs. Breast cancer considered the most common invasive cancer in women. In USA, it is considered to be second leading cause of mortality among women and the most common cause of mortality in the age group 40 to 55 years women (3). The effectiveness of early detection has been proven to reduce a lot of mortality among patients with breast cancer (4).
There are three classical methods available for detecting breast cancer: physical exam, mammography and biopsy including Fine needle aspiration biopsy (FNAB or FNAC), Core needle biopsy, Surgical biopsy, Lymph node biopsy (5).
Mammography is one of the most used methods to detect the breast cancer. In literature, radiologists show considerable variation in interpreting a mammography (6). Accuracy of mammography varies from 68 % to 79% (7). When mammography detects a tumour, biopsy is required to determine its malignancy. The accuracy of surgical biopsy is nearly 100% but it is costly, invasive, time consuming and painful. FNAC is also widely adopted in the diagnosis of breast cancer. The accuracy of FNAC with visual interpretation varies from 35% to 95% depending on the experience of a doctor (8). So, it is necessary to develop better identification methods to recognize the breast cancer. These identification methods can help to assign patients to either a ‘benign’ group that does not have breast cancer or a ‘malignant’ group who has strong evidence of having breast cancer.
Malignant tumours generally are more serious than benign tumours. As mentioned, early detection of breast cancer leads to much higher chances of successful treatment. In order to reach this goal, it is necessary to have diagnostic systems with high levels of accuracy and reliability that help doctors to distinguish between benign breast tumours and malignant ones.
One of the problems in diagnostic systems is the multiplicity of features. Irrelevancy and redundancy in these features increase the confusion of classification algorithm and decrease learning precision (9, 10). Feature selection is one of the methods that can cope with this problem and plays an important role in classification. Feature selection is one of the pre-processing techniques in data mining and extensively used in the fields of statistics, pattern recognition and medical domain.
There are three approaches for feature selection including Wrapper, Filter and Embedded (11). In wrapper approach the goodness of selected subset of features determined by learning and evaluating a classifier using only the variables included in the proposed subset. Filter approach uses some techniques to score the selected subset, ignoring classifier algorithm. In other word goodness of selected subset of features determined by using only intrinsic properties of the data (12). In embedded approach, selecting the best subset of features is performed during the model construction process.
A good amount of research on breast cancer datasets using feature selection methods is found in literature such as ant colony algorithm (13), a discrete particle swarm optimization method (14), wrapper approach with genetic algorithm (15), support vector-based feature selection using fisher’s linear discriminate and support vector machine (16), fast correlation based feature selection (FCBF), multi thread based FCBF feature selection and decision dependent-decision independent correlation (DDC- DIC) (17), Rough set K-Means Clustering (18), modification correlation rough set feature selection (MCRSFS) (19).
In this study a wrapper feature selection method is proposed based on genetic algorithm based feature selection. This model employed particle swarm optimization algorithm based classifier (PS-classifier) as fitness function. The model evaluated on Wisconsin breast cancer databases.
Materials and Methods
Dataset Description (Wisconsin breast cancer databases)
In this study, the Wisconsin breast cancer datasets from UCI Machine Learning Repository is used (20). They have been collected by Dr. William H. Wolberg (1989–1991) at the University of Wisconsin–Madison Hospitals. The detail of these datasets is shown in table 1.
Table 1.
Wisconsin breast cancer datasets (18)
| Dataset | No. of attribute | No. of instances | No. of class |
|---|---|---|---|
| Wisconsin breast cancer (WBC) | 11 | 699 | 2 |
| Wisconsin diagnosis breast cancer (WDBC) | 32 | 569 | 2 |
| Wisconsin prognosis breast cancer (WPBC) | 34 | 198 | 2 |
In WBC dataset there are 699 records that each record has nine attributes expect of id number and class. These nine attributes are graded on an interval scale from a normal state of 1–10, with 10 being the most abnormal state (Table 2). In this database, 241 (65.5%) records are malignant and 458 (34.5%) records are benign.
Table 2.
Wisconsin breast cancer (WBC) Attribute (20)
| # Attribute | Domain | |
|---|---|---|
| 1 | Sample code number | Id number |
| 2 | Clump thickness | 1 – 10 |
| 3 | Uniformity of cell size | 1 – 10 |
| 4 | Uniformity of cell shape | 1 – 10 |
| 5 | Marginal adhesion | 1 – 10 |
| 6 | Single epithelial cell size | 1 – 10 |
| 7 | Bare nuclei | 1 – 10 |
| 8 | Bland chromatin | 1 – 10 |
| 9 | Normal nucleoli | 1 – 10 |
| 10 | Mitoses | 1 – 10 |
| 11 | Class | (2 for benign, 4 for malignant) |
In WDBC there are 569 records that each record has thirty attributes expect of id number and class. Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image.
Ten real-valued features are computed for each cell nucleus:
“radius (mean of distances from center to points on the perimeter)
texture (standard deviation of gray-scale values)
perimeter
area
smoothness (local variation in radius lengths)
compactness (perimeter^2 / area - 1.0)
concavity (severity of concave portions of the contour)
concave points (number of concave portions of the contour)
symmetry
fractal dimension (“coastline approximation”- 1)” (20).
The mean, standard error, and “worst” or largest (mean of the three largest values) of these features were computed for each image, resulting in 30 features. For instance, field 3 is Mean Radius, field 13 is Radius SE and field 23 is Worst Radius.
The WPBC and WDBC have the same features yet the WPBC has two additional features as follows:
Tumour size that is the diameter of the excised tumour in centimeters and lymph node status that is number of positive axillary lymph nodes observed at time of surgery.
Feature selection
Feature selection is a process that reduces the number of attributes and selects a subset of original features. Feature selection is often used in data pre-processing to identify relevant features that are often unknown previous and removes irrelevant or redundant features which do not have significance in classification task. Feature selection aims to improve the classification accuracy (9).
Genetic algorithm
Genetic algorithm (GA), originally developed by Holland, is a computational optimization paradigm modelled on the concept of biological evolution (21). The GA is an optimization procedure that operates in binary search spaces and manipulates a population of potential solutions. A point in the search space is represented by a finite sequence of 0s and 1s, called a chromosome. The quality of possible solutions is evaluated by a fitness function. The probability of survival is proportional to the chromosome’s fitness value. In GA, the initial population is randomly generated by three operators: selection, crossover, and mutation. The selection operator selects elites to transfer directly to next generation. The crossover operator randomly swaps a portion of chromosomes between two chosen parents to produce offspring chromosomes. The mutation operator randomly alerts a bit in chromosomes.
In this work GA is used to eliminate insignificant features. In order to reach this purpose, we defined chromosomes as a mask for features. In other word, each chromosome is a subset of features. The size of chromosome (number of genes) is equal to the number of features that represent the specification of a cancer patient. As mentioned, a chromosome is represented in form of binary string that is 0 or 1. 1 means the corresponding feature is selected and 0 means it is not selected (Figure 1).
Figure 1.
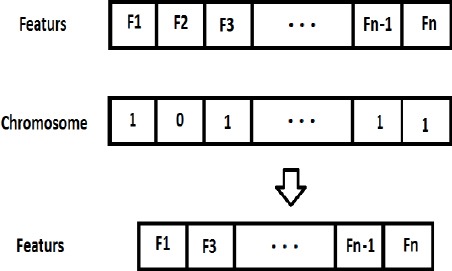
Generating initial population
Evaluation function
The goal of the proposed model is selecting the best subset of features that can produce the highest classification accuracy for diagnosis and prognosis the breast cancer. Therefore, the best subset of features should be selected. For selecting the best subset, a function is needed to evaluate the result of selecting each subset of features (chromosome).
In this work we used a classifier based on the particle swarm optimization algorithm (PS-classifier) which is a novel classifier that proposed by Zahiri and Seyedin (22).
The particle swarm optimization developed by Kennedy and Eberhart (23). This optimization method is based on the behaviour of swarm of bees or flock of birds while searching for food. In PSO, the particles fly through the problem space by following the optimal particles. Each particle remembers the best position that it has visited (Pbest) and also best position among all the particles in the population (Gbest). The position of each particle changes according to the Pbest and Gbest in the problem space.
In PS-classifier, PSO algorithm is used to find the decision hyper planes between the different classes. Decision hyper planes are employed to divide feature space into individual regions. Each region is assigned to a specific class.
A general hyper plane is in the form of
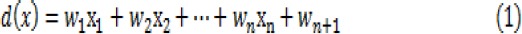
where X=(x1, x2, …, xn) and W=(w1, w2, …, wn+1) are called the augmented feature and weight vector, respectively. n is the feature space dimension.
In a general case, there are a number of hyper planes that separate the feature space to different regions, that each region distinguishes an individual class (Figure 2).
Figure 2.
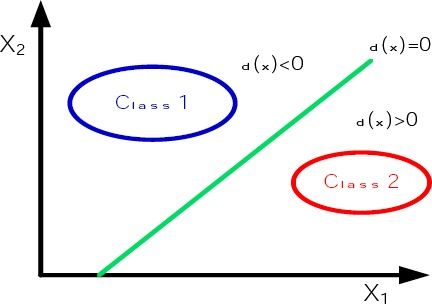
Separating two classes with one hyper plane
The PS-classifier must find Wj (j=1, 2, …, H) in solution space, where H is the necessary number of decision hyper planes.
Fitness function of PS-classifier is defined as follow:

where Miss is the number of misclassified data points by W.
Feature selection process
The feature selection process is represented in Figure 3. It is observed that GA selects subset of features as chromosomes and each chromosome is sent to the PS-classifier for calculating fitness value. PS-classifier uses each chromosome as mask for features. So that each gene on chromosome determines the corresponding feature should be used in PS-classifier or not. PS-classifier determines a fitness value for each chromosomes and GA uses these fitness values to the process of chromosome evolution. Finally GA finds an optimal subset of features.
Figure 3.
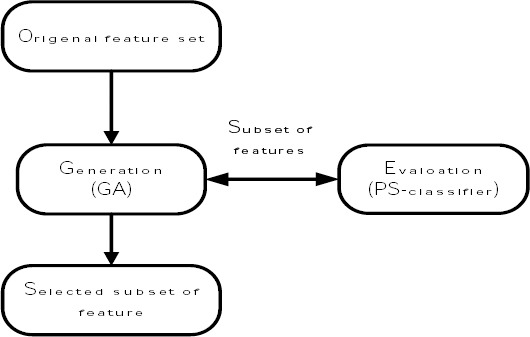
Proposed feature selection flowchart
In proposed model, the number of chromosomes in each population (size of population) is 150 and maximum iteration is 300. The mutation rate is 0.4 and crossover is 0.5 and elite rate is 0.1. Also for PS-classifier, swarm size of 150 was selected and initial inertia weight was chosen 0.7.
Prediction models
In this study we used different classifier algorithms namely artificial neural network (ANN), PS-classifier and GA-classifier as subset evaluating mechanism on Wisconsin breast cancer datasets (WBCD).
In this work we build three 3-layer neural networks by using nprtool in Matlab software. Artificial neural networks are a computational tool, based on the properties of biological neural systems. GA-classifier is another classifier that is used to evaluate proposed method and it is presented by Bandyopadhyay et al (24). The number of chromosomes in each population (size of population) is 150 and maximum iteration is 300. The mutation rate is 0.4 and crossover is 0.5 and elite rate is 0.1. The third selected classifier is PS-classifier that was described before.
In order to evaluate the classification efficiency, three main metrics including accuracy, sensitivity and specificity have been computed for the classifiers. These metrics are calculated from:
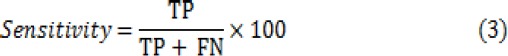
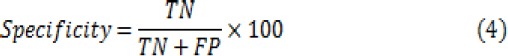
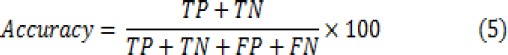
Where TN is number of True Negatives, TP is number of True Positives, FN is number of False Negatives and FP is number of False Positives.
Our training and testing was iterated 30 times for each classifier and average of results was expressed as the final result. 80% of data is allocated to training set and the remaining 20% is allocated to test set (in case of ANN, 20% of data allocated to validating set).
It should be noted that parameters tuning of the classifiers are equal before and after feature selection.
Results
Proposed feature selection method was applied on Wisconsin breast cancer databases and Table 3 shows selected relevant features.
Table 3.
Selected features after applying feature selection method
| Dataset | Selected features |
|---|---|
| WBC | 3,6,8,9 |
| WDBC | 1,2,6,8,12,14,18,19,21,22,25,26,27,29 |
| WPBC | 1,4,5,6,7,10,11,13,15,16,18,23,24,25,28,29 |
In neural network, the layers include an input layer of 9, 30 and 33 discrete variables with WBC, WDBC, WPBC datasets, respectively without feature selection. After feature selection we build layers include an input layer of 4, 14 and 16 discrete variables. In all networks we considered a hidden layer with 5 nodes and an output layer with 2 nodes.
Wisconsin breast cancer dataset (WBC)
We used classifiers with and without feature selection with WBC dataset. Results are summarized in the Table 4.
Table 4.
The Sensitivity, specificity and accuracy of 3 classifiers with and without feature selection (FS) using WBC dataset
| Accuracy | Specificity | Sensitivity | ||||
|---|---|---|---|---|---|---|
| Without FS | With FS | Without FS | With FS | Without FS | With FS | |
| PSO | 96.2 | 96.9 | 96.4 | 97.5 | 96.5 | 97.7 |
| GA | 96 | 96.6 | 96.5 | 96.6 | 96.5 | 97.1 |
| ANN | 96.8 | 96.7 | 95.2 | 97.2 | 94.9 | 97.2 |
Wisconsin diagnosis breast cancer (WDBC)
We employed described classifiers on WDBC. The comparison of average accuracies for the three classifiers (ANN, PS-classifier, GA-classifier) with and without feature selection is shown in Table 5.
Table 5.
The Sensitivity, specificity and accuracy of 3 classifiers with and without feature selection (FS) using WDBC dataset
| Accuracy | Specificity | Sensitivity | ||||
|---|---|---|---|---|---|---|
| Without FS | With FS | Without FS | With FS | Without FS | With FS | |
| PSO | 96.4 | 97.2 | 93.1 | 95.6 | 98.6 | 98 |
| GA | 96.1 | 96.6 | 92.9 | 93.7 | 97.8 | 97.5 |
| ANN | 96.5 | 97.3 | 96 | 95.1 | 98.2 | 98.4 |
Wisconsin prognosis breast cancer (WPBC)
Results of employing three described classifiers on WPBC are summarized in the Table 6.
Table 6.
The Sensitivity, specificity and accuracy of 3 classifiers with and without feature selection (FS) using WPBC dataset
| Accuracy | Specificity | Sensitivity | ||||
|---|---|---|---|---|---|---|
| Without FS | With FS | Without FS | With FS | Without FS | With FS | |
| PSO | 77.8 | 78.2 | 88.5 | 92.9 | 32.0 | 33.3 |
| GA | 76.3 | 78.1 | 90.2 | 92.8 | 26.9 | 31.0 |
| ANN | 77.4 | 79.2 | 94.4 | 96.3 | 28.3 | 33 |
Discussion
In this study a feature selection model with GA-based on feature selection is designed to identify relevant features. GA has more recently developed in compare to different feature selection algorithms. GA can be useful to feature selection when the problem has exponential search space. There are many advantages of the GAs for feature selection that have published in various literatures (25, 26).
The comparison of average accuracies for the three classifiers (ANN, PS-classifier, GA-classifier) with and without feature selection on WBC dataset showed that without feature selection the accuracy of ANN (96.8%) is the best and the accuracy obtained by PS-classifier is better than that produced by GA-classifier (96.2 vs. 96.08). It is observed that feature selection improved the accuracy of all classifiers expect of ANN and the best accuracy with feature selection achieved by PS-classifier (96.9%). Also it is apparent from results obtained that specificity and sensitivity has been approximately improved by feature selection.
Table 7 shows a comparison between classification accuracies of other published studies which used different feature selection methods and the accuracies obtained by ANN, PS-classifier and GA-classifier in this work on WBC dataset.
Table 7.
Comparison of experimental results of proposed method and other papers in WBC
For WDBC dataset, ANN classifier shows the best accuracy (96.5%). From Table 5 it is obvious that the ANN accuracy with WDBC is well than PS-classifier and GA-classifier accuracies respectively (96.4 vs. 96.1). Results show feature selection improved accuracy of all three classifiers and the best accuracy with feature selection achieved by ANN (97.3%). Also Table 5 shows that specificity and sensitivity can improve after feature selection.
Table 8 shows a comparison between classification accuracies of other published studies which used different feature selection methods and the accuracies obtained in this work on WDBC dataset.
Table 8.
Comparison of experimental results of proposed method and other papers in WDBC
The comparison of average accuracies for the described classifiers with and without feature selection on WPBC showed that without feature selection the accuracy of PS-classifier (77.8%) is the best and the accuracy obtained by ANN is better than that produced by GA-classifier (77.4 vs. 76.3). It is clear that feature selection improved the accuracy of all three classifiers and the best accuracy with feature selection achieved by ANN (79.2%). Also as can be seen from the table 8, the specificity and sensitivity improved after feature selection. The result of this dataset is comparable with other studies (35).
Table 9 shows a comparison between classi-fication accuracies of other published studies which used different feature selection methods and the accuracies obtained by three different classifiers in this work on WPBC dataset.
Table 9.
Comparison of experimental results of proposed method and other papers in WPBC
It should be noted while data mining can facilitate analysing of large databases and help medical staff in decision making we should consider the limitations of what it can do. data mining techniques can discover pattern buried in data but it can’t replace physician’s insights (36). Also sometimes the increase in the number of features leads to the decrease in the speed of the algorithm. Therefore identifying patterns may be time consuming.
Conclusion
In this paper, we proposed a feature selection method using GA for selecting the best subset of features for breast cancer diagnosis system.
ANN, PS-classifier and GA-classifier were used to evaluate proposed feature selection method on Wisconsin Breast Cancer Datasets. In WBC, the classification using PS-classifier is superior to other classification. In WDBC and WPBC, ANN achieved the best accuracy. The results show that feature selection can improve accuracy of classifiers. Result of this study is comparable with the other studies on Wisconsin breast cancer datasets.
Acknowledgements
We thank Dr William H Wolberg at the University of Wisconsin for supporting us with the breast cancer dataset which we have used in our experiments.
References
- 1.Sarbaz M, Pournik O, Ghalichi L, Kimiafar K, Razavi AR. Designing a Human T-Lymphotropic Virus Type 1 (HTLV-I) Diagnostic Model Using the Complete Blood Count. Iran J Basic Med Sci. 2013;16:247. [PMC free article] [PubMed] [Google Scholar]
- 2.Tayarani A, Baratian A, Sistani MB, Saberi MR, Tehranizadeh Z. Artificial neural networks analysis used to evaluate the molecular interactions between selected drugs and human cyclooxygenas e2 receptor. Iran J Basic Med Sci. 2013;16:1196. [PMC free article] [PubMed] [Google Scholar]
- 3.Breastcancer.org: Knowing your risk can save your life [Internet] Breastcancer.org. 2016. [cited 12 May 2016]. Available from: http://www.breastcancer.org .
- 4.Basha SS, Prasad KS. Automatic detection of breast cancer mass in mammograms using morphological operators and fuzzy c--means clustering. J Theor Appl Inf Technol. 2009:5. [Google Scholar]
- 5.How is breast cancer diagnosed? [Internet] Cancer.org. 2016. [cited 12 May 2016]. Available from: http://www.cancer.org/cancer/breastcancer/detail-edguide/breast-cancer-diagnosis .
- 6.Elmore JG, Wells CK, Lee CH, Howard DH, Feinstein AR. Variability in radiologists’ interpretations of mammograms. N Engl J Med. 1994;331:1493–1499. doi: 10.1056/NEJM199412013312206. [DOI] [PubMed] [Google Scholar]
- 7.Fletcher SW, Black W, Harris R, Rimer BK, Shapiro S. Report of the international workshop on screening for breast cancer. J Nat Cancer Inst. 1993;85:1644–1656. doi: 10.1093/jnci/85.20.1644. [DOI] [PubMed] [Google Scholar]
- 8.Willems SM, Van Deurzen CH, Van Diest PJ. Diagnosis of breast lesions: fine-needle aspiration cytology or core needle biopsy? A review. J clin pathol. 2012;65:287–292. doi: 10.1136/jclinpath-2011-200410. [DOI] [PubMed] [Google Scholar]
- 9.Kohavi R, John GH. Wrappers for feature subset selection. Artif Intell. 1997;97:273–324. [Google Scholar]
- 10.Abe N, Kudo M, Toyama J, Shimbo M. A divergence criterion for classifier-independent feature selection. Advances in Pattern Recognition: Springer; 2000:668–676. [Google Scholar]
- 11.Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- 12.Bermejo P, Gámez JA, Puerta JM. A GRASP algorithm for fast hybrid (filter-wrapper) feature subset selection in high-dimensional datasets. Pattern Recognit Lett. 2011;32:701–711. [Google Scholar]
- 13.Aghdam MH, Ghasem-Aghaee N, Ehsan Basiri M. Application of ant colony optimization for feature selection in text categorization. Evolutionary Computation, 2008 CEC 2008 (IEEE World Congress on Computational Intelligence) IEEE Congress on; 2008: IEEE [Google Scholar]
- 14.Unler A, Murat A. A discrete particle swarm optimization method for feature selection in binary classification problems. Eur J Oper Res. 2010;206:528–539. [Google Scholar]
- 15.Karegowda AG, Jayaram M, Manjunath A. Feature subset selection problem using wrapper approach in supervised learning. Int J Comput Appl. 2010;1:13–17. [Google Scholar]
- 16.Youn E, Koenig L, Jeong MK, Baek SH. Support vector-based feature selection using Fisher’s linear discriminant and Support Vector Machine. Exp Syst Appl. 2010;37:6148–6156. [Google Scholar]
- 17.Deisy C, Subbulakshmi B, Baskar S, Ramaraj N. Efficient dimensionality reduction approaches for feature selection. Conference on Computational Intelligence and Multimedia Applications, 2007 International Conference on; 2007: IEEE [Google Scholar]
- 18.Sridevi T, Murugan A. An intelligent classifier for breast cancer diagnosis based on K-Means clustering and rough set. Int J Comput Appl. 2014;85:38–42. [Google Scholar]
- 19.Sridevi T, Murugan A. A novel feature selection method for effective breast cancer diagnosis and prognosis. Int J Comput Appl. 2014;88:28–33. [Google Scholar]
- 20.UCI Machine Learning Repository: Breast Cancer Wisconsin (Diagnostic) Data Set [Internet] Archive.ics.uci.edu. 2016. [cited 12 May 2016]. Available from: http://archive.ics.uci.edu/ml/-datasets/Breast+Cancer+Wisconsin+%28Diagnostic%29 .
- 21.Holland JH. Adaptation in natural and artificial systems: An introductory analysis with applications to biology, control, and artificial intelligence. U Michigan Press; 1975. [Google Scholar]
- 22.Zahiri SH, Seyedin SA. Swarm intelligence based classifiers. J Franklin Inst. 2007;344:362–3676. [Google Scholar]
- 23.Kennedy J, Eberhart R. Particle swarm optimization. Proceedings of the IEEE International Conference on Neural Networks. 1995 [Google Scholar]
- 24.Bandyopadhyay S, Murthy CA, Pal SK. Theoretical performance of genetic pattern classifier. J Franklin Inst. 1999;336:387–422. [Google Scholar]
- 25.Oh IS, Lee JS, Moon BR. Hybrid genetic algorithms for feature selection. IEEE Trans Pattern Anal Mach Intell. 2004;26:1424–1437. doi: 10.1109/TPAMI.2004.105. [DOI] [PubMed] [Google Scholar]
- 26.Hadizadeh F, Vahdani S, Jafarpour M. Quantitative Structure-Activity Relationship Studies of 4-Imidazolyl-1, 4-dihydropyridines as Calcium Channel Blockers. Iran J Basic Med Sci. 2013;16:910–916. [PMC free article] [PubMed] [Google Scholar]
- 27.Lavanya D, Rani DK. Analysis of feature selection with classification: Breast cancer datasets. Indian Journal of Computer Science and Engineering (IJCSE) 2011;2:756–763. [Google Scholar]
- 28.Karabatak M, Ince MC. An expert system for detection of breast cancer based on association rules and neural network. Exp Syst Appl. 2009;36:3465–3469. [Google Scholar]
- 29.Chen HL, Yang B, Liu J, Liu DY. A support vector machine classifier with rough set-based feature selection for breast cancer diagnosis. Exp Syst Appl. 2011;38:9014–9022. [Google Scholar]
- 30.Senturk ZK, Kara R. Breast Cancer Diagnosis via Data Mining: Performance Analysis of Seven different algorithms. Computer Science & Engineering. 2014;4:35. [Google Scholar]
- 31.Noruzi A, Sahebi H. A graph-based feature selection method for improving medical diagnosis. Adv Comput Sci. 2015;4:36–40. [Google Scholar]
- 32.Zhao JY, Zhang ZL. Fuzzy rough neural network and its application to feature selection. Advanced Computational Intelligence (IWACI), 2011 Fourth International Workshop on 2011: IEEE [Google Scholar]
- 33.Liu Y, Zheng YF. FS_SFS: A novel feature selection method for support vector machines. Pattern Recognit. 2006;39:1333–1345. [Google Scholar]
- 34.Dumitru D. Prediction of recurrent events in breast cancer using the Naive Bayesian classification. Annals of the University of Craiova-Mathematics and Computer Science Series. 2009;36:92–96. [Google Scholar]
- 35.Jacob SG, Ramani RG. Efficient classifier for classification of prognostic breast cancer data through data mining techniques. Proceedings of the World Congress on Engineering and Computer Science. 2012 [Google Scholar]
- 36.Richards G, Rayward-Smith VJ, Sonksen PH, Carey S, Weng C. Data mining for indicators of early mortality in a database of clinical records. Artif Intell Med. 2001;22:215–231. doi: 10.1016/s0933-3657(00)00110-x. [DOI] [PubMed] [Google Scholar]


